Comparative Genomic Analysis Discloses Differential Distribution of Antibiotic Resistance Determinants between Worldwide Strains of the Emergent ST213 Genotype of Salmonella Typhimurium
Abstract
1. Introduction
2. Results
2.1. Distribution of the Genomes of the Strains Analyzed by Country, Type of Sample and Year of Isolation
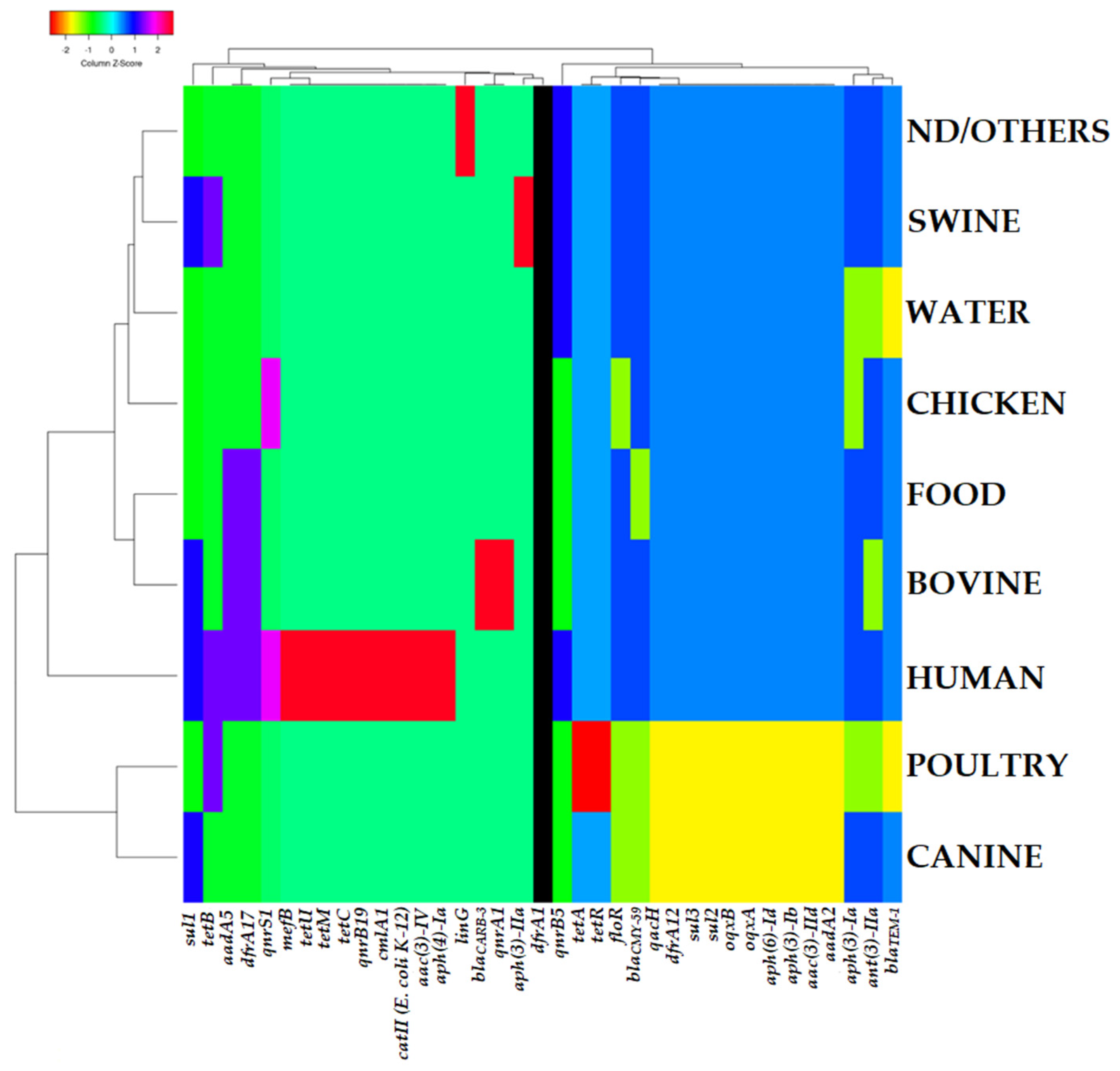
2.2. Presence of Antibiotic Resistance Genes
2.3. Antibiotic Resistance Mutations
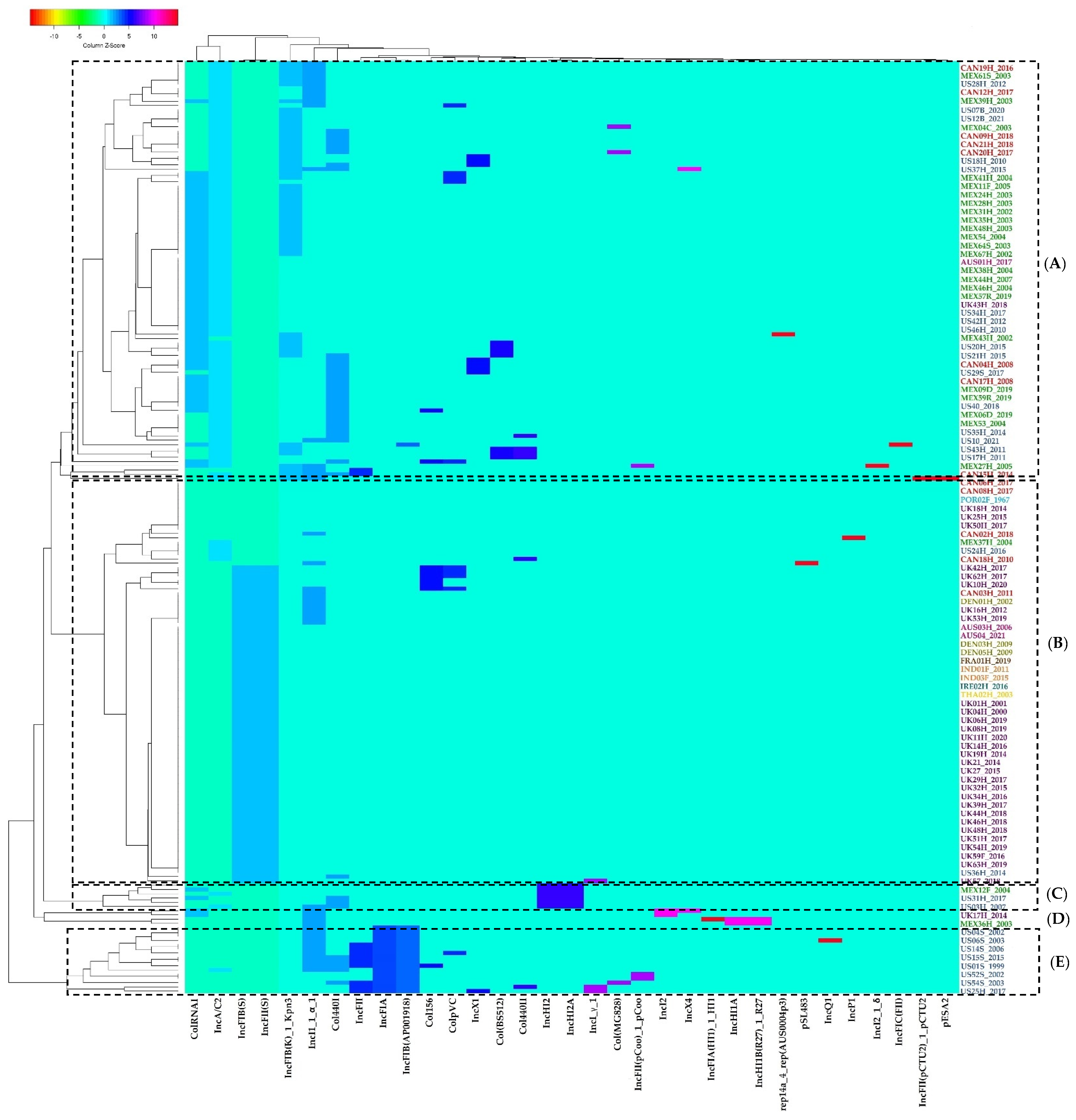
2.4. Plasmid Replicons Detection
2.5. Grouping Patterns
3. Discussion
4. Materials and Methods
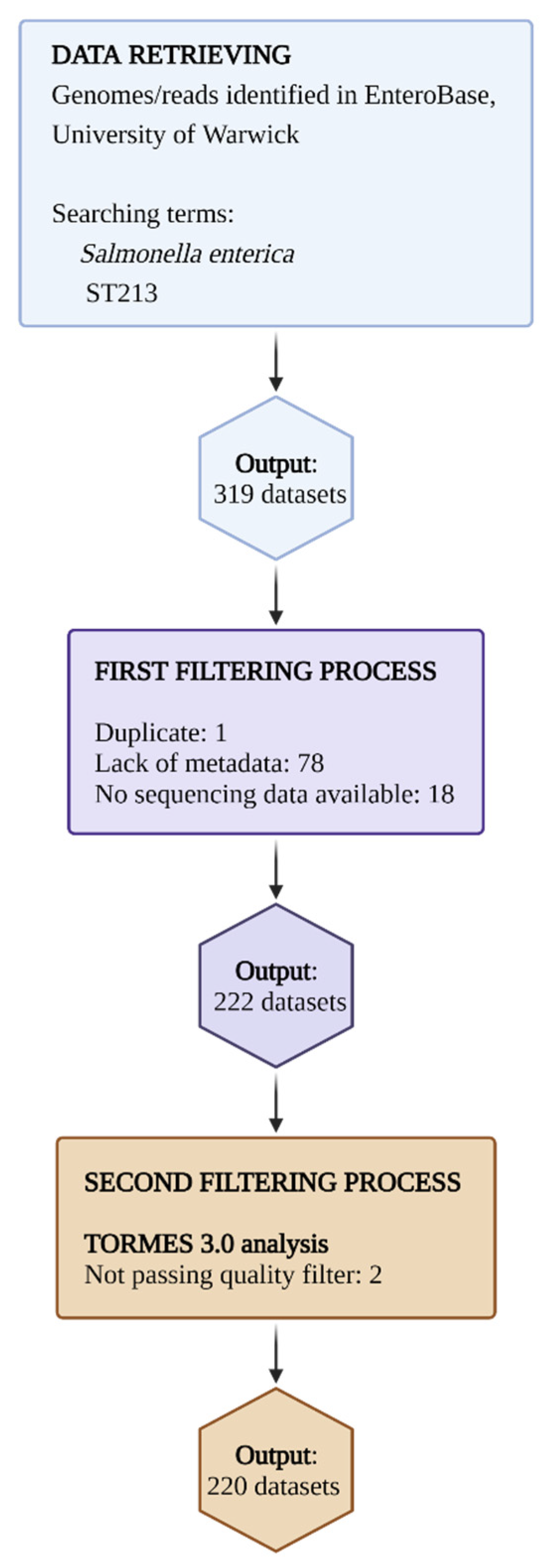
4.1. Data Retrieval
4.2. Bioinformatic Analysis
4.3. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alby, K.; Nachamkin, I. Gastrointestinal infections. Microbiol. Spectr. 2016, 4, 44. [Google Scholar] [CrossRef] [PubMed]
- Eng, S.K.; Pusparajah, P.; Ab Mutalib, N.S.; Ser, H.L.; Chan, K.G.; Lee, L.H. Salmonella: A review on pathogenesis, epidemiology and antibiotic resistance. Front. Life Sci. 2015, 8, 284–293. [Google Scholar] [CrossRef]
- Mafi, N.; Orenstein, R. Salmonella. In Encyclopedia of Gastroenterology, 2nd ed.; Johnson, L.R., Ed.; Elsevier: New York, NY, USA, 2020; pp. 384–391. [Google Scholar] [CrossRef]
- St John-Brooks, R. The Genus Salmonella Lignieres, 1900: Issued by the Salmonella Subcommittee of the Nomenclature Committee of the International Society for Microbiology. J. Hyg. 1934, 34, 333–350. [Google Scholar]
- Grimont, P.A.; Weill, F.X. Antigenic Formulae of the Salmonella Serovars, 9th ed.; WHO Collaborating Centre for Reference and Research on Salmonella; WHO: Paris, France, 2007; Volume 9, pp. 1–166. [Google Scholar]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; De Pinna, E.; Nair, S.; Fields, I.P.; Weill, F.X. Supplement 2008–2010 (no. 48) to the white–Kauffmann–Le minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, R.S.; Vieira, A.R.; Karlsmose, S.; Lo Fo Wong, D.M.; Jensen, A.B.; Wegener, H.C.; Aarestrup, F.M. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: Results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 2011, 8, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.G.; Rosario, D.K.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Inns, T.; Ashton, P.M.; Herrera-Leon, S.; Lighthill, J.; Foulkes, S.; Jombart, T.; Rehman, Y.; Fox, A.; Dallman, T.; de Pinna, E.; et al. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella enteritidis. Epidemiol. Infect. 2017, 145, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Leekitcharoenphon, P.; Nielsen, E.M.; Kaas, R.S.; Lund, O.; Aarestrup, F.M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS ONE 2014, 9, e87991. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Trost, E.; Bender, J.; Fuchs, S.; Malorny, B.; Rabsch, W.; Prager, R.; Tietze, E.; Flieger, A. Evaluation of WGS based approaches for investigating a food-borne outbreak caused by Salmonella enterica serovar Derby in Germany. Food Microbiol. 2018, 71, 46–54. [Google Scholar] [CrossRef]
- Deng, X.; den Bakker, H.C.; Hendriksen, R.S. Genomic epidemiology: Whole-genome-sequencing–powered surveillance and outbreak investigation of foodborne bacterial pathogens. Annu. Rev. Food Sci. Technol. 2016, 7, 353–374. [Google Scholar] [CrossRef]
- Tang, P.; Croxen, M.A.; Hasan, M.R.; Hsiao, W.W.; Hoang, L.M. Infection control in the new age of genomic epidemiology. Am. J. Infect. Control. 2017, 45, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Kathariou, S.; Winkler, A.; Skandamis, P.; Saint-Cyr, M.J.; Rouzeau-Szynalski, K.; Amézquita, A. Next generation microbiological risk assessment: Opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int. J. Food Microbiol. 2018, 287, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Achtman, M.; Wain, J.; Weill, F.X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1009040. [Google Scholar] [CrossRef]
- Lan, R.; Reeves, P.R.; Octavia, S. Population structure, origins and evolution of major Salmonella enterica clones. Infect. Genet. Evol. 2009, 9, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Feijao, P.; Yao, H.T.; Fornika, D.; Gardy, J.; Hsiao, W.; Chauve, C.; Chindelevitch, L. MentaLiST—A fast MLST caller for large MLST schemes. Microb. Genom. 2018, 4, e000146. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Ianzano, A.; Iurescia, M.; Diaconu, E.L.; ENGAGE-EURL-AR Network Study Group; et al. Molecular epidemiology of Salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb Genomics. 2020, 6, e000365. [Google Scholar] [CrossRef] [PubMed]
- Bonifait, L.; Thépault, A.; Baugé, L.; Rouxel, S.; Le Gall, F.; Chemaly, M. Occurrence of Salmonella in the cattle production in France. Microorganisms 2021, 9, 872. [Google Scholar] [CrossRef]
- Ashton, P.M.; Owen, S.V.; Kaindama, L.; Rowe, W.P.; Lane, C.R.; Larkin, L.; Nair, S.; Jenkins, C.; de Pinna, E.M.; Feasey, N.A.; et al. Public health surveillance in the UK revolutionises our understanding of the invasive Salmonella Typhimurium epidemic in Africa. Genome Med. 2017, 9, 92. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Msefula, C.L.; Thomson, N.R.; Kariuki, S.; Holt, K.E.; Gordon, M.A.; Harris, D.; Clarke, L.; Whitehead, S.; Sangal, V.; et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009, 19, 2279–2287. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Barquist, L.; Connor, T.R.; Harris, S.R.; Clare, S.; Stevens, M.P.; Arends, M.J.; Hale, C.; Kane, L.; Pickard, D.J.; et al. Signatures of adaptation in human invasive Salmonella Typhimurium ST313 populations from sub-Saharan Africa. PLOS Negl. Trop. Dis. 2015, 9, e0003611. [Google Scholar] [CrossRef]
- Hammarlöf, D.L.; Kröger, C.; Owen, S.V.; Canals, R.; Lacharme-Lora, L.; Wenner, N.; Schager, A.E.; Wells, T.J.; Henderson, I.R.; Wigley, P.; et al. Role of a single noncoding nucleotide in the evolution of an epidemic African clade of Salmonella. Proc. Natl. Acad. Sci. USA 2018, 115, E2614–E2623. [Google Scholar] [CrossRef] [PubMed]
- Neuert, S.; Nair, S.; Day, M.R.; Doumith, M.; Ashton, P.M.; Mellor, K.C.; Jenkins, C.; Hopkins, K.L.; Woodford, N.; de Pinna, E.; et al. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 2018, 9, 592. [Google Scholar] [CrossRef]
- Tassinari, E.; Bawn, M.; Thilliez, G.; Charity, O.; Acton, L.; Kirkwood, M.; Petrovska, L.; Dallman, T.; Burgess, C.M.; Hall, N.; et al. Whole-genome epidemiology links phage-mediated acquisition of a virulence gene to the clonal expansion of a pandemic Salmonella enterica serovar Typhimurium clone. Microb. Genom. 2020, 6, mgen000456. [Google Scholar] [CrossRef]
- Crouse, A.; Schramm, C.; Emond-Rheault, J.G.; Herod, A.; Kerhoas, M.; Rohde, J.; Gruenheid, S.; Kukavica-Ibrulj, I.; Boyle, B.; Greenwood, C.M.T.; et al. Combining Whole-Genome Sequencing and multimodel phenotyping to identify genetic predictors of Salmonella virulence. mSphere 2020, 5, e00293-20. [Google Scholar] [CrossRef] [PubMed]
- Pires, J.; Huisman, J.S.; Bonhoeffer, S.; Van Boeckel, T.P. Multidrug resistance dynamics in Salmonella in food animals in the United States: An analysis of genomes from public databases. Microbiol. Spectr. 2021, 9, e00495-21. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Fu, Y.; Tate, H.; Pinto, C.; Dudley, E.G.; M’ikanatha, N.M. Genomic analysis of Salmonella Typhimurium from humans and food sources accurately predicts phenotypic multi-drug resistance. Food Microbiol. 2022, 103, 103957. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Kingsley, R.A.; Connor, T.R.; Harris, S.R.; Parry, C.M.; Al-Mashhadani, M.N.; Kariuki, S.; Msefula, C.L.; Gordon, M.A.; de Pinna, E.; et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 2012, 44, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.H.Y.; Yan, M.; Chan, E.W.C.; Liu, L.Z.; Kan, B.; Chen, S. Expansion of Salmonella enterica serovar Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob. Agents Chemother. 2013, 57, 4599–4601. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Ke, B.; Huang, Y.; He, D.; Li, X.; Liang, Z.; Ke, C. The molecular epidemiological characteristics and genetic diversity of Salmonella Typhimurium in Guangdong, China, 2007–2011. PLoS ONE 2014, 9, e113145. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, M.B.; Calva, J.J.; Estrada-Garcia, M.T.; Leon, V.; Vazquez, G.; Figueroa, G.; Lopez, E.; Contreras, J.; Abbott, J.; Zhao, S.; et al. Integrated food chain surveillance system for Salmonella spp. in Mexico. Emerg. Infect. Dis. 2008, 14, 429–435. [Google Scholar] [CrossRef] [PubMed]
- Ambrose, S.J.; Harmer, C.J.; Hall, R.M. Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 2018, 96–97, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, M.; Calva, E.; Fernández-Mora, M.; Cevallos, M.A.; Campos, F.; Zaidi, M.B.; Silva, C. Salmonella Typhimurium ST213 is associated with two types of IncA/C plasmids carrying multiple resistance determinants. BMC Microbiol. 2011, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Enterobase. Quality Assessment Evaluation—EnteroBase Documentation. 2018. Available online: https://enterobase.readthedocs.io/en/latest/pipelines/backend-pipeline-qaevaluation.html (accessed on 31 January 2022).
- Lee, K.Y.; Atwill, E.R.; Pitesky, M.; Huang, A.; Lavelle, K.; Rickard, M.; Shafii, M.; Hung-Fan, M.; Li, X. Antimicrobial resistance profiles of non-typhoidal Salmonella from retail meat products in California, 2018. Front. Microbiol. 2022, 13, 835699. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer MS, M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable plasmids of Salmonella enterica associated with antibiotic resistance genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef]
- Lima, T.; Domingues, S.; Da Silva, G.J. Plasmid-mediated colistin resistance in Salmonella enterica: A review. Microorganisms 2019, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Portes, A.B.; Rodrigues, G.; Leitão, M.P.; Ferrari, R.; Conte Junior, C.A.; Panzenhagen, P. Global distribution of plasmid-mediated colistin resistance mcr gene in Salmonella: A systematic review. J. Appl. Microbiol. 2022, 132, 872–889. [Google Scholar] [CrossRef]
- Jain, P.; Sudhanthirakodi, S.; Chowdhury, G.; Joshi, S.; Anandan, S.; Ray, U.; Mukhopadhyay, A.; Dutta, S. Antimicrobial resistance, plasmid, virulence, multilocus sequence typing and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium clinical and environmental isolates from India. PLoS ONE 2018, 13, e0207954. [Google Scholar] [CrossRef]
- Elnekave, E.; Hong, S.L.; Lim, S.; Boxrud, D.; Rovira, A.; Mather, A.E.; Perez, A.; Alvarez, J. Transmission of multidrug-resistant Salmonella enterica subspecies enterica 4,[5], 12: I:- Sequence Type 34 between Europe and the United States. Emerg. Infect. Dis. 2020, 26, 3034–3038. [Google Scholar] [CrossRef] [PubMed]
- Mather, A.E.; Phuong TL, T.; Gao, Y.; Clare, S.; Mukhopadhyay, S.; Goulding, D.A.; Hoang, N.T.D.; Tuyen, H.T.; Lan NP, H.; Thompson, C.N.; et al. New variant of multidrug-resistant Salmonella enterica serovar Typhimurium associated with invasive disease in immunocompromised patients in Vietnam. mBio 2018, 9, e01056-18. [Google Scholar] [CrossRef]
- Wiesner, M.; Zaidi, M.B.; Calva, E.; Fernández-Mora, M.; Calva, J.J.; Silva, C. Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains. BMC Microbiol. 2009, 9, 131. [Google Scholar] [CrossRef] [PubMed]
- Holley, R.A. Food safety challenges within North American free trade agreement (NAFTA) partners. Compr. Rev. Food Sci. 2011, 10, 131–142. [Google Scholar] [CrossRef]
- Johnson, L.R.; Gould, L.H.; Dunn, J.R.; Berkelman, R.; Mahon, B.E.; The FoodNet Travel Working Group. Salmonella infections associated with international travel: A Foodborne Diseases Active Surveillance Network (FoodNet) study. Foodborne Pathog. Dis. 2011, 8, 1031–1037. [Google Scholar] [CrossRef]
- Tighe, M.K.; Savage, R.; Vrbova, L.; Toolan, M.; Whitfield, Y.; Varga, C.; Lee, B.; Allen, V.; Maki, A.; Walton, R.; et al. The epidemiology of travel-related Salmonella Enteritidis in Ontario, Canada, 2010–2011. BMC Public Health 2012, 12, 310. [Google Scholar] [CrossRef] [PubMed]
- Coipan, C.E.; Westrell, T.; van Hoek, A.H.; Alm, E.; Kotila, S.; Berbers, B.; de Keersmaecker, S.C.J.; Ceyssens, P.J.; Borg, M.L.; Chattaway, M.; et al. Genomic epidemiology of emerging ESBL-producing Salmonella Kentucky bla CTX-M-14b in Europe. Emerg. Microbes Infect. 2020, 9, 2124–2135. [Google Scholar] [CrossRef] [PubMed]
- Waldram, A.; Dolan, G.; Ashton, P.M.; Jenkins, C.; Dallman, T.J. Epidemiological analysis of Salmonella clusters identified by whole genome sequencing, England and Wales 2014. Food Microbiol. 2018, 71, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.A.; Lane, C.R.; Easton, M.; Valcanis, M.; Strachan, J.; Veitch, M.G.; Kirk, M.D.; Howden, B.P. Increasing antimicrobial resistance in nontyphoidal Salmonella isolates in Australia from 1979 to 2015. Antimicrob. Agents Chemother. 2018, 62, e02012–e02017. [Google Scholar] [CrossRef] [PubMed]
- Medalla, F.; Gu, W.; Friedman, C.R.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Increased incidence of antimicrobial-resistant nontyphoidal Salmonella infections, United States, 2004–2016. Emerg. Infect. Dis. 2021, 27, 1662. [Google Scholar] [CrossRef]
- Varga, C.; John, P.; Cooke, M.; Majowicz, S.E. Spatial and space-time clustering and demographic characteristics of human nontyphoidal Salmonella infections with major serotypes in Toronto, Canada. PLoS ONE 2020, 15, e0235291. [Google Scholar] [CrossRef]
- Zaidi, M.B.; Campos, F.D.; Estrada-García, T.; Gutierrez, F.; León, M.; Chim, R.; Calva, J.J. Burden and transmission of zoonotic foodborne disease in a rural community in Mexico. Clin. Infect. Dis. 2012, 55, 51–60. [Google Scholar] [CrossRef]
- Parmley, E.J.; Pintar, K.; Majowicz, S.; Avery, B.; Cook, A.; Jokinen, C.; Gannon, V.; Lapen, D.R.; Topp, E.; Edge, T.A.; et al. A Canadian application of One Health: Integration of Salmonella data from various Canadian surveillance programs (2005–2010). Foodborne Pathog. Dis. 2013, 10, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Flockhart, L.; Pintar, K.; Cook, A.; McEwen, S.; Friendship, R.; Kelton, D.; Pollari, F. Distribution of Salmonella in humans, production animal operations and a watershed in a FoodNet Canada sentinel site. Zoonoses Public Health 2017, 64, 41–52. [Google Scholar] [CrossRef]
- Marus, J.R.; Magee, M.J.; Manikonda, K.; Nichols, M.C. Outbreaks of Salmonella enterica infections linked to animal contact: Demographic and outbreak characteristics and comparison to foodborne outbreaks—United States, 2009–2014. Zoonoses Public Health 2019, 66, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, S.; Gu, W.; den Bakker, H.; Boxrud, D.; Taylor, A.; Roe, C.; Driebe, E.; Engelthaler, D.M.; Allard, M.; et al. Zoonotic source attribution of Salmonella enterica serotype Typhimurium using genomic surveillance data, United States. Emerg. Infect. Dis. 2019, 25, 82–91. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization (WHO): Geneva, Switzerland, 2018. [Google Scholar]
- Ruiz, J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, e00007–e00019. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Poeta, P.; Hébraud, M.; Capelo, J.L.; Igrejas, G. Mechanisms of quinolone action and resistance: Where do we stand? J. Med. Microbiol. 2017, 66, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Suárez, E.J.; Ortíz-López, R.; Gebreyes, W.A.; Allard, M.W.; Barona-Gómez, F.; Rubio-Lozano, M.S. Genomic surveillance links livestock production with the emergence and spread of multi-drug resistant non-typhoidal Salmonella in Mexico. J. Microbiol. 2019, 57, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Suárez, E.J.; Palós-Guitérrez, T.; Ruíz-López, F.A.; Hernández Pérez, C.F.; Ballesteros-Nova, N.E.; Soberanis-Ramos, O.; Méndez-Medina, R.D.; Allard, M.W.; Rubio-Lozano, M.S. Genomic surveillance of antimicrobial resistance shows cattle and poultry are a moderate source of multi-drug resistant non-typhoidal Salmonella in Mexico. PLoS ONE 2021, 16, e0243681. [Google Scholar] [CrossRef]
- Zaidi, M.B.; Estrada-García, T.; Campos, F.D.; Chim, R.; Arjona, F.; Leon, M.; Michell, A.; Chaussabel, D. Incidence, clinical presentation, and antimicrobial resistance trends in Salmonella and Shigella infections from children in Yucatan, Mexico. Front. Microbiol. 2013, 4, 288. [Google Scholar] [CrossRef]
- McDermott, P.F.; Zhao, S.; Tate, H. Antimicrobial resistance in nontyphoidal Salmonella. Microbiol. Spectr. 2018, 6, 780–790. [Google Scholar] [CrossRef]
- Sjölund-Karlsson, M.; Folster, J.P.; Pecic, G.; Joyce, K.; Medalla, F.; Rickert, R.; Whichard, J.M. Emergence of plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates from humans in the United States. Antimicrob. Agents Chemother. 2009, 53, 2142–2144. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sjölund-Karlsson, M.; Howie, R.; Rickert, R.; Krueger, A.; Tran, T.T.; Zhao, S.; Ball, T.; Haro, J.; Pecic, G.; Joyce, K.; et al. Plasmid-mediated quinolone resistance among non-typhi Salmonella enterica isolates, USA. Emerg. Infect. Dis. 2010, 16, 1789–1791. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, W.L.; Jacobs, J.; Wong, V.; Klemm, E.J.; Deborggraeve, S.; Van Puyvelde, S. Fluoroquinolone resistance in Salmonella: Insights by whole-genome sequencing. Microbial. Genomics. 2018, 4, e000195. [Google Scholar] [CrossRef]
- Kim, J.; Han, X.; Bae, J.; Chui, L.; Louie, M.; Finley, R.; Mulvey, M.R.; Ferrato, C.J.; Jeon, B. Prevalence of plasmid-mediated quinolone resistance (PMQR) genes in non-typhoidal Salmonella strains with resistance and reduced susceptibility to fluoroquinolones from human clinical cases in Alberta, Canada, 2009–2013. J. Antimicrob. Chemother. 2016, 71, 2988–2990. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Li, J.; Meng, Y.; Ma, Y.; Hu, C.; Jin, S.; Zhang, Q.; Ding, H.; Cui, S. Joint effects of topoisomerase alterations and plasmid-mediated quinolone-resistant determinants in Salmonella enterica Typhimurium. Microb. Drug. Resist. 2011, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.C.; Folster, J.P.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between phenotypic and in silico detection of antimicrobial resistance in Salmonella enterica in Canada using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; Le Hello, S.; Doublet, B.; Granier, S.A.; Hendriksen, R.S.; Fricke, W.F.; Ceyssens, P.J.; Gomart, C.; Billman-Jacobe, H.; Holt, K.E.; et al. Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microbial. Genomics. 2019, 5, e000269. [Google Scholar] [CrossRef] [PubMed]
- Van Hoek, A.H.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA; Available online: http://em100.edaptivedocs.net/dashboard.aspx (accessed on 1 May 2022).
- Van Duijkeren, E.; Schwarz, C.; Bouchard, D.; Catry, B.; Pomba, C.; Baptiste, K.E.; Moreno, M.A.; Rantala, M.; Ružauskas, M.; Sanders, P.; et al. The use of aminoglycosides in animals within the EU: Development of resistance in animals and possible impact on human and animal health: A review. J. Antimicrob. Chemother. 2019, 74, 2480–2496. [Google Scholar] [CrossRef]
- Cox, G.W.; Parmley, E.J.; Avery, B.P.; Irwin, R.J.; Reid-Smith, R.J.; Deckert, A.E.; Finley, R.L.; Daignault, D.; Alexander, D.C.; Allen, V.; et al. A One-Health genomic investigation of gentamicin resistance in Salmonella from human and chicken sources in Canada, 2014 to 2017. Antimicrob. Agents Chemother. 2021, 65, e00966-21. [Google Scholar] [CrossRef]
- Nyirabahizi, E.; Tyson, G.H.; Tate, H.; Strain, E. The Western United States has greater antibiotic resistance among Salmonella recovered from intestinal cecal samples of food animals. J. Food Protect. 2020. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Wencewicz, T.A. Tetracycline-inactivating enzymes. Front. Microbiol. 2018, 9, 1058. [Google Scholar] [CrossRef] [PubMed]
- Nunes, O.C.; Manaia, C.M.; Kolvenbach, B.A.; Corvini PF, X. Living with sulfonamides: A diverse range of mechanisms observed in bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 10389–10408. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.; Sellera, F.P.; Lopes, R.; Keelara, S.; Landgraf, M.; Greene, S.; Fedorka-Cray, P.J.; Thakur, S. Class 1 integron-borne cassettes harboring bla CARB-2 gene in multidrug-resistant and virulent Salmonella Typhimurium ST19 strains recovered from clinical human stool samples, United States. PLoS ONE 2020, 15, e0240978. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Latifi, T.; Groisman, E.A. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar typhimurium. Mol. Microbiol. 2006, 59, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Li, J.; Zhang, X.; Gao, F.; Benton, C.S.; Andam, C.P. Diverse lineages of multidrug resistant clinical Salmonella enterica and a cryptic outbreak in New Hampshire, USA revealed from a year-long genomic surveillance. Infect. Genet. Evol. 2021, 87, 104645. [Google Scholar] [CrossRef]
- Rahmati, S.; Yang, S.; Davidson, A.L.; Zechiedrich, E.L. Control of the AcrAB multidrug efflux pump by quorum-sensing regulator SdiA. Mol. Microbiol. 2002, 43, 677–685. [Google Scholar] [CrossRef]
- Tezel, U.; Pavlostathis, S.G. Quaternary ammonium disinfectants: Microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015, 33, 296–304. [Google Scholar] [CrossRef]
- OIE. OIE Strategy on Antimicrobial Resistance and the Prudent Use of Antimicrobials. 2016. Available online: http://www.oie.int/fileadmin/Home/eng/Media_Center/docs/pdf/PortailAMR/EN_OIE-AMRstrategy.pdf (accessed on 20 May 2022).
- Monte, D.F.; Lincopan, N.; Fedorka-Cray, P.J.; Landgraf, M. Current insights on high priority antibiotic-resistant Salmonella enterica in food and foodstuffs: A review. Curr. Opin. Food Sci. 2019, 26, 35–46. [Google Scholar] [CrossRef]
- Elbediwi, M.; Tang, Y.; Shi, D.; Ramadan, H.; Xu, Y.; Xu, S.; Li, Y.; Yue, M. Genomic investigation of antimicrobial-resistant Salmonella enterica isolates from dead chick embryos in China. Front. Microbiol. 2021, 12, 684400. [Google Scholar] [CrossRef]
- Diaconu, E.L.; Alba, P.; Feltrin, F.; Di Matteo, P.; Iurescia, M.; Chelli, E.; Donati, V.; Marani, I.; Giacomi, A.; Franco, A.; et al. Emergence of IncHI2 plasmids with mobilized colistin resistance (mcr)-9 gene in esbl-producing, multidrug-resistant Salmonella Typhimurium and its monophasic variant ST34 from food-producing animals in Italy. Front. Microbiol. 2021, 12, 705230. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.; Lincopan, N.; Berman, H.; Cerdeira, L.; Keelara, S.; Siddhartha Thakur, S.; Fedorka-Cray, P.; Landgraf, M. Genomic features of high-priority Salmonella enterica serovars circulating in the food production chain, Brazil, 2000–2016. Sci. Rep. 2019, 9, 11058. [Google Scholar] [CrossRef] [PubMed]
- Ball, T.; Monte, D.; Aidara-Kane, A.; Matheu, J.; Ru, H.; Thakur, S.; Ejobi, F.; Fedorka-Cray, P. International lineages of Salmonella enterica serovars isolated from chicken farms, Wakiso District, Uganda. PLoS ONE 2020, 15, e0220484. [Google Scholar] [CrossRef] [PubMed]
- Moon, D.C.; Kim, S.-J.; Mechesso, A.F.; Kang, H.Y.; Song, H.-J.; Choi, J.-H.; Yoon, S.-S.; Lim, S.-K. Mobile colistin resistance gene mcr-1 detected on an IncI2 plasmid in Salmonella Typhimurium Sequence Type 19 from a healthy pig in South Korea. Microorganisms 2021, 9, 398. [Google Scholar] [CrossRef]
- Wu, B.; Ed-Dra, A.; Pan, H.; Dong, C.; Jia, C.; Yue, M. Genomic investigation of Salmonella isolates recovered from a pig slaughtering process in Hangzhou, China. Front. Microbiol. 2021, 12, 704636. [Google Scholar] [CrossRef]
- Viana, C.; Grossi, J.L.; Sereno, M.J.; Yamatogi, R.S.; Bersot, L.D.S.; Call, D.R.; Nero, L.A. Phenotypic and genotypic characterization of non-typhoidal Salmonella isolated from a Brazilian pork production chain. Food Res. Int. 2020, 137, 109406. [Google Scholar] [CrossRef]
- Carroll, L.M.; Wiedmann, M.; den Bakker, H.; Siler, J.; Warchocki, S.; Kent, D.; Lyalina, S.; Davis, M.; Sischo, W.; Besser, T.; et al. Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington states reveals source and geographic associations. Appl. Environ. Microbiol. 2017, 83, e00140-17. [Google Scholar] [CrossRef]
- Jensen, L.B.; Garcia-Migura, L.; Valenzuela, A.J.; Løhr, M.; Hasman, H.; Aarestrup, F.M. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods. 2010, 80, 25–43. [Google Scholar] [CrossRef]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Salmonella Whole Genome Sequencing Implementation Group. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef]
- Chattaway, M.A.; Dallman, T.J.; Larkin, L.; Nair, S.; McCormick, J.; Mikhail, A.; Hartman, H.; Godbole, G.; Powell, D.; Day, M.; et al. The transformation of reference microbiology methods and surveillance for Salmonella with the use of whole genome sequencing in England and Wales. Public. Health Front. 2019, 7, 317. [Google Scholar] [CrossRef]
- Van Goethem, N.; Van Den Bossche, A.; Ceyssens, P.J.; Lajot, A.; Coucke, W.; Vernelen, K.; Roosens, N.C.H.; De Keersmaecker, S.C.J.; Van Cauteren, D.; Mattheus, W. Coverage of the national surveillance system for human Salmonella infections, Belgium, 2016–2020. PLoS ONE 2021, 16, e0256820. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Dessai, U.; McGarry, S.; Gerner-Smidt, P. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.; da Silva, A.G.; Jennison, A.V.; Williamson, D.A.; Howden, B.P.; Seemann, T. AusTrakka: Fast-tracking nationalized genomics surveillance in response to the COVID-19 pandemic. Nat. Commun. 2022, 13, 865. [Google Scholar] [CrossRef] [PubMed]
- Goig, G.A.; Blanco, S.; Garcia-Basteiro, A.L.; Comas, I. Contaminant DNA in bacterial sequencing experiments is a major source of false genetic variability. BMC Biol. 2020, 18, 1–15. [Google Scholar] [CrossRef]
- Zwe, Y.H.; Chin, S.F.; Kohli, G.S.; Aung, K.T.; Yang, L.; Yuk, H.G. Whole genome sequencing (WGS) fails to detect antimicrobial resistance (AMR) from heteroresistant subpopulation of Salmonella enterica. Food Microbiol. 2020, 91, 103530. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E. Comparison of the web tools ARG-ANNOT and ResFinder for detection of resistance genes in bacteria. Antimicrob. Agents Chemother. 2014, 58, 4986. [Google Scholar] [CrossRef]
- Achtman, M.; Zhou, Z.; Alikhan, N.F.; Tyne, W.; Parkhill, J.; Cormican, M.; Chiou, C.S.; Torpdahl, M.; Litrup, E.; Prendergast, D.M.; et al. Genomic diversity of Salmonella enterica—The UoWUCC 10 K genomes project. Wellcome Open Res. 2021, 5, 223. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M.; Brown, D.; Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An automated pipeline for whole bacterial genome analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Wajid, B.; Serpedin, E. Do it yourself guide to genome assembly. Brief. Funct. Genomics. 2016, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, A.; Hendriksen, R.S.; Aaresturp, F.M.; Ussery, D.W.; Friis, C. The Salmonella enterica pan-genome. Microb. Ecol. 2011, 62, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]





| Antibiotic Group/Resistance Gene(s) | Encoded 1 | % Frequency 2 |
|---|---|---|
| Aminoglycosides | ||
| aph(4)-Ia, aac(3)-IV, ant(3)-IIa, aac(3)-IId | P (IncHI2) | 0.4 (n = 1), 0.9 (n = 2), 10.9 (n = 24), 29 (n = 64) |
| aph(3)-IIa, aph(3)-Ia | T | 0.9 (n = 2), 9.5 (n = 21) |
| aac(6)-Iaa | C | 100 (n = 220) |
| aadA2 | P/I (IncHI2/IncHI2A) | 40 (n = 89) |
| aadA5 | P/T/I | 1.8 (n = 4) |
| aph(3)-Ib | P/T/C (IncC, IncFII/IA/IB,) | 50 (n = 111) |
| aph(6)-Id | P/CG-I (IncC, IncFII/IA/IB,) | 50 (n = 110) |
| Cephamicin | ||
| blaCMY-59 | P (IncC) | 36.8 (n = 81) |
| Diaminopyrimidines | ||
| dfrA1, dfrA12, dfrA17 | I | 0.4 (n = 1), 37.2 (n = 82), 1.8 (n = 4) |
| Penam | ||
| blaCARB-3 | P | 0.4 (n = 1) |
| Penam, penem, cephalosporin, monobactam | ||
| blaTEM-1 | C/P (IncHI2/IncHI2A) | 14 (n = 31) |
| Disinfecting agents and intercalating dyes | ||
| qacH | P | 26.3 (n = 58) |
| Fluoroquinolones | ||
| mdtK | C | 100 (n = 220) |
| qnrA1, qnrB5, qnrB19, qnrS1 | P (IncHI2/IncHI2A) | 0.4 (n = 1), 10 (n = 22), 0.4 (n = 1), 0.9 (n = 2) |
| Lincosamides | ||
| linG | I-agc, with aadA2 | 0.4 (n = 1) |
| Macrolides | ||
| mefB | P, located in the sul3 vicinity | 0.4 (n = 1) |
| Sulfonamides | ||
| sul1 | C-1 I | 8.6 (n = 19) |
| sul2 | SP (IncHI2, IncC, IncFII/IA/IB) | 49.5 (n = 109) |
| sul3 | P (IncHI2/IncHI2A) | 27.2 (n = 60) |
| Tetracyclines | ||
| tetA, tetB, tetR | C/P (IncHI2, IncC, IncFII/IA/IB) | 56.8 (n = 125), 1.8 (n = 4), 55.9 (n = 123) |
| tetC | P (IncHI2/IncHI2A/IncI1_I_γ) | 1.3 (n = 3) |
| tetU | P (pKQ10) | 0.4 (n = 1) |
| tetM | T | 0.4 (n = 1) |
| Phenicol | ||
| catII (E. coli K-12), cmlA1, floR | C/P (IncHI2/ IncHI2A/ IncI1_I_γ/IncQ1, IncC) | 0.4 (n = 1), 0.9 (n = 2), 47.7 (n = 105) |
| Phenicol, β-lactams, diaminopyrimidines, fluoroquinolones, glycyl-cyclines, nitrofuran and tetracyclines, rifamycin, triclosan. | ||
| golS, mdsA, mdsB, mdsC | C | 100 (n = 220) |
| oqxA, oqxB | C/P (IncHI2) | 24.5 (n = 54) |
| sdiA | C/P | 100 (n = 220) |
| Mutation | Codon Change | Amino Acid Change | Genome | % of Genomes 1 |
|---|---|---|---|---|
| gyrA p.D87G | GAC → GGC | D → G | MEX04C_2003 | 3.4 (n = 1) |
| parC p.S80I | AGC → ATC | S → I | US37H_2015 | 3.4 (n = 1) |
| gyrA p.S83F | TCC → TTC | S → F | MEX22F_2008 | 6.8 (n = 2) |
| MEX51H_2011 | ||||
| gyrA p.D87N | GAC → AAC | D → N | US17H_2011 | 10.3 (n = 3) |
| US19H_2011 | ||||
| US43H_2011 | ||||
| gyrA p.S83Y | TCC → TAC | S → Y | CAN20H_2017 | 79.3 (n = 23) |
| MEX14F_2009 | ||||
| MEX15F_2009 | ||||
| MEX16F_2009 | ||||
| MEX18F_2008 | ||||
| MEX34H_2003 | ||||
| MEX56R_2019 | ||||
| THA01C_2019 | ||||
| US07B_2020 | ||||
| US12B_2021 | ||||
| US18H_2010 | ||||
| US20H_2015 | ||||
| US21H_2015 | ||||
| US22H_2015 | ||||
| US24H_2016 | ||||
| US28H_2012 | ||||
| US30H_2017 | ||||
| US37H_2015 | ||||
| US38H_2017 | ||||
| US44H_2010 | ||||
| US46H_2010 | ||||
| US47H_2017 | ||||
| US49B_2019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Díaz, E.A.; Vázquez-Garcidueñas, M.S.; Negrete-Paz, A.M.; Vázquez-Marrufo, G. Comparative Genomic Analysis Discloses Differential Distribution of Antibiotic Resistance Determinants between Worldwide Strains of the Emergent ST213 Genotype of Salmonella Typhimurium. Antibiotics 2022, 11, 925. https://doi.org/10.3390/antibiotics11070925
Hernández-Díaz EA, Vázquez-Garcidueñas MS, Negrete-Paz AM, Vázquez-Marrufo G. Comparative Genomic Analysis Discloses Differential Distribution of Antibiotic Resistance Determinants between Worldwide Strains of the Emergent ST213 Genotype of Salmonella Typhimurium. Antibiotics. 2022; 11(7):925. https://doi.org/10.3390/antibiotics11070925
Chicago/Turabian StyleHernández-Díaz, Elda Araceli, Ma. Soledad Vázquez-Garcidueñas, Andrea Monserrat Negrete-Paz, and Gerardo Vázquez-Marrufo. 2022. "Comparative Genomic Analysis Discloses Differential Distribution of Antibiotic Resistance Determinants between Worldwide Strains of the Emergent ST213 Genotype of Salmonella Typhimurium" Antibiotics 11, no. 7: 925. https://doi.org/10.3390/antibiotics11070925
APA StyleHernández-Díaz, E. A., Vázquez-Garcidueñas, M. S., Negrete-Paz, A. M., & Vázquez-Marrufo, G. (2022). Comparative Genomic Analysis Discloses Differential Distribution of Antibiotic Resistance Determinants between Worldwide Strains of the Emergent ST213 Genotype of Salmonella Typhimurium. Antibiotics, 11(7), 925. https://doi.org/10.3390/antibiotics11070925






