Genetic Diversity and Virulence Profiling of Multi-Drug Resistant Escherichia coli of Human, Animal, and Environmental Origins
Abstract
1. Introduction
2. Results
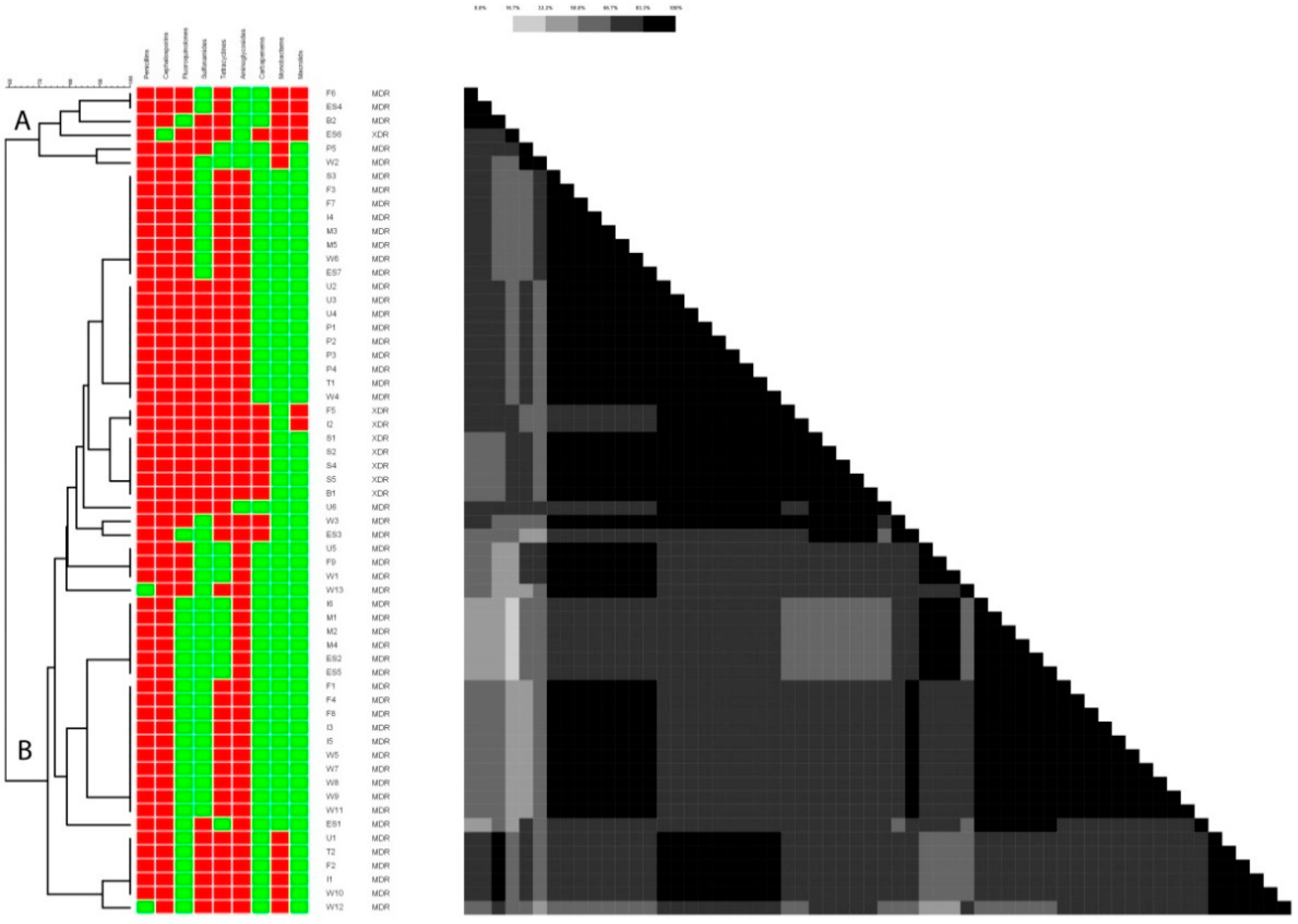
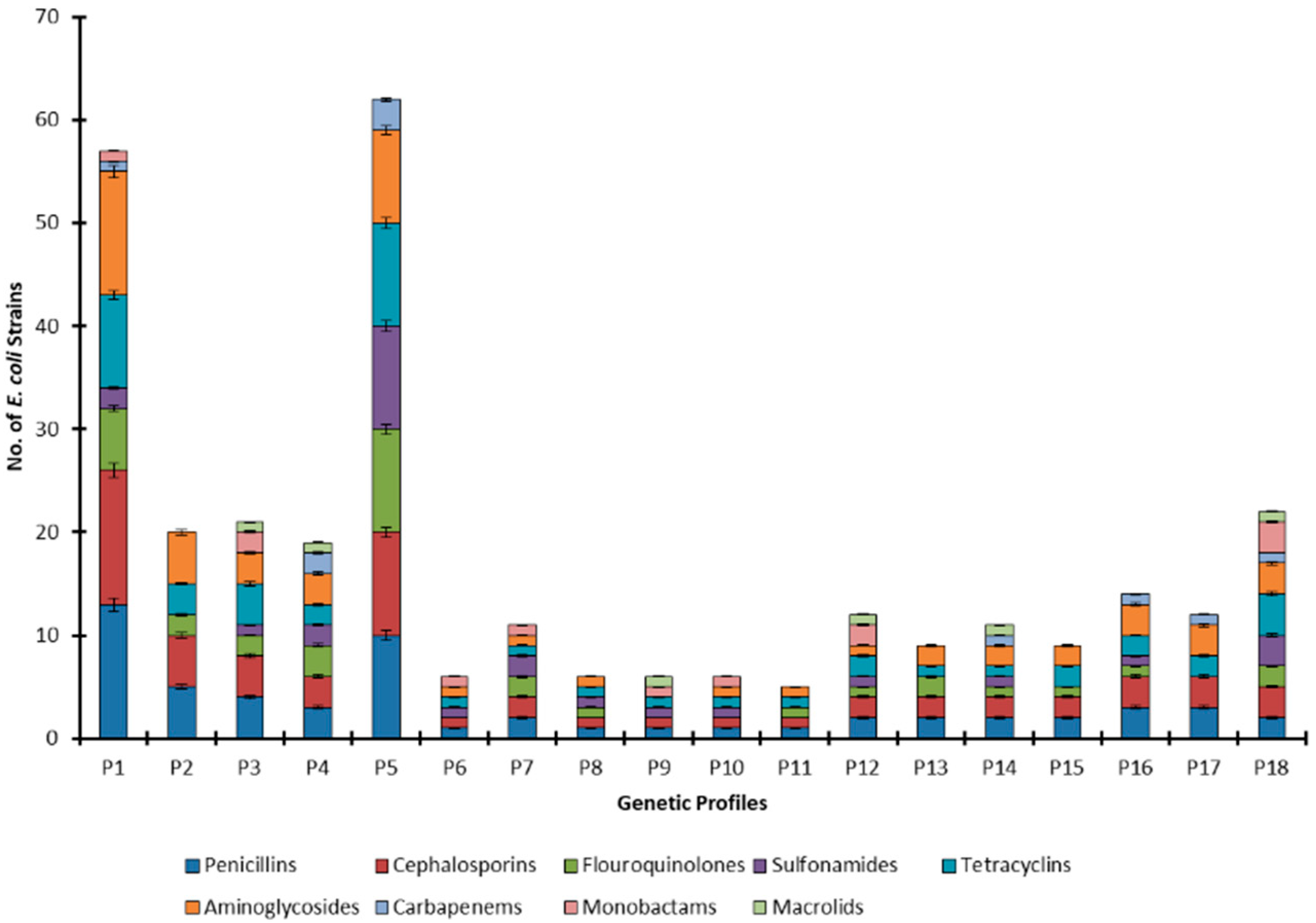
2.1. Antibiotic Resistance Profiling
2.2. In-Vitro Biofilm Production
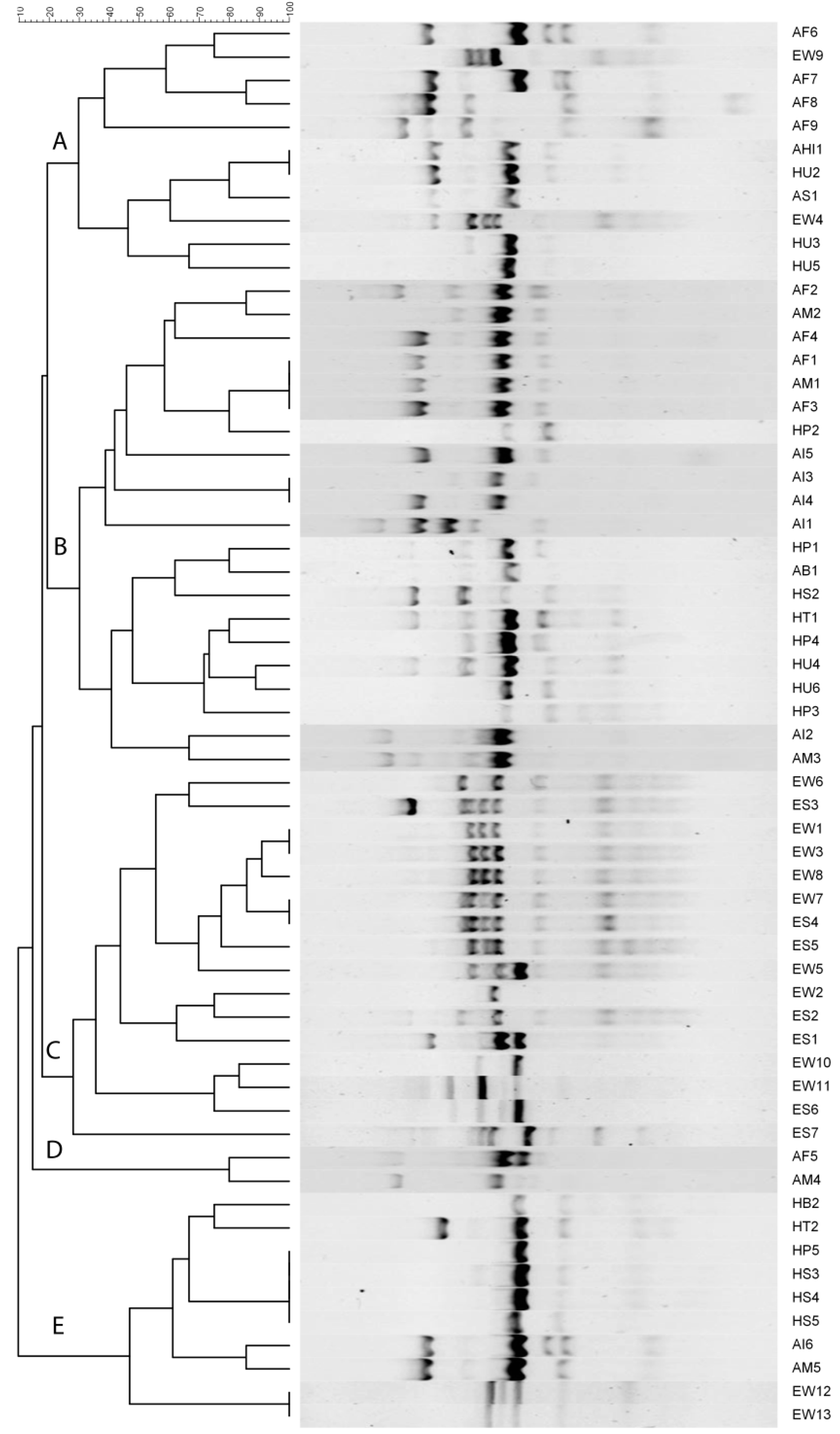
2.3. ERIC-PCR Based Fingerprinting
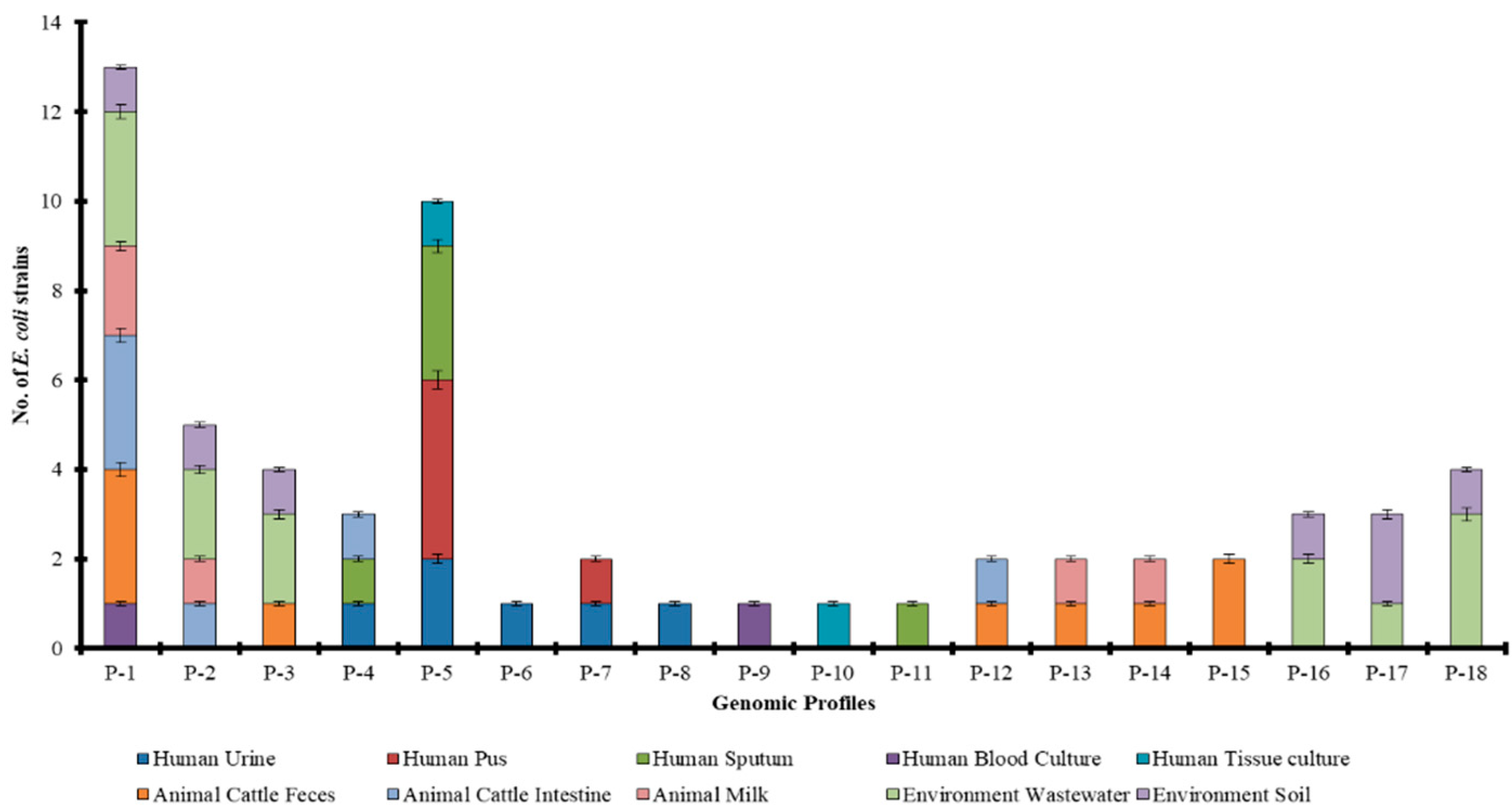
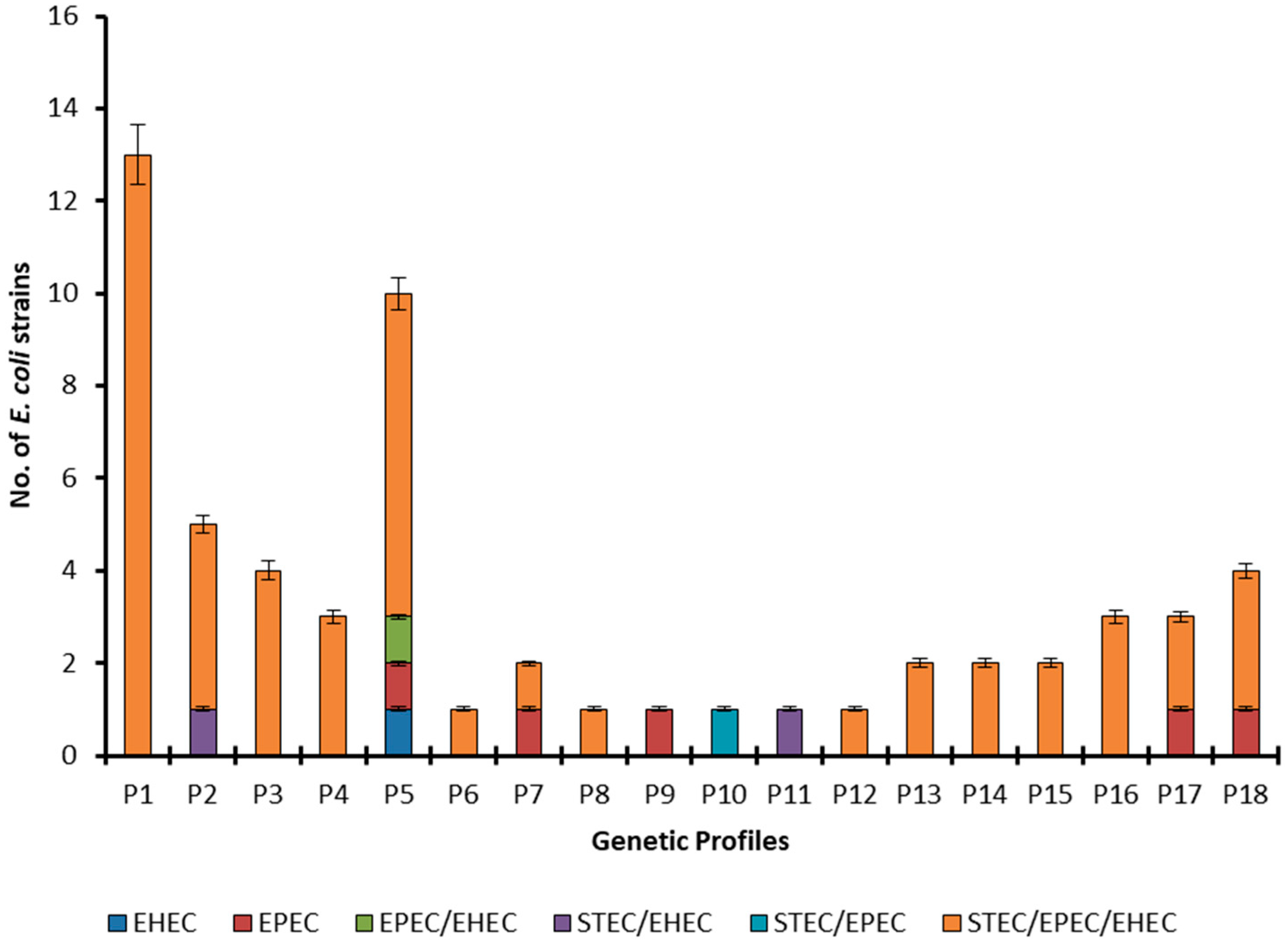
2.4. Source Wise Genetic Diversity
2.5. ERIC-PCR-Based Clustering
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. Biochemical and Molecular Characterization of E. coli
4.3. Antibiotic Resistance Pattern
4.4. Detection of Biofilm Formation
4.5. Detection of Virulence Genes
4.6. Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR)
4.7. E. coli Genetic Diversity Evaluation
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; El Zowalaty, M.E.; Rahman, A.T.; Ashour, H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef]
- Marrana, M. Chapter 3—Epidemiology of disease through the interactions between humans, domestic animals, and wildlife. In One Health; Prata, J.C., Ribeiro, A.I., Rocha-Santos, T., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 73–111. [Google Scholar]
- Rehman, A.; Jingdong, L.; Chandio, A.A.; Hussain, I. Livestock production and population census in Pakistan: Determining their relationship with agricultural GDP using econometric analysis. Inf. Process. Agric. 2017, 4, 168–177. [Google Scholar] [CrossRef]
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J.; et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404. [Google Scholar] [CrossRef] [PubMed]
- Boutayeb, A. The double burden of communicable and non-communicable diseases in developing countries. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 191–199. [Google Scholar] [CrossRef]
- Remoundou, K.; Koundouri, P. Environmental effects on public health: An economic perspective. Int. J. Environ. Res. Public Health 2009, 6, 2160–2178. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with antibiotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Ward, M.J. Sources of antimicrobial resistance. Science 2013, 341, 1460–1461. [Google Scholar] [CrossRef] [PubMed]
- Sabir, S.; Anjum, A.A.; Ijaz, T.; Ali, M.A. Isolation and antibiotic susceptibility of E. coli from urinary tract infections in a tertiary care hospital. Pak. J. Med. Sci. 2014, 30, 389. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Fahrenfeld, N.; Knowlton, K.; Krometis, L.A.; Hession, W.C.; Xia, K.; Lipscomb, E.; Libuit, K.; Green, B.L.; Pruden, A. Effect of manure application on abundance of antibiotic resistance genes and their attenuation rates in soil: Field-scale mass balance approach. Environ. Sci. Technol. 2014, 48, 2643–2650. [Google Scholar] [CrossRef] [PubMed]
- Ungureanu, N.; Vlăduț, V.; Voicu, G. Water scarcity and wastewater reuse in crop irrigation. Sustainability 2020, 12, 9055. [Google Scholar] [CrossRef]
- Ajmal, A.W.; Saroosh, S.; Mulk, S.; Hassan, M.N.; Yasmin, H.; Jabeen, Z.; Nosheen, A.; Shah, S.M.U.; Naz, R.; Hasnain, Z. Bacteria isolated from wastewater irrigated agricultural soils adapt to heavy metal toxicity while maintaining their plant growth promoting traits. Sustainability 2021, 13, 7792. [Google Scholar] [CrossRef]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Martínez-Vázquez, A.V.; Rivera-Sánchez, G.; Lira-Méndez, K.; Reyes-López, M.; Bocanegra-García, V. Prevalence, antimicrobial resistance and virulence genes of Escherichia coli isolated from retail meat in Tamaulipas, Mexico. J. Glob. Antimicrob. Resist. 2018, 14, 266–272. [Google Scholar] [CrossRef] [PubMed]
- May, M.K. Characterizing Bacterial Antibiotic Resistance, Prevalence, and Persistence in the Marine Environment; Massachusetts Institute of Technology: Cambridge, MA, USA, 2019. [Google Scholar]
- Martin, B.; Humbert, O.; Camara, M.; Guenzi, E.; Walker, J.; Mitchell, T.; Andrew, P.; Prudhomme, M.; Alloing, G.; Hakenbeck, R.; et al. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992, 20, 3479–3483. [Google Scholar] [CrossRef]
- Ramazanzadeh, R.; Zamani, S.; Zamani, S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)-PCR technique in Sanandaj hospitals. Iran J. Microbiol. 2013, 5, 126–131. [Google Scholar]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef]
- Byappanahalli, M.; Fowler, M.; Shively, D.; Whitman, R. Ubiquity and persistence of Escherichia coli in a Midwestern coastal stream. Appl. Environ. Microbiol. 2003, 69, 4549–4555. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, L.; Spangholm, D.J.; Pedersen, K.; Jensen, L.B.; Emborg, H.D.; Agersø, Y.; Aarestrup, F.M.; Hammerum, A.M.; Frimodt-Møller, N. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int. J. Food Microbiol. 2010, 142, 264–272. [Google Scholar] [CrossRef]
- Lagerstrom, K.M.; Hadly, E.A. The under-investigated wild side of Escherichia coli: Genetic diversity, pathogenicity and antimicrobial resistance in wild animals. Proc. Biol. Sci. 2021, 288, 20210399. [Google Scholar] [CrossRef]
- Gavrilescu, M. Water, Soil, and Plants Interactions in a Threatened Environment. Water 2021, 13, 2746. [Google Scholar] [CrossRef]
- Tahamtan, Y.; Hayati, M.; Namavari, M. Prevalence and distribution of the stx, stx genes in Shiga toxin producing E. coli (STEC) isolates from cattle. Iran J. Microbiol. 2010, 2, 8–13. [Google Scholar]
- Blanco, M.; Blanco, J.E.; Mora, A.; Dahbi, G.; Alonso, M.P.; González, E.A.; Bernárdez, M.I.; Blanco, J. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae-xi). J. Clin. Microbiol. 2004, 42, 645–651. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Liu, Z.; Liu, P.; Shi, Z.; Wei, J.; Shao, D.; Li, B.; Ma, Z. Molecular Characterization of Enterohemorrhagic E. coli O157 Isolated from Animal Fecal and Food Samples in Eastern China. Sci. World J. 2014, 2014, 946394. [Google Scholar]
- El-Leithy, M.; El-Shatoury, E.H.; El-Senousy, W.; Abou-Zeid, M.; El-Taweel, G.E. Detection of six E. coli O157 virulence genes in water samples using multiplex PCR. Egypt. J. Microbiol. 2012, 47, 171–188. [Google Scholar]
- El-Shatoury, E.H.; El-Leithy, M.A.; Abou-Zeid, M.A.; El-Taweel, G.E.; El-Senousy, W.M. Antibiotic susceptibility of Shiga toxin Producing E. coli O157: H7 isolated from different water sources. Open Conf. Proc. 2015, 6, 30–34. [Google Scholar] [CrossRef][Green Version]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-PLoSkonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, P.; Natarajan, V.; Sevanan, M. In Vitro biofilm formation by uropathogenic Escherichia coli and their antimicrobial susceptibility pattern. Asian Pac. J. Trop. Med. 2012, 5, 210–213. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar]
- Khurshid, M.; Rasool, M.H.; Ashfaq, U.A.; Aslam, B.; Waseem, M.; Xu, Q.; Zhang, X.; Guo, Q.; Wang, M. Dissemination of bla(OXA-23)-harbouring carbapenem-resistant Acinetobacter baumannii clones in Pakistan. J. Glob. Antimicrob. Resist. 2020, 21, 357–362. [Google Scholar] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
| Resistance Profiles | Antibiotic Combinations | Number of E. coli Strains (n) | ||
|---|---|---|---|---|
| Human Origin | Animal Origin | Environmental Origin | ||
| R1 | Penicillin/Cephalosporins/Sulfonamides/ Tetracycline/Aminoglycosides/Monobactams | 2 | 2 | 1 |
| R2 | Penicillin/Cephalosporins/Fluoroquinolones/ Sulfonamides/Tetracycline/Aminoglycosides | 8 | - | 1 |
| R3 | Cephalosporins/Sulfonamides/Tetracycline/ Aminoglycoside/Monobactams | - | - | 1 |
| R4 | Cephalosporins/Fluoroquinolones/ Tetracycline/Aminoglycosides | - | - | 1 |
| R5 | Penicillin/Fluoroquinolones/Sulfonamides/ Tetracycline/Carbapenems/Monobactams/ Macrolides | - | - | 1 |
| R6 | Penicillin/Cephalosporins/Sulfonamides/ Aminoglycosides | 1 | - | 1 |
| R7 | Penicillin/Cephalosporins/Fluoroquinolones/ Sulfonamides/Tetracycline | 1 | - | - |
| R8 | Penicillin/Cephalosporins/Fluoroquinolones/ Sulfonamides/Monobactams | 1 | - | - |
| R9 | Penicillin/Cephalosporins/Fluoroquinolones/ Sulfonamides/Tetracycline/Aminoglycosides/ Carbapenems | 5 | - | - |
| R10 | Penicillin/Cephalosporins/Sulfonamides/ Tetracycline/Monobactams/Macrolides | 1 | - | - |
| R11 | Penicillin/Cephalosporins/Sulfonamides/ Tetracycline/Aminoglycosides | 1 | 5 | 2 |
| R12 | Penicillin/Cephalosporins/Tetracycline/ Aminoglycosides | - | 5 | 5 |
| R13 | Penicillin/Cephalosporins/Fluoroquinolones/ Sulfonamides/Tetracycline/Aminoglycosides/ Carbapenems/Macrolides | - | 2 | - |
| R14 | Penicillin/Cephalosporins/Fluoroquinolones/ Tetracycline/Monobactams/Macrolides | - | 1 | 1 |
| R15 | Penicillin/Cephalosporins/Fluoroquinolones/ Aminoglycosides | - | 1 | 1 |
| R16 | Penicillin/Cephalosporins/Aminoglycosides | - | 4 | 2 |
| R17 | Penicillin/Cephalosporins/Fluoroquinolones/ Monobactams | - | - | 1 |
| R18 | Penicillin/Cephalosporins/Fluoroquinolones/ Tetracycline/Aminoglycosides/Carbapenems | - | - | 1 |
| R19 | Penicillin/Cephalosporins/Tetracycline/ Aminoglycosides/Carbapenems | - | - | 1 |
| Virulence Genes | E. coli Strains | |||
|---|---|---|---|---|
| Human (n = 20) | Animal (n = 20) | Environment (n = 20) | Total (n = 60) | |
| stx1 | 0 | 5 | 5 | 10 |
| stx2 | 15 | 18 | 18 | 51 |
| eaeA | 18 | 18 | 20 | 56 |
| hlyA | 16 | 19 | 18 | 53 |
| Antibiotic Groups | Biofilm | |||
|---|---|---|---|---|
| Non-Producer | Producer | |||
| Resistant n (%) | Sensitive n (%) | Resistant n (%) | Sensitive n (%) | |
| Penicillins | 11 (92) | 01 (8) | 47 (98) | 1 (2) |
| Cephalosporins | 12 (100) | 0 (0) | 47 (98) | 1(2) |
| Fluoroquinolones | 7 (58) | 5 (42) | 28 (58) | 20 (42) |
| Sulfonamides | 4 (33) | 8 (67) | 23 (48) | 25 (52) |
| Tetracycline | 7 (58) | 5 (42) | 41 (85) | 7 (15) |
| Aminoglycosides | 10 (83) | 2 (17) | 43 (89) | 5 (10) |
| Carbapenems | 0 (0) | 12 (100) | 10 (21) | 38 (79) |
| Monobactams | 2 (17) | 10 (83) | 10 (21) | 38 (79) |
| Macrolides | 1 (8) | 11 (92) | 5 (10) | 43 (89) |
| Origins and Sources | Shannon (H) | Simpson (1-D) | Evenness (E) | Richness (S) |
|---|---|---|---|---|
| Human | 1.7 | 0.7 | 0.6 | 9.0 |
| Urine | 1.5 | 0.7 | 0.9 | 5.0 |
| Pus | 0.5 | 0.3 | 0.8 | 2.0 |
| Sputum | 0.9 | 0.5 | 0.8 | 3.0 |
| Blood Culture | 0.6 | 0.5 | 1.0 | 2.0 |
| Tissue culture | 0.6 | 0.5 | 1.0 | 2.0 |
| Animal | 1.8 | 0.7 | 0.7 | 8.0 |
| Cattle Faeces | 1.6 | 0.7 | 0.8 | 6.0 |
| Cattle Intestine | 1.2 | 0.6 | 0.8 | 4.0 |
| Cattle Milk | 1.3 | 0.7 | 0.9 | 4.0 |
| Environment | 1.7 | 0.8 | 0.9 | 6.0 |
| Wastewater | 1.7 | 0.8 | 0.9 | 6.0 |
| Soil | 1.7 | 0.8 | 0.9 | 6.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yar, A.; Choudary, M.A.; Rehman, A.; Hussain, A.; Elahi, A.; ur Rehman, F.; Waqar, A.B.; Alshammari, A.; Alharbi, M.; Nisar, M.A.; et al. Genetic Diversity and Virulence Profiling of Multi-Drug Resistant Escherichia coli of Human, Animal, and Environmental Origins. Antibiotics 2022, 11, 1061. https://doi.org/10.3390/antibiotics11081061
Yar A, Choudary MA, Rehman A, Hussain A, Elahi A, ur Rehman F, Waqar AB, Alshammari A, Alharbi M, Nisar MA, et al. Genetic Diversity and Virulence Profiling of Multi-Drug Resistant Escherichia coli of Human, Animal, and Environmental Origins. Antibiotics. 2022; 11(8):1061. https://doi.org/10.3390/antibiotics11081061
Chicago/Turabian StyleYar, Asfand, Muhammad Adil Choudary, Abdul Rehman, Abid Hussain, Amina Elahi, Farooq ur Rehman, Ahmed Bilal Waqar, Abdulrahman Alshammari, Metab Alharbi, Muhammad Atif Nisar, and et al. 2022. "Genetic Diversity and Virulence Profiling of Multi-Drug Resistant Escherichia coli of Human, Animal, and Environmental Origins" Antibiotics 11, no. 8: 1061. https://doi.org/10.3390/antibiotics11081061
APA StyleYar, A., Choudary, M. A., Rehman, A., Hussain, A., Elahi, A., ur Rehman, F., Waqar, A. B., Alshammari, A., Alharbi, M., Nisar, M. A., Khurshid, M., & Khan, Z. (2022). Genetic Diversity and Virulence Profiling of Multi-Drug Resistant Escherichia coli of Human, Animal, and Environmental Origins. Antibiotics, 11(8), 1061. https://doi.org/10.3390/antibiotics11081061









