Abstract
Studies have previously described the occurrence of multidrug-resistant (MDR) Escherichia coli in human and veterinary medical settings, livestock, and, to a lesser extent, in the environment and food. While they mostly analyzed foodborne E. coli regarding phenotypic and sometimes genotypic antibiotic resistance and basic phylogenetic classification, we have limited understanding of the in vitro and in vivo virulence characteristics and global phylogenetic contexts of these bacteria. Here, we investigated in-depth an E. coli strain (PBIO3502) isolated from a pork sausage in Germany in 2021. Whole-genome sequence analysis revealed sequence type (ST)58, which has an internationally emerging high-risk clonal lineage. In addition to its MDR phenotype that mostly matched the genotype, PBIO3502 demonstrated pronounced virulence features, including in vitro biofilm formation, siderophore secretion, serum resilience, and in vivo mortality in Galleria mellonella larvae. Along with the genomic analysis indicating close phylogenetic relatedness of our strain with publicly available, clinically relevant representatives of the same ST, these results suggest the zoonotic and pathogenic character of PBIO3502 with the potential to cause infection in humans and animals. Additionally, our study highlights the necessity of the One Health approach while integrating human, animal, and environmental health, as well as the role of meat products and food chains in the putative transmission of MDR pathogens.
1. Introduction
Multidrug-resistant (MDR) Enterobacterales, such as Escherichia coli, which are non-susceptible to more than three classes of antibiotics [1], have been on the rise for years. They are not only found in human and veterinary clinics but also in livestock and related animal products. Therefore, they are an important component of the One Health approach [2,3], which aims at increasing and/or preserving the well-being of human, animal, and environmental health. While most One Health reports in this research area focus on the distribution of antibiotic resistance and phylogenetic backgrounds, such as sequence types (STs), little is known about phenotypic virulence traits in vitro and in vivo [4]. The latter is particularly important when considering the zoonotic nature of MDR E. coli from animal sources, which have the potential to infect humans, and vice versa. These pathogenic E. coli are divided into extraintestinal pathogenic E. coli (ExPEC) and intestinal pathogenic representatives (InPEC) [5], with the latter exhibiting obligate pathogenicity, and foodborne isolates potentially causing outbreaks, such as in Germany in 2011 [6,7]. In contrast, ExPEC strains usually colonize the intestinal tract of humans and animals asymptomatically but frequently cause infection outside the gut. ExPEC possess several virulence factors (VFs) that enable adhesion, biofilm formation, resilience and, thus, infection in human and/or animal hosts, and are critical for bacterial pathogenicity. However, the exact contribution of foodborne MDR ExPEC to human and animal infection and colonization has not yet been established beyond doubt, as the differentiation between commensal E. coli and facultative ExPEC strains remains difficult when performing either geno- or phenotypic characterization alone [8].
Here, we report an extended-spectrum β-lactamase (ESBL)-producing E. coli isolated from a pork sausage in Germany that we investigated in-depth by whole-genome sequencing (WGS) and functional analyses. The aim of this study was to (i) identify the phylogenetic background of the food-derived strain and explore its phylogenetic relationship to other genomes of the same sequence type, and (ii) examine the repertoire of genotypic and phenotypic resistance- and virulence-associated features combining bioinformatics with multiple in vitro and in vivo assays.
2. Results
2.1. Genomic Characterization
As part of a small screening study for which we analyzed various meat products (including fresh pork sausage) from several grocery stores in Germany in May 2021, we isolated the ESBL-producing E. coli strain PBIO3502, which was first verified by MALDI-TOF MS and subsequently analyzed in detail by WGS. WGS revealed that PBIO3502 belonged to ST58 (serotype O65:H16) and, thus, phylogroup B1, which includes mostly commensal but also some ExPEC and InPEC strains [9]. PBIO3502 carried plasmids with the incompatibly groups (Inc)FIA and IncI1, as well as Col plasmids. Plasmid multilocus sequence typing [10] revealed that the IncI1 plasmid was ST3 (clonal complex 3).
The presence of resistance genes was investigated using AMRFinderPlus [11] and revealed the presence of the ESBL gene blaCTX-M-1 (Ambler class A) and a BlaEC family class C β-lactamase blaEC (98.67% identity to EC-15) in addition to genes conferring resistance to macrolides (mdf(A), mef(C), and mph(G)), streptomycin (aadA5), sulfonamides (sul2), and diaminopyrimidines (dfrA17). Our phenotypic analysis of antibiotic resistance was mostly consistent with the genotype (Table 1) and showed that PBIO3502 had an MDR phenotype [1]. Additionally, we noticed the presence of heavy metal resistance-associated genes (terWZABCDE) that enable resistance to tellurite. Usually, E. coli strains exhibit minimum inhibitory concentrations of tellurite of 1 µg mL−1 [12], but PBIO3502 showed a 64-fold increased tolerance.

Table 1.
Phenotypic and genotypic resistance profile of PBIO3502.
Among the nearly 300 genes associated with E. coli virulence provided by the virulence factor database (VFDB) [13], PBIO3502 harbored several gene clusters mainly associated with adherence (CFA/I fimbriae, cfaABCD/E; E. coli common pilus, ecpABCDER; factor adherence E. coli, fdeC; Hsp60, htpB; immunogenic lipoprotein A, ilpA; type I fimbriae, fimABCDEFGHIZ; type IV pili, vfr), biofilm formation (alginate, algU; autoinducer-2, luxS; curli fibers, csgABCDEFG), invasion (ibeBC, ompA, traJ), iron uptake (aerobactin, iucABCD, iutA; enterobactin, entABCDEFS, fepABCDEG, fes; salmochelin, iroB), and motility (peritrichous flagella, cheBRWYZ, flgBCDFGHI, flhABCD, fliAGIMPQR, motA; polar flagella, flmH, nueA). Genes encoding Shiga toxin were not present.
2.2. Phenotypic Assessment of Virulence-Associated Traits
Given the potential zoonotic background of PBIO3502 and this large genomic set of VFs, we further characterized PBIO3502 in several phenotypic virulence-associated experiments (Figure 1).
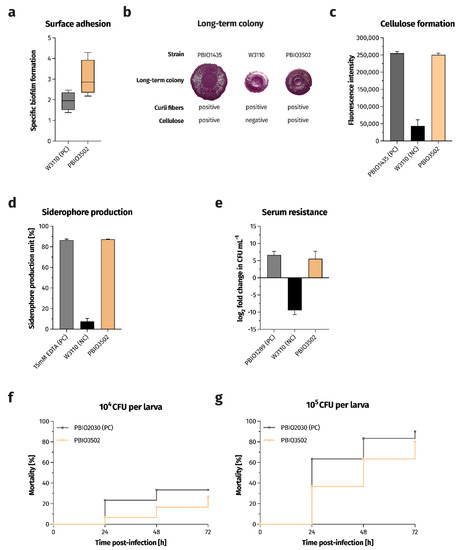
Figure 1.
PBIO3205 exhibits a highly virulent phenotype. (a) Biofilm formation on polystyrene surfaces as determined by crystal violet staining (n = 4). Results are expressed as growth-adjusted specific biofilm formation. The line within the box marks the median value, while the boxplot represents the 25th to 75th percentile of the data set. The whiskers mark the minimum and maximum values, respectively. (b) Morphology of long-term colonies examining the expression of the biofilm-associated extracellular matrix components curli fibers and cellulose. Congo red was used to specifically stain curli fibers, and cellulose was visually interpreted based on the textured surface. PBIO1435 and W3110 were used as references. (c) Cellulose formation was examined by staining with Calcofluor and measuring the fluorescence intensity of bound Calcofluor (n = 3). Results are given as mean values of fluorescence intensity and standard errors. (d) The extent of secreted siderophores is expressed as the mean of the percent unit of siderophore production and standard error (n = 3–5). (e) Survival in 50% human serum (n = 3–5). Results are given as means and standard errors of log2 fold change in CFU mL−1 after 4 h of incubation in the presence of human serum. (f,g) Kaplan–Meier plot of mortality in the Galleria mellonella larvae infection model (n = 3). Results are expressed as mean percent mortality after injection of 104 CFU per larva (g) and 105 CFU per larva, respectively. NC, negative control. PC, positive control.
Some bacterial populations are able to protect themselves from external stressors (e.g., UV radiation and antibiotic therapy) by forming a biofilm [14]. In addition, there is evidence that biofilm formation is a critical factor in infection development [15]. Therefore, we investigated the ability of PBIO3502 to form biofilms and analyzed whether the strain formed curli fibers (bacterial amyloid fibers) and/or cellulose (polysaccharide structures), which are important biofilm- and, thus, virulence-associated features [16]. When compared to the strong biofilm producer E. coli W3110 (specific biofilm formation (SBF) median, 1.96), PBIO3502 showed high adhesiveness to the plastic surface of a 96-well plate (SBF median, 2.86; Figure 1a), which likely promotes biofilm formation on catheters [17], for example. In addition, PBIO3502 showed both curli and cellulose formation in the long-term colony. The dye was used to assess the morphology of long-term colony stains only the curli fibers, but not the cellulose structures [18]. The latter are interpreted visually based on the structured surface (Figure 1b). Calcofluor, which has a high affinity for cellulose [19], was used to obtain a (semi-) quantitative result on the cellulose formation of PBIO3502. Our strain showed pronounced cellulose formation indicated by high fluorescence intensities (Figure 1c).
Iron is an important element for pathogenic bacteria, especially during infection, but also for general growth and fitness [20]. Since bacteria cannot produce iron themselves, they have to acquire it from their surroundings. Therefore, pathogens produce low-molecular iron-binding molecules (so-called siderophores) that chelate free extracellular iron(III) and are subsequently internalized by membrane receptors. We examined whether and to what extent PBIO3502 produced siderophores, and as shown in Figure 1d, PBIO3502 secreted large amounts of siderophores (mean, 87.25%). The extent of siderophores was 11.5-fold higher when compared to the negative control W3110.
Highly virulent E. coli representatives are capable of causing bacteremia [21] and, therefore, must compete with the host defense system (e.g., the complement system). Gram-negative bacteria use several different mechanisms to deal with the immune response provided by various mechanisms, such as capsule expression [22], down-regulation of envelope stress response pathways [23], and outer membrane surface modifications [24]. Therefore, we challenged this foodborne strain by incubating it in 50% human serum for 4 h. PBIO3502 was not inhibited by the complement-containing serum and was able to double its cell number (Figure 1e). This result again highlights the highly virulent and clinically relevant nature of PBIO3502.
In addition, we performed an infection model using larvae of the greater wax moth Galleria mellonella to investigate the ability of PBIO3502 to cause in vivo mortality. We injected 104 or 105 colony forming units (CFU) per larva. When 104 CFU were injected, a mortality rate of 6.7% was obtained after 24 h (Figure 1f). This mortality rate increased to 26.7% 72 h after infection. The 10-fold increase in inoculation size resulted in an increase in the mortality rate to 36.7% at 24 h and 80.0% at 72 h of infection (Figure 1g). These results were comparable to the mortality rates of a hypervirulent Klebsiella pneumoniae ST420 strain PBIO2030 isolated from a wound [25]. In conclusion, the findings demonstrate the virulent potential of PBIO3502.
2.3. Phylogenetics of ST58
Finally, to classify our strain in a more global perspective, and again evaluate the zoonotic potential, we performed a phylogenetic comparison of an overall 159 closely related, publicly available ST58 genomes and PBIO3502 (Figure 2).
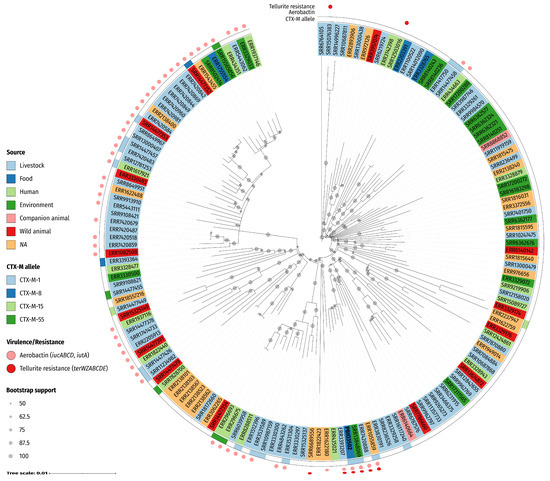
Figure 2.
Phylogenetics reveals close relationship of PBIO3502 with publicly available genomes (n = 159). Included genomes were selected based on most shared k-mers using Mash. The phylogenetic tree was constructed using a maximum likelihood-based approach and is based on a core singe-nucleotide polymorphism alignment (4984 sites). The circular tree was midpoint-rooted and the circles on the branches indicate bootstrap support of ≥50% from 1000 replicates. The labels indicate the accession number (except PBIO3502) and are colored according to their source/host, as indicated in the legend. Annotations indicate (from inner to outer circle): encoded CTX-M (cefotaximase) allele, presence of genes encoding aerobactin and tellurite resistance. NA, not applicable (i.e., no metadata provided).
The phylogenetic tree shows that the genomes grouped into multiple clades with several subclades. Dispersed across the clades, genomes originated from humans, livestock, companion and wild animals, food, and the environment. Note that PBIO3502 clustered with three genomes from livestock (ERR3393207, ERR7420888) and environmental sources (SRR13848069), respectively. Interestingly, these closely related strains were sampled from different European locations in different years and were mainly associated with pigs. For example, ERR7420888 (designated MSG45-C09) was isolated from a healthy pig on a commercial pig farm in the Midlands of the United Kingdom in 2015 [26], while SRR13848069 (designated 20-MO00084-0) originated from scalding water from a pig slaughterhouse in Germany in 2020 [27]. All genomes assigned to the “PBIO3502 and ERR1055859 subclade” encoded aerobactin and tellurite resistance genes, a combination that was unique in the phylogenetic tree. Although PBIO3502 did not show immediate phylogenetic relatedness to human-derived strains and was in a clade primarily composed of strains from domestic and wild animals and the environment, our analysis demonstrates that phylogenetically related ST58 strains from humans were present in other clades. This again indicates the zoonotic character of the ST58 lineage and highlights the complex connections within the One Health continuum.
3. Discussion
A growing body of studies shows the global spread of MDR E. coli and underlines the clinical relevance of this pathogen, with nearly 60,000 deaths caused by antibiotic-resistant E. coli in 2019 alone [28]. In addition, MDR E. coli strains are commonly found in the environment, in animals (pets and livestock), and in meat products [2,29,30,31,32,33,34]. Due to increasing worldwide meat consumption and pressure on the prices of processed foods, slaughterhouses have to operate in an economically-oriented high throughput, potentially leading to the contamination of meat products and the water used with enteric pathogens of animal origin [35]. Therefore, it is not surprising that we and others have recently detected MDR E. coli strains in the wastewater of German slaughterhouses that are even resistant to the last-resort drug colistin [27,36]. Moreover, contaminated meat products might foster the transmission of pathogens from animals to humans through the food chain, of which a confirmed MDR ExPEC outbreak in London (United Kingdom) in the mid-1980s [37,38,39] and the above-mentioned outbreak in Germany in 2011 are good examples. This underlines the interdependence of animal and human health and, thus, the need for the One Health approach. Although direct consumption of unprocessed fresh pork and poultry products is uncommon in Germany, putative foodborne ExPEC transmission involves the ingestion of inadequately cooked meat or cross-contamination in the kitchen during food handling, potentially, for example, resulting in wound infection [40,41].
The establishment and frequent application of next-generation sequencing technologies have demonstrated that the spread of MDR E. coli appears to be driven by a limited number of pandemic STs such as ST131, ST648, or ST410 [42,43,44]. These pathogenic clonal lineages successfully combine multidrug resistance and high virulence. However, other successful clonal ExPEC lineages, such as ST58 (which belongs to clonal complex 87 [45]), increasingly emerge as globally disseminated pathogens. PBIO3502 not only exhibited numerous virulence-associated characteristics, such as strong biofilm formation, extensive siderophore secretion, and serum resistance as well as in vivo mortality, but also demonstrated a MDR phenotype, supporting the above-mentioned previous findings. Additionally note that the phenotypic results were largely consistent with predictions from our genotypic analysis.
Unlike most pandemic ExPEC lineages, which are mostly associated with phylogroups B2 or D, ST58 belongs to phylogroup B1. Interestingly, this phylogroup contains only a small number of phylogroup-specific core genes compared to other phylogroups, indicating high diversity within this phylogroup [46]. Here, we describe a foodborne E. coli ST58 strain that exhibits pronounced in vitro and in vivo virulence properties required for pathogenicity and bacterial survival, indicating the great potential to actually cause infection in humans and animals upon successful transmission. While the direct transfer of E. coli ST58 strains via the food chain has not yet been confirmed beyond doubt [4,47], and is also not within the scope of this study, it has been previously suggested that ST58 shows high clonality among strains originating from humans, animals, and the environment [48,49,50]. Our phylogenetic analysis comparing global ST58 genomes supports the latter and reiterates the clinical and zoonotic relevance of our strain. However, we used a limited number of genomes in this study. Future investigations will need to address this further and provide a more comprehensive phylogenetic and epidemiologic picture of this (putatively) successful, emerging clonal lineage.
Many resistance genes, but especially ESBL enzymes, are often encoded on plasmids, which allows rapid resistance transfer among different bacteria and, thus, widespread dissemination [51]. Interestingly, PBIO3502 carried an IncI1 plasmid, which appears to be a common plasmid type of InPEC strains, but also, to a lesser extent, occurs in ExPEC [52]. Moreover, ST3 IncI1 plasmids are not only frequently found in strains isolated from humans but also in pathogens of animal origin [53]. This is further evidence of the zoonotic character of PBIO3502.
4. Materials and Methods
4.1. Strain Origin and General Methods
As part of a small screening study, packaged fresh meat samples from various grocery stores were tested for different MDR bacteria. Briefly, the meat samples were opened under sterile conditions, two pieces of approximately 5 mm³ were cut out with a sterile scalpel (Braun, Melsungen, Germany) and transferred to 5 mL of tryptic soy broth (Carl Roth, Karlsruhe, Germany). The bacteria were then enriched under shaking conditions (130 rpm) at 37 °C overnight. Then, 100 µL of the bacterial suspension was plated onto different chromogenic selection plates and incubated overnight at 37 °C. The strain 27-ESBL-EC (internal designation PBIO3502) was cultivated on a CHROM-ESBL selection plate (Mast Diagnostica GmbH, Reinfeld, Germany) and the species E. coli was confirmed using MALDI-TOF MS (Bruker, Bremen, Germany). It was derived from a fresh bratwurst purchased from a German grocery store. All strains were stored at −80 °C in lysogeny broth (LB; Carl Roth, Karlsruhe, Germany) supplemented with 20% (v/v) glycerol (anhydrous; Merck, Darmstadt, Germany). Prior to use, one single colony of fresh overnight cultures on LB agar plates was inoculated in 5 mL of LB and grown under shaking conditions (130 rpm) at 37 °C overnight.
4.2. Whole-Genome Sequencing
Total DNA was extracted using the MasterPure DNA Purification Kit for Blood, v. 2 (Lucigen, Middleton, WI, USA), according to the manufacturer’s instructions. The isolated DNA was quantified fluorometrically using the Qubit 4 fluorometer and the corresponding dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). DNA was shipped to the Microbial Genome Sequencing Center (MiGS), now SeqCenter (Pittsburgh, PA, USA), and after library preparation using the Illumina DNA Prep Kit and IDT 10 bp UDI indices (Illumina, San Diego, CA, USA) sequenced on an Illumina NextSeq 2000, producing 2 × 151 bp reads. Demultiplexing, quality control, and adapter trimming were performed using bcl-convert v. 3.9.3 [54].
4.3. Sequence Assembly and Genomic Analyses
Short-read data were processed using BBDuk from BBTools v. 38.95 (https://sourceforge.net/projects/bbmap/, accessed on 3 March 2022) to trim the adapters, filter possible PhiX contaminants, and do further quality- and polymer-trimming. Quality control (QC) of provided (raw) and processed (trimmed) reads was performed using FastQC v. 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, accessed on 3 March 2022). Trimmed reads were assembled using shovill v. 1.1.0 (https://github.com/tseemann/shovill, accessed on 3 March 2022) with SPAdes v. 3.15.3 [55]. An additional polishing step (besides the one implemented in the shovill assembly pipeline) was performed by first mapping the trimmed reads to the assembly using BWA v. 0.7.17 [56]. The alignment files were then converted to binary format, sorted, and duplicate reads were marked with SAMtools v. 1.14 [57]. Finally, the draft contigs were corrected using Pilon v. 1.24 [58]. Genome completeness and contamination were assessed using CheckM v. 1.1.3 [59]. Prokka v. 1.14.6 [60] was used to automatically annotate the draft assembly. Genomic analyses including in silico multilocus sequence typing, serotype prediction, and antibiotic resistance and virulence feature detection were performed using mlst v. 2.19.0 (https://github.com/tseemann/mlst, accessed on 3 March 2022; with the PubMLST [61] database and Enterobase [62]), ABRicate v. 1.0.0 (https://github.com/tseemann/abricate, accessed on 3 March 2022; with ResFinder [63], PlasmidFinder [10], VFDB [13], BacMet2 [64], and EcOH [65] databases), and AMRFinderPlus v. 3.10.30 with database v. 2022-05-26.1 [11]. Assignment to the phylogroup was performed using ClermonTyping v. 20.03 [66].
4.4. Phylogeny
For the creation of a core single-nucleotide polymorphism (SNP)-based phylogeny of closely related isolates, Enterobase [62] was searched for E. coli ST58 strains (n = 2199), for which paired Illumina read data were available (n = 2186). Data were downloaded from the public European Nucleotide Archive SRA FTP server (ftp://ftp.sra.ebi.ac.uk/vol1/, accessed on 20 June 2022). Mash v. 2.3 [67] was used to generate sketches (k-mer size, 21; sketch size, 1,000,000; minimum number of k-mers for reads, 3) of the reference sequence (draft genome of PBIO3502) and the public raw read data. Afterwards, the distance between the reference sketch and the query raw read sketches was estimated and the 200 read sets with the shortest distance (most shared k-mers) selected. For these accessions, read trimming, genome assembly, and analyses were performed as described above. The trimmed reads were mapped against the draft genome of PBIO3502 using snippy v. 4.6.0 (https://github.com/tseemann/snippy, accessed on 22 June 2022) to create a whole-genome alignment with 201 sequences. The alignment was processed using Gubbins v. 3.2.1 [68] to filter out regions with SNPs that are likely the result of recombination. In this step, two isolates were removed due to alignment gaps or missing information at more than 25% of the alignment sites. The alignment of the remaining sequences (n = 199) was processed using snp-sites v. 2.5.1 [69] to retain only alignment positions containing A, C, G, or T exclusively. A maximum likelihood tree was inferred with RAxML-NG v. 1.1.0 [70] using GTR+G by first parsing the alignment and excluding 40 sequences that were completely identical to another sequence. The final alignment (containing 159 sequences and 4984 sites) was then processed by searching 500 parsimony and 500 random starting trees and performing 1000 bootstrap repeats. The best-scoring maximum likelihood tree was midpoint-rooted in iTOL v. 6.5.7 [71] and visualized with bootstrap support values and metadata. The exported graphic was post-processed using Affinity Designer v. 1.10.5.1342 (https://affinity.serif.com/en-us/designer/, accessed on 22 June 2022).
4.5. Minimum Inhibitory Concentration
Phenotypic antimicrobial susceptibility testing was performed in collaboration with the IMD Laboratory Greifswald (Germany) using the automated VITEK 2 system (bioMérieux, Marcy l’Etoile, France). Testing was performed using the AST-N389 card, according to the manufacturer’s instructions. MIC values of gentamicin (Carl Roth, Karlsruhe, Germany), streptomycin (Sigma-Aldrich, St. Louis, MO, USA), tetracycline (Carl Roth, Karlsruhe, Germany), and tellurite (Alfa Aesar, Haverhill, MA, USA) were determined by broth microdilution according to ISO Standard 20776-1 [72]. Briefly, several single colonies were resuspended in a 0.9% (w/v) NaCl solution until the corresponding suspensions had an OD600 equal to 0.5 McFarland standard turbidity. The bacterial suspensions were then diluted 1:230 in cation-adjusted Mueller–Hinton broth 2 (MH-2; Sigma-Aldrich, St. Louis, MO, USA), corresponding to approximately 105 CFU mL−1. Serial 2-fold dilutions were performed in a 96-well plate (Sarstedt, Nümbrecht, Germany) with concentrations ranging from 64 to 0.5 µg mL−1 for the antibiotics gentamicin, streptomycin, and tetracycline, and from 512 to 0.5 µg mL−1 for the heavy metal tellurite, respectively. Finally, the bacterial suspensions were added and incubated at 37 °C for 18 ± 2 h. The MIC represents the lowest concentration of antimicrobial agent that inhibits visible bacterial growth.
4.6. Biofilm Formation
Biofilm formation on polystyrene surfaces was determined by crystal violet (CV) staining, as previously described [73,74] with some adjustments. First, overnight cultures were diluted 100-fold in 5 mL of fresh M9 minimal salt medium (MP Biomedicals, Irvine, CA, USA) containing 0.4% (w/v) glucose (Carl Roth, Karlsruhe, Germany) and incubated at 37 °C and 130 rpm until OD600 reached 0.5 McFarland standard turbidity. The bacterial suspensions were then diluted 1:10 and 200 µL were then transferred in triplicate to a 96-well flat-bottomed polystyrene plate (Nunc, Thermo Fisher Scientific, Waltham, MA, USA). In addition, three control wells were filled with 200 µL of sterile medium. The plates were hermetically sealed and incubated at 28 °C for 24 h without shaking. Subsequently, OD600 was measured using CLARIOstar Plus (BMG LABTECH GmbH, Ortenberg, Germany), planktonic cells were removed by washing three times with deionized water, and the microtiter plates were air-dried for 10 min. After fixation with 250 µL of 99% (v/v) methanol (Merck, Darmstadt, Germany) for 15 min and air-drying, the cells were stained with 250 µL of a 0.1% (w/v) aqueous CV solution (Sigma-Aldrich, St. Louis, MO, USA) for 30 min. The staining solution was then discarded, and unbound dye was removed by washing three times with deionized water. After air-drying for 10 min, the bound CV was dissolved with a mixture of 80 parts ethanol (99.8% (v/v); Carl Roth, Karlsruhe, Germany) and 20 parts acetone (Merck, Darmstadt, Germany) at room temperature (rt) with horizontal shaking at 200 rpm for 30 min. After complete dissolution, 125 µL were transferred to a new microtiter plate and optical density was measured at a wavelength of 570 nm using the plate reader. The strength of biofilm formation was expressed as specific biofilm formation (SBF). The SBF was calculated according to the following formula [75]: SBF = (B−NC)/G, where B is the OD570 of the stained bacteria, NC is the OD570 of the stained control wells to eliminate the fraction of CV adhering to the polystyrene surface due to abiotic factors, and G is the OD600 representing the density of cells grown in the media.
The expression of biofilm-associated extracellular matrix components such as cellulose and curli fibers was tested using macrocolony assays as previously described [73]. Five microliters of overnight culture was dropped onto span agar plates (Hellmuth Carroux, Hamburg, Germany). The plates were hermetically sealed and incubated at 28 °C for 5 days. For detection of curli fimbriae, 0.005% Congo red and 0.0025% Coomassie Brilliant Blue G-250 (both chemicals were purchased from Carl Roth, Karlsruhe, Germany) were added to the span agar and evaluated visually. For cellulose staining, the span agar contained 0.004% Calcofluor (Sigma-Aldrich, St. Louis, MO, USA), and the extent of cellulose formation was measured by fluorescence intensity using the plate reader (excitation, 400–415 nm, emission, 480–520 nm).
4.7. Siderophore Production Assay
Quantification of the extent of siderophores secreted by PBIO3502 was performed according to a previously published protocol [23]. Briefly, the adjusted bacterial suspensions were diluted and grown in 5 mL of chelated M9 minimal medium supplemented with casamino acids (c-M9-CA) at 37 °C and 130 rpm for 24 h. The c-M9-CA consisted of the following compounds: M9 minimal salt medium (MP Biomedicals, Irvine, CA, USA), 2 mM MgSO4 (Carl Roth, Karlsruhe, Germany), 200 µM 2,2′-dipyridyl (Carl Roth, Karlsruhe, Germany), and 0.3% (w/v) casamino acids (BD, Franklin Lakes, NJ, USA). After incubation, 1 mL of the bacterial cultures were centrifuged (4900× g for 20 min at rt) and 100 µL of the siderophore-containing supernatant were transferred in triplicate to 96-well microtiter plates (Nunc, Thermo Fisher Scientific, Waltham, MA, USA) with 100 µL of chrome azurol S shuttle solution (composition according to [76]) already in the wells. Wells containing only fresh medium and 15 mM EDTA (Carl Roth, Karlsruhe, Germany) served as a blank and positive control, respectively. The non-siderophore producer W3110 served as a negative control. All mixtures were incubated in the dark at rt for 30 min. Finally, absorbance was measured at a wavelength of 630 nm using the plate reader. Secretion of siderophores was calculated as previously described [77] and expressed as a percentage unit of siderophore production.
4.8. Serum Resistance
Determination of survival in 50% human serum was performed as previously described [23]. Briefly, overnight cultures were diluted 1:100 in 5 mL of fresh LB and incubated with shaking at 37 °C until OD600 reached 0.5 McFarland standard turbidity. Then, 1000 µL of the bacterial suspension was pelleted (7500× g for 5 min at rt) and resuspended in 1 mL of phosphate-buffered saline (PBS; Thermo Fisher Scientific, Waltham, MA, USA). One hundred microliters of the sample were seeded in a 96-well microtiter plate containing 100 µL of human serum (United States origin; Sigma-Aldrich, St. Louis, MO, USA) per well (resulting in a final concentration of 50% human serum and approximately 108 CFU mL−1). Following this, 20 µL of each sample was withdrawn and serial dilutions were plated on LB agar plates and incubated overnight at 37 °C to determine the size of the inoculum. The inoculated microtiter plates were incubated for 4 h at 37 °C without agitation. Thereafter, the number of surviving CFU mL−1 was determined by plating out serial dilutions and incubating at 37 °C overnight. The positive control in each experiment was the serum-resistant PBIO1289 (initially designated as IMT10740 [78,79]). The serum-sensitive W3110 served as the negative control. Serum resistance was expressed as log2 fold change in CFU mL−1 after treatment with respect to inoculum size.
4.9. Infection of Galleria mellonella Larvae
Infections of larvae of the greater wax moth G. mellonella were performed as previously described [23]. Overnight cultures were diluted 1:100 in 30 mL of fresh LB and incubated with shaking at 37 °C to an OD600 of 1.0. Then, 1000 µL of the bacterial suspension were pelleted (16,000× g for 5 min at rt) and washed twice with PBS. The bacterial suspensions were diluted to 106 CFU mL−1 and 107 CFU mL−1, respectively. Larvae (proinsects, Minden, Germany) were randomly divided into groups of 10 individuals each and 10 µL of the adjusted bacterial suspensions were injected into the left proleg. In addition, 10 µL of PBS was injected into a group of larvae to ensure that death was not due to trauma from the injection. Each group was placed in 90 mm glass Petri dishes, kept at 37 °C in the dark, and death was recorded every 24 h. Individuals were considered dead when they no longer responded to physical stimuli and showed pigmentation. The results of three independent tests were pooled for each strain to generate Kaplan–Meier plots of mortality rates [80].
4.10. Data Visualization and Analysis
Data visualization was performed using GraphPad Prism v. 9.3.1 for macOS (GraphPad Software, San Diego, CA, USA). All experiments were performed with three or more independent biological replicates. Unless otherwise specified, data were expressed as mean and standard error.
5. Conclusions
The finding of a highly virulent and MDR, foodborne E. coli ST58, which was phylogenetically related to clinical strains of the same ST, suggests its zoonotic and pathogenic potential and the relevance of the One Health approach. Additionally, our study highlights the potential role of food (chains) in the spread and putative transmission of MDR pathogens.
Author Contributions
Conceptualization, E.E. and K.S.; methodology, E.E. and S.E.H.; software, S.E.H.; validation, E.E.; formal analysis, C.H., D.Z., E.E., K.S., S.E.H., T.H.-B., M.D., M.P. and V.B.; investigation, E.E., M.D., M.P. and V.B.; resources, C.H., D.Z. and T.H.-B.; data curation, E.E. and M.D.; writing—original draft preparation, E.E.; writing—review and editing, K.S.; visualization, E.E. and S.E.H.; supervision, K.S.; project administration, K.S.; funding acquisition, K.S. and T.H.-B. All authors have read and agreed to the published version of the manuscript.
Funding
The APC was funded by Land Schleswig-Holstein within the funding programme Open Access Publikationsfonds.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB53849 (https://www.ebi.ac.uk/ena/browser/view/PRJEB53849, uploaded on 24 June 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Hughes, J.M. Critical importance of a One Health approach to antimicrobial resistance. Ecohealth 2019, 16, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Buchholz, U.; Bernard, H.; Werber, D.; Bohmer, M.M.; Remschmidt, C.; Wilking, H.; Delere, Y.; an der Heiden, M.; Adlhoch, C.; Dreesman, J.; et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 2011, 365, 1763–1770. [Google Scholar] [CrossRef]
- Bielaszewska, M.; Mellmann, A.; Zhang, W.; Köck, R.; Fruth, A.; Bauwens, A.; Peters, G.; Karch, H. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: A microbiological study. Lancet Infect. Dis. 2011, 11, 671–676. [Google Scholar] [CrossRef]
- Day, M.; Hopkins, K.; Wareham, D.; Toleman, M.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Moller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.E. Bacterial tellurite resistance. Trends Microbiol. 1999, 7, 111–115. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Van Houdt, R.; Michiels, C.W. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 2005, 156, 626–633. [Google Scholar] [CrossRef]
- Ciofu, O.; Moser, C.; Jensen, P.O.; Hoiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022. [Google Scholar] [CrossRef]
- Bokranz, W.; Wang, X.; Tschape, H.; Romling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef]
- Wilks, S.A.; Koerfer, V.V.; Prieto, J.A.; Fader, M.; Keevil, C.W. Biofilm development on urinary catheters promotes the appearance of viable but nonculturable bacteria. mBio 2021, 12, e03584-20. [Google Scholar] [CrossRef]
- Reichhardt, C.; Jacobson, A.N.; Maher, M.C.; Uang, J.; McCrate, O.A.; Eckart, M.; Cegelski, L. Congo red interactions with curli-producing E. coli and native curli amyloid fibers. PLoS ONE 2015, 10, e0140388. [Google Scholar] [CrossRef]
- Serra, D.O.; Richter, A.M.; Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 2013, 195, 5540–5554. [Google Scholar] [CrossRef]
- Page, M.G.P. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin. Infect. Dis. 2019, 69, S529–S537. [Google Scholar] [CrossRef]
- Denamur, E.; Condamine, B.; Esposito-Farese, M.; Royer, G.; Clermont, O.; Laouenan, C.; Lefort, A.; de Lastours, V.; Galardini, M.; COLIBAFI and SEPTICOLI groups. Genome wide association study of Escherichia coli bloodstream infection isolates identifies genetic determinants for the portal of entry but not fatal outcome. PLoS Genet. 2022, 18, e1010112. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Eger, E.; Schwabe, M.; Schulig, L.; Hübner, N.-O.; Bohnert, J.A.; Bornscheuer, U.T.; Heiden, S.E.; Müller, J.U.; Adnan, F.; Becker, K.; et al. Extensively drug-resistant Klebsiella pneumoniae counteracts fitness and virulence costs that accompanied ceftazidime-avibactam resistance acquisition. Microbiol. Spectr. 2022, 10, e0014822. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.D.; Peters, K.M.; Sarkar, S.; Lukowski, S.W.; Allsopp, L.P.; Gomes Moriel, D.; Achard, M.E.; Totsika, M.; Marshall, V.M.; Upton, M.; et al. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet. 2013, 9, e1003834. [Google Scholar] [CrossRef] [PubMed]
- Eger, E.; Heiden, S.E.; Becker, K.; Rau, A.; Geisenhainer, K.; Idelevich, E.A.; Schaufler, K. Hypervirulent Klebsiella pneumoniae sequence type 420 with a chromosomally inserted virulence plasmid. Int. J. Mol. Sci. 2021, 22, 9196. [Google Scholar] [CrossRef]
- AbuOun, M.; O’Connor, H.M.; Stubberfield, E.J.; Nunez-Garcia, J.; Sayers, E.; Crook, D.W.; Smith, R.P.; Anjum, M.F. Characterizing antimicrobial resistant Escherichia coli and associated risk factors in a cross-sectional study of pig farms in Great Britain. Front. Microbiol. 2020, 11, 861. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Kreyenschmidt, J.; Schmithausen, R.M.; Sib, E.; Schmoger, S.; Kasbohrer, A.; Hammerl, J.A. Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in Germany. Microorganisms 2021, 9, 698. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 2012, 55, 712–719. [Google Scholar] [CrossRef]
- Ibrahim, D.R.; Dodd, C.E.; Stekel, D.J.; Ramsden, S.J.; Hobman, J.L. Multidrug resistant, extended spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol. Ecol. 2016, 92, fiw013. [Google Scholar] [CrossRef]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Kasbohrer, A.; Schwarz, S.; Rosler, U.; Kreienbrock, L.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef] [PubMed]
- Bergspica, I.; Kaprou, G.; Alexa, E.A.; Prieto, M.; Alvarez-Ordonez, A. Extended spectrum beta-lactamase (ESBL) producing Escherichia coli in pigs and pork meat in the European Union. Antibiotics 2020, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Eger, E.; Heiden, S.E.; Korolew, K.; Bayingana, C.; Ndoli, J.M.; Sendegeya, A.; Gahutu, J.B.; Kurz, M.S.E.; Mockenhaupt, F.P.; Müller, J.; et al. Circulation of extended-spectrum beta-lactamase-producing Escherichia coli of pandemic sequence types 131, 648, and 410 among hospitalized patients, caregivers, and the community in Rwanda. Front. Microbiol. 2021, 12, 662575. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.P.; Dreyer, S.; Heppelmann, M.; Schaufler, K.; Homeier-Bachmann, T.; Bachmann, L. Prevalence and risk factors for ESBL/AmpC-E. coli in pre-weaned dairy calves on dairy farms in Germany. Microorganisms 2021, 9, 2135. [Google Scholar] [CrossRef]
- Sofos, J.N. Challenges to meat safety in the 21st century. Meat Sci. 2008, 78, 3–13. [Google Scholar] [CrossRef]
- Homeier-Bachmann, T.; Heiden, S.E.; Lubcke, P.K.; Bachmann, L.; Bohnert, J.A.; Zimmermann, D.; Schaufler, K. Antibiotic-resistant Enterobacteriaceae in wastewater of abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- Phillips, I.; King, A.; Rowe, B.; Gross, R.J.; Eykyn, S.; Gransden, W.R.; Frost, J.A. Epidemic multiresistant Escherichia coli infection in West Lambeth Health District. Lancet 1988, 1, 1038–1041. [Google Scholar] [CrossRef]
- Waghorn, D.J.; Kelly, T.W.; Gibbins, W. Epidemic multi-resistant Escherichia coli infection in south London. J. Hosp. Infect. 1988, 11, 192–193. [Google Scholar] [CrossRef]
- Wright, E.D.; Perinpanayagam, R.M. Multiresistant invasive Escherichia coli Infection in South London. Lancet 1987, 1, 556–557. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R. Reservoirs of extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 2015, 3, 415–421. [Google Scholar] [CrossRef]
- Hara-Kudo, Y.; Takatori, K. Contamination level and ingestion dose of foodborne pathogens associated with infections. Epidemiol. Infect. 2011, 139, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Grobbel, M.; Stamm, I.; Kopp, P.A.; Diehl, I.; Semmler, T.; Fruth, A.; Beutlich, J.; Guerra, B.; Wieler, L.H.; et al. Emergence of human pandemic O25:H4-ST131 CTX-M-15 extended-spectrum-beta-lactamase-producing Escherichia coli among companion animals. J. Antimicrob. Chemother. 2010, 65, 651–660. [Google Scholar] [CrossRef]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Trott, D.J.; Pitout, J.; Peirano, G.; Bonnedahl, J.; Dolejska, M.; Literak, I.; Fuchs, S.; et al. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob. Agents Chemother. 2019, 63, e00243-00219. [Google Scholar] [CrossRef] [PubMed]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Wohrmann, M.; Baddam, R.; Ahmed, N.; Muller, K.; Kola, A.; Fruth, A.; Ewers, C.; et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410-Another successful pandemic clone? FEMS Microbiol. Ecol. 2016, 92, fiv155. [Google Scholar] [CrossRef] [PubMed]
- Skurnik, D.; Clermont, O.; Guillard, T.; Launay, A.; Danilchanka, O.; Pons, S.; Diancourt, L.; Lebreton, F.; Kadlec, K.; Roux, D.; et al. Emergence of antimicrobial-resistant Escherichia coli of animal origin spreading in humans. Mol. Biol. Evol. 2016, 33, 898–914. [Google Scholar] [CrossRef][Green Version]
- Abram, K.; Udaondo, Z.; Bleker, C.; Wanchai, V.; Wassenaar, T.M.; Robeson, M.S., 2nd; Ussery, D.W. Mash-based analyses of Escherichia coli genomes reveal 14 distinct phylogroups. Commun. Biol. 2021, 4, 117. [Google Scholar] [CrossRef]
- Lazarus, B.; Paterson, D.L.; Mollinger, J.L.; Rogers, B.A. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin. Infect. Dis. 2015, 60, 439–452. [Google Scholar] [CrossRef]
- Reid, C.J.; Cummins, M.L.; Borjesson, S.; Brouwer, M.S.M.; Hasman, H.; Hammerum, A.M.; Roer, L.; Hess, S.; Berendonk, T.; Nesporova, K.; et al. A role for ColV plasmids in the evolution of pathogenic Escherichia coli ST58. Nat. Commun. 2022, 13, 683. [Google Scholar] [CrossRef]
- Fuentes-Castillo, D.; Navas-Suarez, P.E.; Gondim, M.F.; Esposito, F.; Sacristan, C.; Fontana, H.; Fuga, B.; Piovani, C.; Kooij, R.; Lincopan, N.; et al. Genomic characterization of multidrug-resistant ESBL-producing Escherichia coli ST58 causing fatal colibacillosis in critically endangered Brazilian merganser (Mergus octosetaceus). Transbound Emerg. Dis. 2021, 68, 258–266. [Google Scholar] [CrossRef]
- Borges, C.A.; Tarlton, N.J.; Riley, L.W. Escherichia coli from commercial broiler and backyard chickens share sequence types, antimicrobial resistance profiles, and resistance genes with human extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2019, 16, 813–822. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Int. J. Med. Microbiol. 2011, 301, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Shepard, S.M.; Rivet, B.; Danzeisen, J.L.; Carattoli, A. Comparative genomics and phylogeny of the IncI1 plasmids: A common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid 2011, 66, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.; Bossers, A.; Harders, F.; Wu, G.; Woodford, N.; Schwarz, S.; Guerra, B.; Rodriguez, I.; van Essen-Zandbergen, A.; Brouwer, M.; et al. Characterization of epidemic IncI1-Iγ plasmids harboring Ambler class A and C genes in Escherichia coli and Salmonella enterica from animals and humans. Antimicrob. Agents Chemother. 2015, 59, 5357–5365. [Google Scholar] [CrossRef]
- Illumina. Bcl-Convert: A Proprietary Illumina Software for the Conversion of Bcl Files to Basecalls. Available online: https://support-docs.illumina.com/SW/BCL_Convert/Content/SW/FrontPages/BCL_Convert.htm (accessed on 7 March 2022).
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Rensing, C.; Kristiansson, E.; Larsson, D.G. BacMet: Antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014, 42, D737–D743. [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar] [CrossRef] [PubMed]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- ISO 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: London, UK, 2019.
- Schaufler, K.; Semmler, T.; Pickard, D.J.; de Toro, M.; de la Cruz, F.; Wieler, L.H.; Ewers, C.; Guenther, S. Carriage of extended-spectrum beta-lactamase-plasmids does not reduce fitness but enhances virulence in some strains of pandemic E. coli lineages. Front. Microbiol. 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed]
- Naves, P.; del Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Dahbi, G.; Blanco, M.; Ponte Mdel, C.; Soriano, F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 2008, 45, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Gilbert, E.S. Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 2004, 70, 6951–6956. [Google Scholar] [CrossRef] [PubMed]
- Himpsl, S.D.; Mobley, H.L.T. Siderophore detection using chrome azurol S and cross-feeding assays. Methods Mol. Biol. 2019, 2021, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 381. [Google Scholar] [CrossRef]
- Ewers, C.; Antão, E.M.; Diehl, I.; Philipp, H.C.; Wieler, L.H. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 2009, 75, 184–192. [Google Scholar] [CrossRef]
- Schierack, P.; Heiden, S.E.; Khan, M.M.; Nikolaus, L.; Kolenda, R.; Stubbe, M.; Lkhagvasuren, D.; Rodiger, S.; Guenther, S.; Schaufler, K. Genomic and phenotypic analysis of an ESBL-producing E. coli ST1159 clonal lineage from wild birds in Mongolia. Front. Microbiol. 2020, 11, 1699. [Google Scholar] [CrossRef]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).