Revealing Genome-Based Biosynthetic Potential of Streptomyces sp. BR123 Isolated from Sunflower Rhizosphere with Broad Spectrum Antimicrobial Activity
Abstract
1. Introduction
2. Materials and Methods
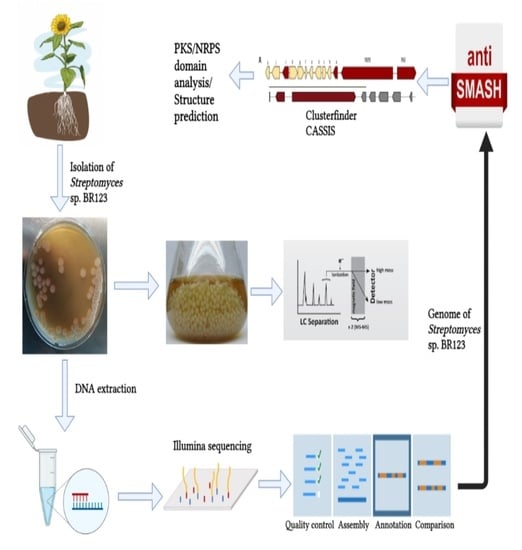
2.1. Isolation and Cultivation Conditions of Streptomyces sp. BR123
2.2. Sequencing and Assembly of the Genome
2.3. Annotation of Genome and Bioinformatics Analysis
2.4. Amplification of NRPS and PKS Genes by PCR
2.5. Assessment of Antimicrobial Potential
2.6. Analysis of Metabolites through HPLC-MS from Streptomyces sp. BR123
2.6.1. Sample Preparation
2.6.2. Analysis of Metabolites
2.7. Comparative Genome Analysis
2.8. Accession Number of Genome Sequence
3. Results and Discussion
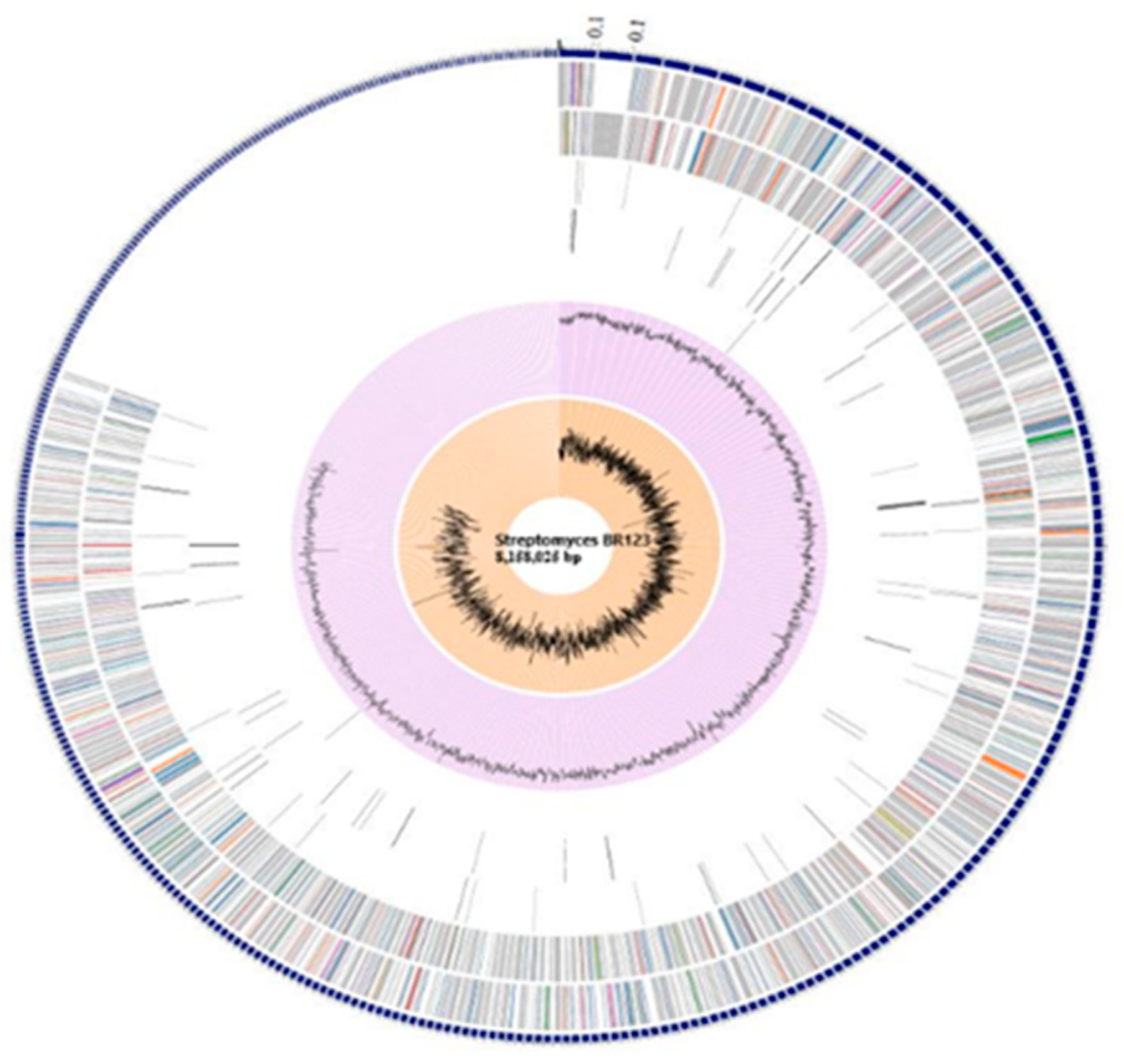
3.1. General Genomic Characteristics and Phylogenetic Analysis of Streptomyces sp. BR123
3.2. Annotation and Assembly of Genome Sequence
3.3. Biosynthetic Secondary Metabolite Gene Clusters of Streptomyces sp. BR123
3.4. Detection of NRPS and PKS Genes in Streptomyces sp. BR123
3.5. Production of Secondary Metabolites by Streptomyces sp. BR123
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jakubiec-Krzesniak, K.; Rajnisz-Mateusiak, A.; Guspiel, A.; Ziemska, J.; Solecka, J. Secondary metabolites of actinomycetes and their antibacterial, antifungal and antiviral properties. Pol. J. Microbiol. 2018, 67, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Falzone, M.; Crespo, E.; Jones, K.; Khan, G.; Korn, V.L.; Patel, A.; Patel, M.; Patel, K.; Perkins, C.; Siddiqui, S.; et al. Nutritional control of antibiotic production by Streptomyces platensis MA7327: Importance of L-aspartic acid. J. Antibiot. 2017, 70, 828–831. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Metsämuuronen, S.; Sirén, H. Bioactive phenolic compounds, metabolism and properties: A review on valuable chemical compounds in Scots pine and Norway spruce. Phytochem. Rev. 2019, 18, 623–664. [Google Scholar] [CrossRef]
- Park, S.R.; Yoon, Y.J.; Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Reynolds, J.M. A review of the microbial production of bioactive natural products and biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar]
- Dholakiya, R.N.; Kumar, R.; Mishra, A.; Mody, K.H.; Jha, B. Antibacterial and antioxidant activities of novel actinobacteria strain isolated from Gulf of Khambhat, Gujarat. Front. Microbiol. 2017, 8, 2420. [Google Scholar] [CrossRef]
- Das, R.; Romi, W.; Das, R.; Sharma, H.K.; Thakur, D. Antimicrobial potentiality of actinobacteria isolated from two microbiologically unexplored forest ecosystems of Northeast India. BMC Microbiol. 2018, 18, 71. [Google Scholar] [CrossRef]
- Guerrero-Garzón, J.F.; Zehl, M.; Schneider, O.; Rückert, C.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Streptomyces spp. from the marine sponge Antho dichotoma: Analyses of secondary metabolite biosynthesis gene clusters and some of their products. Front. Microbiol. 2020, 11, 437. [Google Scholar] [CrossRef]
- Ziemert, N.; Lechner, A.; Wietz, M.; Millán-Aguiñaga, N.; Chavarria, K.L.; Jensen, P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc. Natl. Acad. Sci. USA 2014, 111, E1130–E1139. [Google Scholar] [CrossRef]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Igarashi, Y.; Tamura, T. Diversity of nonribosomal peptide synthetase and polyketide synthase gene clusters among taxonomically close Streptomyces strains. Sci. Rep. 2018, 8, 6888. [Google Scholar] [CrossRef]
- Xu, L.; Ye, K.-X.; Dai, W.-H.; Sun, C.; Xu, L.-H.; Han, B.-N. Comparative Genomic Insights into Secondary Metabolism Biosynthetic Gene Cluster Distributions of Marine Streptomyces. Mar. Drugs 2019, 17, 498. [Google Scholar] [CrossRef]
- Betancur, L.A.; Forero, A.M.; Vinchira-Villarraga, D.M.; Cardenas, J.D.; Romero-Otero, A.; Chagas, F.O.; Pupo, M.T.; Castellanos, L.; Ramos, F.A. NMR-based metabolic profiling to follow the production of anti-phytopathogenic compounds in the culture of the marine strain Streptomyces sp. PNM-9. Microbiol. Res. 2020, 239, 126507. [Google Scholar] [CrossRef]
- Rebets, Y.; Brötz, E.; Tokovenko, B.; Luzhetskyy, A. Actinomycetes biosynthetic potential: How to bridge in silico and in vivo? J. Ind. Microbiol. Biotechnol. 2014, 41, 387–402. [Google Scholar] [CrossRef]
- Ziemert, N.; Alanjary, M.; Weber, T. The evolution of genome mining in microbes—A review. Nat. Prod. Rep. 2016, 33, 988–1005. [Google Scholar] [CrossRef]
- Lacey, H.J.; Rutledge, P.J. Recently discovered secondary metabolites from Streptomyces species. Molecules 2022, 27, 887. [Google Scholar] [CrossRef]
- Mohseni, M.; Norouzi, H.; Hamedi, J.; Roohi, A. Screening of antibacterial producing actinomycetes from sediments of the Caspian Sea. Int. J. Mol. Cell. Med. 2013, 2, 64–71. [Google Scholar]
- Ashraf, N.; Bechthold, A.; Anwar, M.A.; Ghauri, M.A.; Anjum, M.S.; Khan, A.N.; Akhtar, K.; Khaliq, S. Production of a broad spectrum streptothricin like antibiotic from halotolerant Streptomyces fimbriatus isolate G1 associated with marine sediments. Folia Microbiol. 2021, 66, 639–649. [Google Scholar] [CrossRef]
- Ashraf, N.; Bechthold, A.; Anwar, M.A.; Khaliq, S. Draft Genome Sequence of Streptomyces sp. Strain BR123, Endowed with Broad-Spectrum Antimicrobial Potential. Microbiol. Resour. Announc. 2020, 9, e00972-20. [Google Scholar] [CrossRef]
- Akhtar, N.; Ghauri, M.A.; Akhtar, K.; Parveen, S.; Farooq, M.; Ali, A.; Schierack, P. Comparative analysis of draft genome sequence of Rhodococcus sp. Eu-32 with other Rhodococcus species for its taxonomic status and sulfur metabolism potential. Curr. Microbiol. 2019, 76, 1207–1214. [Google Scholar] [CrossRef]
- Escalante-Réndiz, D.; de-la-Rosa-García, S.; Tapia-Tussell, R.; Martín, J.; Reyes, F.; Vicente, F.; Gamboa-Angulo, M. Molecular identification of selected Streptomyces strains isolated from Mexican tropical soils and their anti-Candida activity. Int. J. Environ. Res. Public Health 2019, 16, 1913. [Google Scholar] [CrossRef]
- Asma Azmani, A.; Lemriss, S.; Barakate, M.; Souiri, A.; Dhiba, D.; Hassani, L.; Hamdali, H. Screening and characterization of Streptomyces spp. isolated from three moroccan ecosystems producing a potential. BioTech 2022, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Chaves, J.V.; Ojeda, C.P.O.; da Silva, I.; de Lima Procopio, R. Identification and Phylogeny of Streptomyces Based on Gene Sequences. Res. J. Microbiol. 2017, 13, 13–20. [Google Scholar]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90 K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Castro, J.F.; Razmilic, V.; Chandra, G.; Andrews, B.; Asenjo, J.A.; Bibb, M.J. The Streptomyces leeuwenhoekii genome: De novo sequencing and assembly in single contigs of the chromosome, circular plasmid pSLE1 and linear plasmid pSLE2. BMC Genom. 2015, 16, 485. [Google Scholar] [CrossRef]
- Subramaniam, G.; Thakur, V.; Saxena, R.K.; Vadlamudi, S.; Purohit, S.; Kumar, V.; Rathore, A.; Chitikineni, A.; Varshney, R.K. Complete genome sequence of sixteen plant growth promoting Streptomyces strains. Sci. Rep. 2020, 10, 10294. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Ma, J.; Hou, Q.; Liu, K.; Ding, Y.; Du, B. Screening and whole-genome sequencing of two Streptomyces species from the rhizosphere soil of peony reveal their characteristics as plant growth-promoting rhizobacteria. BioMed Res. Int. 2018, 2018, 2419686. [Google Scholar] [CrossRef]
- Ser, H.-L.; Tan, W.-S.; Ab Mutalib, N.-S.; Yin, W.-F.; Chan, K.-G.; Goh, B.-H.; Lee, L.-H. Genome sequence of Streptomyces mangrovisoli MUSC 149T isolated from intertidal sediments. Braz. J. Microbiol. 2018, 49, 13–15. [Google Scholar] [CrossRef]
- Franco, C.M.; Adetutu, E.M.; Le, H.X.; Ballard, R.A.; Araujo, R.; Tobe, S.S.; Paul, B.; Mallya, S.; Satyamoorthy, K. Complete genome sequences of the endophytic Streptomyces sp. strains LUP30 and LUP47B, isolated from lucerne plants. Genome Announc. 2017, 5, e00556-17. [Google Scholar] [CrossRef]
- Alam, M.T.; Merlo, M.E.; Takano, E.; Breitling, R. Genome-based phylogenetic analysis of Streptomyces and its relatives. Mol. Phylogenet. Evol. 2010, 54, 763–772. [Google Scholar] [CrossRef]
- Heinsch, S.C.; Hsu, S.-Y.; Otto-Hanson, L.; Kinkel, L.; Smanski, M.J. Complete genome sequences of Streptomyces spp. isolated from disease-suppressive soils. BMC Genom. 2019, 20, 994. [Google Scholar] [CrossRef]
- Majer, H.M.; Ehrlich, R.L.; Ahmed, A.; Earl, J.P.; Ehrlich, G.D.; Beld, J. Whole genome sequencing of Streptomyces actuosus ISP-5337, Streptomyces sioyaensis B-5408, and Actinospica acidiphila B-2296 reveals secondary metabolomes with antibiotic potential. Biotechnol. Rep. 2021, 29, e00596. [Google Scholar] [CrossRef]
- Vicente, C.M.; Thibessard, A.; Lorenzi, J.-N.; Benhadj, M.; Hôtel, L.; Gacemi-Kirane, D.; Lespinet, O.; Leblond, P.; Aigle, B. Comparative genomics among closely related Streptomyces strains revealed specialized metabolite biosynthetic gene cluster diversity. Antibiotics 2018, 7, 86. [Google Scholar] [CrossRef]
- Qian, P.-Y.; Li, Z.; Xu, Y.; Li, Y.; Fusetani, N. Mini-review: Marine natural products and their synthetic analogs as antifouling compounds: 2009–2014. Biofouling 2015, 31, 101–122. [Google Scholar] [CrossRef]
- Matsumoto, A.; Takahashi, Y. Endophytic actinomycetes: Promising source of novel bioactive compounds. J. Antibiot. 2017, 70, 514–519. [Google Scholar] [CrossRef]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef]
- Malik, A.; Kim, Y.R.; Jang, I.H.; Hwang, S.; Oh, D.-C.; Kim, S.B. Genome-based analysis for the bioactive potential of Streptomyces yeochonensis CN732, an acidophilic filamentous soil actinobacterium. BMC Genom. 2020, 21, 118. [Google Scholar] [CrossRef]
- Siupka, P.; Hansen, F.T.; Schier, A.; Rocco, S.; Sørensen, T.; Seget, Z.P. Antifungal activity and biosynthetic potential of new Streptomyces sp. MW-W600-10 strain isolated from coal mine water. Int. J. Mol. Sci. 2021, 22, 7441. [Google Scholar] [CrossRef]
- Peña, A.; Del Carratore, F.; Cummings, M.; Takano, E.; Breitling, R. Output ordering and prioritisation system (OOPS): Ranking biosynthetic gene clusters to enhance bioactive metabolite discovery. J. Ind. Microbiol. Biotechnol. 2018, 45, 615–619. [Google Scholar] [CrossRef]
- Davis, K.E.R.; Joseph, S.J.; Janssen, P.H. Effects of Growth Medium, Inoculum Size, and Incubation Time on Culturability and Isolation of Soil Bacteria. Appl. Environ. Microbiol. 2005, 71, 826–834. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef]
- Lu, L.; Wang, J.; Xu, Y.; Wang, K.; Hu, Y.; Tian, R.; Yang, B.; Lai, Q.; Li, Y.; Zhang, W. A high-resolution LC-MS-based secondary metabolite fingerprint database of marine bacteria. Sci. Rep. 2014, 4, 6537. [Google Scholar] [CrossRef] [PubMed]
- Tangerina, M.M.; Furtado, L.C.; Leite, V.M.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic study of marine Streptomyces sp.: Secondary metabolites and the production of potential anticancer compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Haltli, B.; Summers, M.; Feng, X.; Hucul, J. Isolation and characterization of meridamycin biosynthetic gene cluster from Streptomyces sp. NRRL 30748. Gene 2006, 377, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hong, H.; Samborskyy, M.; Mironenko, T.; Leadlay, P.F.; Haydock, S.F. Organization of the biosynthetic gene cluster in Streptomyces sp. DSM 4137 for the novel neuroprotectant polyketide meridamycin. Microbiology 2006, 152, 3507–3515. [Google Scholar]
- Liu, M.; Lu, C.; Shen, Y. Four New Meridamycin Congeners Streptomyces sp. SR107. RSC Adv. 2016, 6, 49792–49796. [Google Scholar] [CrossRef]




| Strain | Bio-Project Accession | Size (Mbps) | No. of Contigs | % G + C | CDS | tRNA | rRNA |
|---|---|---|---|---|---|---|---|
| Streptomyces sp. isolate BR123 | PRJNA643667 | 8.16 | 723 | 72.63 | 8103 | 68 | 8 |
| Streptomyces globosus LZH-48 | PRJNA428275 | 7.54 | - | 73.62 | 6524 | 71 | 3 |
| Streptomyces katrae NRRL ISP-5550 | PRJNA238534 | 8.05 | 1874 | 72.69 | 7305 | 56 | 2 |
| Streptomyces virginiae NRRL ISP-5094 | PRJNA238534 | 8.32 | 133 | 72.4 | 7245 | 74 | 13 |
| Streptomyces clavuligerus F1D7 | PRJNA679926 | 7.59 | - | 72.5 | 6122 | 65 | 18 |
| Streptomyces diastaticus NBRC 15402 | PRJDB6184 | 7.85 | 32 | 72.7 | - | 75 | 8 |
| Streptomyces bacillaris ATCC 15855 | PRJNA471017 | 7.89 | - | 72.0 | 6746 | 65 | 18 |
| Streptomyces cyaneofuscatus SID 10855 | PRJNA603111 | 7.88 | 52 | 71.6 | 6755 | 66 | 12 |
| Streptomyces griseus NBRC 13350 | PRJDA20085 | 8.55 | - | 72.2 | 7087 | 67 | 18 |
| Streptomyces lavendulae YAKB-15 | PRJNA526603 | 7.77 | 100 | 72.2 | 7009 | 70 | 21 |
| Query | Reference | ANI Estimate | Matches | Total |
|---|---|---|---|---|
| Streptomyces lavendulae subsp. lavendulae | Streptomyces sp. isolate BR123 | 81.6134 | 1070 | 2300 |
| Streptomyces sp. isolate BR123 | Streptomyces lavendulae subsp. lavendulae | 81.673 | 1050 | 2391 |
| Streptomyces sp. isolate BR123 | Streptomyces virginiae | 86.0723 | 1576 | 2391 |
| Streptomyces virginiae | Streptomyces sp. isolate BR123 | 86.0802 | 1554 | 2721 |
| Streptomyces globosus | Streptomyces sp. isolate BR123 | 87.1686 | 1630 | 2510 |
| Streptomyces sp. isolate BR123 | Streptomyces globosus | 87.3066 | 1626 | 2391 |
| Streptomyces sp. isolate BR123 | Streptomyces katrae | 87.1854 | 1671 | 2391 |
| Streptomyces katrae | Streptomyces sp. isolate BR123 | 87.2335 | 1635 | 2813 |
| Cluster | Size (bp) | Most Similar Known Biosynthetic Gene Cluster | MIBG BGC-ID |
|---|---|---|---|
| Siderophores: | |||
| 3 | 11,590 | - | - |
| 56 | 6349 | - | - |
| 226 | 8264 | Desferrioxamin B (100%) | BGC0000941 |
| 261 | 8036 | Ficellomycin (7%) | BGC0001593 |
| 279 | 6963 | Ficellomycin (7%) | BGC0001593 |
| Terpenes: | |||
| 9 | 16,885 | - | - |
| 11 | 21,676 | Hopene (61%) | BGC0000663 |
| 16 | 21,086 | - | - |
| 19 | 13,165 | - | - |
| 24 | 25,408 | Isorenieratene (63%) | BGC0001227 |
| 69 | 13,506 | Ebelactone (5%) | BGC0001580 |
| PKS: | |||
| 2 (Type I) | 103,249 | Concanamycin A (21%) | BGC0000040 |
| 4 (Type I) | 46,281 | Clifednamide A (30%) | BGC0001553 |
| 94 (Type I) | 23,404 | Tetrocarcin A (8%) | BGC0000162 |
| 129 (Type I) | 19,401 | - | - |
| 320(Type I) | 7593 | - | - |
| 350(Type I) | 6899 | - | - |
| 58 (Type II) | 34,290 | Granaticin (16%) | BGC0000227 |
| 89 (Type III) | 24,296 | Alkylresorcinol (100%) | BGC0000282 |
| 338 (Type III) | 7187 | Flaviolin (75%) | BGC0000902 |
| NRPS: | |||
| 104 | 23,007 | Lactonamycin (5%) | BGC0000238 |
| 271 | 9618 | Griseoviridin/Fijimycin A (8%) | BGC0000459 |
| 239 | 11,133 | - | - |
| 401 | 5437 | Virginiamycin S1 (11%) | BGC0001116 |
| Peptides: | |||
| 59 (Lanthipeptide class II) | 13,149 | - | - |
| 76 (Lanthipeptide class I) | 23,247 | Chejuenolide A/Chejuenolide B (7%) | BGC0001543 |
| Butyrolactones: | |||
| 100 | 6302 | Griseoviridin/Fijimycin A (8%) | BGC0000459 |
| NRPS/PKS-like: | |||
| 221 (NRPS-like) | 12,004 | Lipstatin (14%) | BGC0000382 |
| 429 (NRPS-like) | 4493 | Glycinocin A (4%) | BGC0000379 |
| 243 (PKS-like) | 10,893 | Virginiamycin S1 (33%) | BGC0001116 |
| Hybrids: | |||
| 3 (Melanin, terpene) | 33,435 | Melanin (40%) | BGC0000909 |
| 29 (Lanthipeptide-3, NRPS) | 43,146 | Azicemicin (8%) | BGC0000202 |
| 46 (NRPS, transAT-PKS) | 36,866 | Virgimiamycin S1 (55%) | BGC0001116 |
| 62 (Type I PKS, NRPS-like) | 29,119 | Monensin (26%) | BGC0000100 |
| 98 (Type III PKS, guanidinotides, RiPP-like) | 23,202 | Pheganomycin (52%) | BGC0001148 |
| 149 (Type I PKS, NAPAA) | |||
| 433 (RRE-containing, thiopeptide) | 17,747 | Mediomycin A (34%) | BGC0001661 |
| 4312 | Lactazol (33%) | BGC0000606 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, N.; Zafar, S.; Makitrynskyy, R.; Bechthold, A.; Spiteller, D.; Song, L.; Anwar, M.A.; Luzhetskyy, A.; Khan, A.N.; Akhtar, K.; et al. Revealing Genome-Based Biosynthetic Potential of Streptomyces sp. BR123 Isolated from Sunflower Rhizosphere with Broad Spectrum Antimicrobial Activity. Antibiotics 2022, 11, 1057. https://doi.org/10.3390/antibiotics11081057
Ashraf N, Zafar S, Makitrynskyy R, Bechthold A, Spiteller D, Song L, Anwar MA, Luzhetskyy A, Khan AN, Akhtar K, et al. Revealing Genome-Based Biosynthetic Potential of Streptomyces sp. BR123 Isolated from Sunflower Rhizosphere with Broad Spectrum Antimicrobial Activity. Antibiotics. 2022; 11(8):1057. https://doi.org/10.3390/antibiotics11081057
Chicago/Turabian StyleAshraf, Neelma, Sana Zafar, Roman Makitrynskyy, Andreas Bechthold, Dieter Spiteller, Lijiang Song, Munir Ahmad Anwar, Andriy Luzhetskyy, Ali Nisar Khan, Kalsoom Akhtar, and et al. 2022. "Revealing Genome-Based Biosynthetic Potential of Streptomyces sp. BR123 Isolated from Sunflower Rhizosphere with Broad Spectrum Antimicrobial Activity" Antibiotics 11, no. 8: 1057. https://doi.org/10.3390/antibiotics11081057
APA StyleAshraf, N., Zafar, S., Makitrynskyy, R., Bechthold, A., Spiteller, D., Song, L., Anwar, M. A., Luzhetskyy, A., Khan, A. N., Akhtar, K., & Khaliq, S. (2022). Revealing Genome-Based Biosynthetic Potential of Streptomyces sp. BR123 Isolated from Sunflower Rhizosphere with Broad Spectrum Antimicrobial Activity. Antibiotics, 11(8), 1057. https://doi.org/10.3390/antibiotics11081057