Companion Animals—An Overlooked and Misdiagnosed Reservoir of Carbapenem Resistance
Abstract
1. Introduction
2. Carbapenemase-Producing Bacteria in Companion Animals
| Enzyme | Year | Country | Host | Source | Bacterial Species | Detection Methods | Refs. |
|---|---|---|---|---|---|---|---|
| IMP-4 | 2016 | Australia | Cats | Commensal | Salmonella enterica serovar Typhimurium | AST | [12] |
| KPC-2 | 2018 | Brazil | Dog | Infection (UTI) | Escherichia coli | Imipenem synergy test, modified Hodge testing, PCR | [9] |
| KPC-2 | 2021 | Brazil | Dog | Infection (UTI) | Klebsiella pneumoniae | Imipenem synergy test, AST | [10] |
| KPC-4 | 2018 | USA | Dog | Infection (UTI, SSTI) | Enterobacter xiangfangensis | Biochemical Tests | [11] |
| NDM-1 | 2013 | United States | Dogs, Cats | Infection (SSTI, UTI) | Escherichia coli | AST | [24] |
| NDM-1 | 2017 | China | Dogs | Commensal | Escherichia coli | Selective culture media | [16,25] |
| NDM−1 | 2018 | Italy | Dog | Commensal | Acinetobacter radioresistens | Selective culture media | [23] |
| NDM-5 | 2016 | Algeria | Dogs | Commensal | Escherichia coli | PCR | [17] |
| NDM-5 | 2017 | China | Dogs | Commensal | Escherichia coli | Selective culture media | [16] |
| NDM-5 | 2019 | United Kingdom | Dog | Infection (SSTI) | Escherichia coli | AST | [19] |
| NDM-5 | 2018 | Finland | Dogs | Infection (Otitis externa) | Escherichia coli | AST followed by modified Hodge testing, UV spectrometric detection of imipenem hydrolysis | [18] |
| NDM-5 | 2021 | Italy | Dog | Infection (UTI) | Escherichia coli | Meropenem synergy test | [15] |
| NDM-5 | 2018 | United States | Dog | Infection (URTI) | Escherichia coli | AST | [20] |
| NDM-5 | 2018 | United States | Dogs, Cats | Infection (UTI, URTI) | Escherichia coli | AST | [22] |
| NDM-5 | 2018 | South Korea | Dog, Cat | Commensal | Escherichia coli | AST, PCR | [21] |
| NDM-9 | 2017 | China | Dog | Commensal | Escherichia coli | Selective culture media | [16] |
| OXA-48 | 2009–2010 | Germany | Dogs, Cats, Horses | Infection | Escherichia coli, Klebsiella pneumoniae, Enterobacter cloacae | Selective culture media for cephalosporin resistance, PCR | [36] |
| OXA-48 | 2013 | Germany | Dog | Commensal, Infection (UTI, SSTI, URTI, CRBSI) | Klebsiella pneumoniae, Escherichia coli | AST | [29] |
| OXA-48 | 2016 | United States | Dogs, Cats | Infection (UTI, SSTI, Genital tract) | Escherichia coli | AST | [31] |
| OXA-48 | 2016 | Algeria | Dogs | Commensal | Escherichia coli | PCR | [17] |
| OXA-48 | 2017 | Algeria | Dogs, Cat, Horses, Pet birds | Commensal | Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae | Selective culture media | [32] |
| OXA-48 | 2017 | France | Dog | Commensal | Escherichia coli | Selective culture media | [30] |
| OXA-48 | 2018 | Germany | Dogs, Cats, Horses | Infection (UTI, SSTI, genital tract, otitis, URTI) | Klebsiella pneumoniae, Enterobacter cloacae, Escherichia coli, Klebsiella oxytoca | Selective culture media | [28] |
| OXA-181 | 2018 | Switzerland | Dogs, Cats | Commensal | Escherichia coli | Selective culture media | [26] |
| OXA-181 | 2020 | Portugal | Dog | Commensal | Escherichia coli | Selective culture media and AST | [27] |
| OXA-181 | 2021 | Portugal | Cat | Infection (SSTI) | Klebsiella pneumoniae | Selective culture media and AST | [37] |
| OXA-23 | 2014 | Portugal | Cat | Infection (UTI) | Acinetobacter baumannii | AST | [33] |
| OXA-23 | 2017 | Germany | Dogs, Cats | Infection (UTI, suppurate inflammation) | Acinetobacter baumannii | Selective culture media | [34] |
| OXA−23 | 2018 | Italy | Dogs, Cats | Commensal | Acinetobacter baumanni | Selective culture media | [23] |
| OXA-66 | 2017 | Germany | Dogs, Cats | Infection (UTI, SSTI, URTI, CRBSI, suppurate inflammation) | Acinetobacter baumannii | Selective culture media | [34] |
| VIM-1 | 2016 | Spain | Dog | Commensal | Klebsiella pneumoniae | Selective culture media, Meropenem synergy test | [14] |
| VIM-2 | 2018 | South Korea | Dog | Infection (SSTI) | Pseudomonas aeruginosa | AST | [13] |
3. Phenotypic Characteristics of Carbapenemases and Their Genetic Background in Isolates from Companion Animals
3.1. Serine Carbapenemases
3.2. Metallo-β-Lactamases
3.3. Oxacillinases
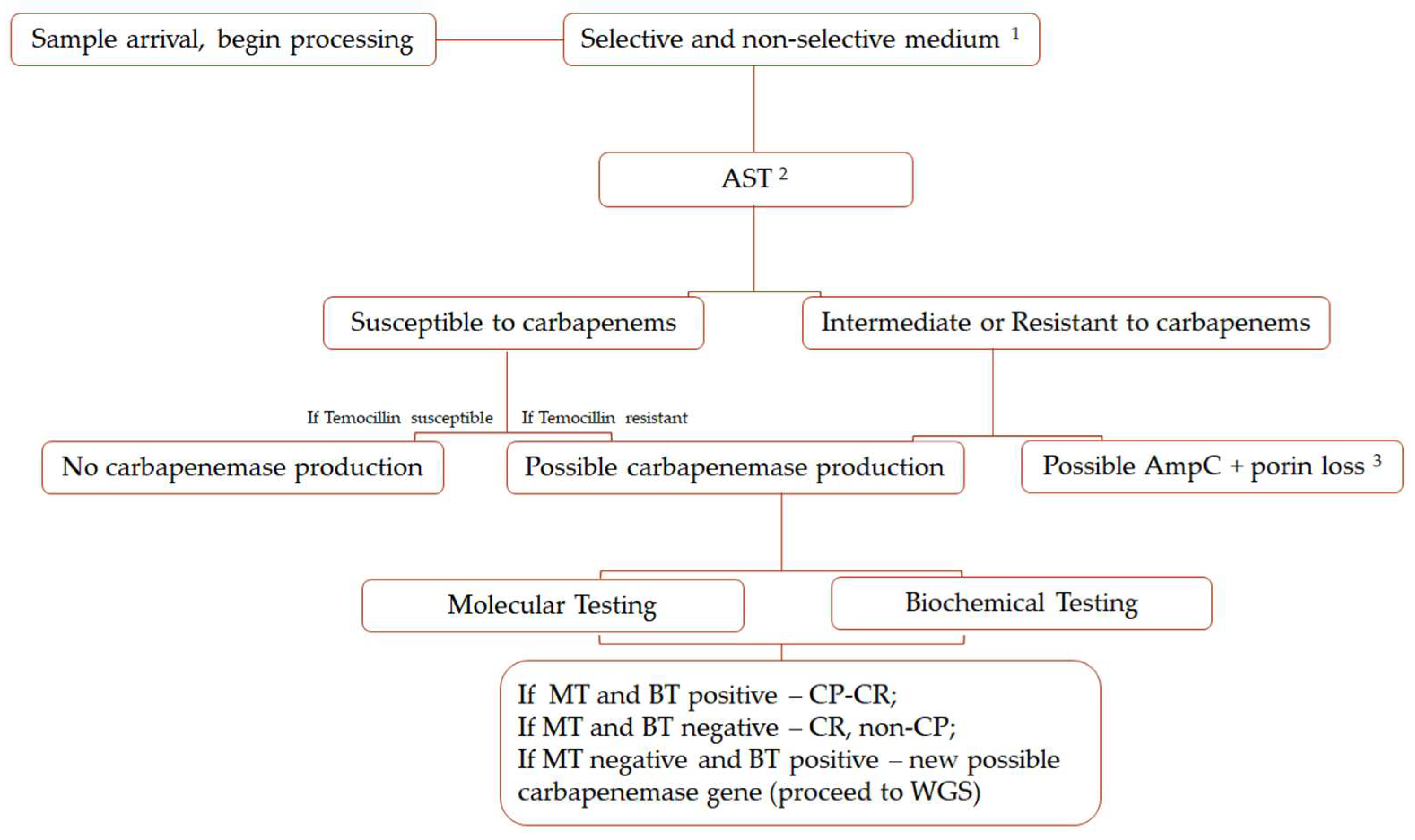
4. Methods for Detection and Identification of Carbapenemases
4.1. Selective Culture Media
4.2. Biochemical Tests
4.3. Disc Diffusion Methods
4.4. Lateral Flow Assays
4.5. Molecular Testing
4.6. Mass Spectrometry Analysis
5. Transmission Potential
- Achieving the principles of prudent use of antibiotics in veterinary practice to ensure that carbapenems are used only in the very few cases that lack other suitable alternatives based on culture and AST;
- Include the systematic screening for carbapenem resistance in veterinary microbiology laboratories;
- Surveillance and monitoring for the presence of genes encoding resistance to critically important antimicrobials, such as carbapenems;
- Appropriate hygiene practices after handling animals both in domestic and health care settings;
- Infection control measures when dealing with companion animals with infections caused by carbapenem-resistant strains that include isolation of infected animals.
6. Conclusions and Final Remarks for Veterinary Medicine
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Meletis, G. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem Resistance in Enterobacteriaceae: Here Is the Storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-β-Lactamases: The Quiet Before the Storm. Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.A.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (OIE). Oie List of Antimicrobial Agents of Veterinary Importance. In Proceedings of the FAO2/OIE/WHO3 Expert Workshop on Non-Human Antimicrobial Usage and Antimicrobial Resistance, Geneva, Switzerland, 1–5 December 2003; Volume Xxviii, pp. 1–10. [Google Scholar]
- European Medicines Agency (EMA). Categorisation of Antibiotics in the European Union; European Medicines Agency: London, UK, 2019. [Google Scholar]
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyörälä, S.; Ružauskas, M.; et al. Public Health Risk of Antimicrobial Resistance Transfer from Companion Animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef]
- Mader, R.; Damborg, P.; Amat, J.P.; Bengtsson, B.; Bourély, C.; Broens, E.M.; Busani, L.; Crespo-Robledo, P.; Filippitzi, M.E.; Fitzgerald, W.; et al. Building the European Antimicrobial Resistance Surveillance Network in Veterinary Medicine (EARS-Vet). Eurosurveillance 2021, 26, 2001359. [Google Scholar] [CrossRef]
- Sellera, F.P.; Fernandes, M.R.; Ruiz, R.; Falleiros, A.C.M.; Rodrigues, F.P.; Cerdeira, L.; Lincopan, N. Identification of KPC-2-Producing Escherichia Coli in a Companion Animal: A New Challenge for Veterinary Clinicians. J. Antimicrob. Chemother. 2018, 73, 2259–2261. [Google Scholar] [CrossRef]
- Sellera, F.P.; Fuga, B.; Fontana, H.; Esposito, F.; Cardoso, B.; Konno, S.; Berl, C.; Cappellanes, M.H.; Cortez, M.; Ikeda, M.; et al. Detection of IncN-PST15 One-Health Plasmid Harbouring BlaKPC-2 in a Hypermucoviscous Klebsiella Pneumoniae CG258 Isolated from an Infected Dog, Brazil. Transbound. Emerg. Dis. 2021, 68, 3083–3088. [Google Scholar] [CrossRef]
- Daniels, J.B.; Chen, L.; Grooters, S.V.; Mollenkopf, D.F.; Mathys, D.A.; Pancholi, P.; Kreiswirth, B.N.; Wittum, T.E. Enterobacter cloacae Complex Sequence Type 171 Isolates Expressing KPC-4 Carbapenemase Recovered from Canine Patients in Ohio. Antimicrob. Agents Chemother. 2018, 62, e01161-18. [Google Scholar] [CrossRef]
- Abraham, S.; O’Dea, M.; Trott, D.J.; Abraham, R.J.; Hughes, D.; Pang, S.; McKew, G.; Cheong, E.Y.L.; Merlino, J.; Saputra, S.; et al. Isolation and Plasmid Characterization of Carbapenemase (IMP-4) Producing Salmonella enterica Typhimurium from Cats. Sci. Rep. 2016, 6, 35527. [Google Scholar] [CrossRef]
- Hyun, J.E.; Chung, T.H.; Hwang, C.Y. Identification of VIM-2 Metallo-β-Lactamase-Producing Pseudomonas aeruginosa Isolated from Dogs with Pyoderma and Otitis in Korea. Vet. Dermatol. 2018, 29, 186-e68. [Google Scholar] [CrossRef] [PubMed]
- González-Torralba, A.; Oteo, J.; Asenjo, A.; Bautista, V.; Fuentes, E.; Alós, J.I. Survey of Carbapenemase-Producing Enterobacteriaceae in Companion Dogs in Madrid, Spain. Antimicrob. Agents Chemother. 2016, 60, 2499–2501. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Taddei, R.; Cordaro, G.; Fontana, M.C.; Toschi, E.; Gaibani, P.; Marani, I.; Giacomi, A.; Diaconu, E.L.; Iurescia, M.; et al. Carbapenemase IncF-Borne BlaNDM-5 Gene in the E. Coli ST167 High-Risk Clone from Canine Clinical Infection, Italy. Vet. Microbiol. 2021, 256, 109045. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive Resistome Analysis Reveals the Prevalence of NDM and MCR-1 in Chinese Poultry Production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef]
- Yousfi, M.; Touati, A.; Mairi, A.; Brasme, L.; Gharout-Sait, A.; Guillard, T.; De Champs, C. Emergence of Carbapenemase-Producing Escherichia Coli Isolated from Companion Animals in Algeria. Microb. Drug Resist. 2016, 22, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Grönthal, T.; Österblad, M.; Eklund, M.; Jalava, J.; Nykäsenoja, S.; Pekkanen, K.; Rantala, M. Sharing More than Friendship—Transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia Coli between Dogs and Humans in a Family, Finland, 2015. Euro Surveill. 2018, 23, 1700497. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.E.; Phan, H.T.T.; George, S.; Hubbard, A.T.M.; Stoesser, N.; Maciuca, I.E.; Crook, D.W.; Timofte, D. Occurrence and Characterization of Escherichia coli ST410 Co-Harbouring BlaNDM-5, BlaCMY-42 and BlaTEM-190 in a Dog from the UK. J. Antimicrob. Chemother. 2019, 74, 1207–1211. [Google Scholar] [CrossRef]
- Tyson, G.H.; Li, C.; Ceric, O.; Reimschuessel, R.; Cole, S.; Peak, L.; Rankin, S.C. Complete Genome Sequence of a Carbapenem-Resistant Escherichia coli Isolate with BlaNDM-5 from a Dog in the United States. Microbiol. Resour. Announc. 2019, 8, 22–23. [Google Scholar] [CrossRef]
- Hong, J.S.; Song, W.; Jeong, S.H. Molecular Characteristics of NDM-5-Producing Escherichia coli from a Cat and a Dog in South Korea. Microb. Drug Resist. 2020, 26, 1005–1008. [Google Scholar] [CrossRef]
- Cole, S.D.; Peak, L.; Tyson, G.H.; Reimschuessel, R.; Ceric, O.; Rankin, S.C. New Delhi Metallo-β-Lactamase-5–Producing Escherichia coli in Companion Animals, United States. Emerg. Infect. Dis. 2020, 26, 381–383. [Google Scholar] [CrossRef]
- Gentilini, F.; Turba, M.E.; Pasquali, F.; Mion, D.; Romagnoli, N.; Zambon, E.; Terni, D.; Peirano, G.; Pitout, J.D.D.; Parisi, A.; et al. Hospitalized Pets as a Source of Carbapenem-Resistance. Front. Microbiol. 2018, 9, 2872. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, B.W.; Nayak, R.; Boothe, D.M. Emergence of a New Delhi Metallo-β-Lactamase (NDM-1)-Encoding Gene in Clinical Escherichia coli Isolates Recovered from Companion Animals in the United States. Antimicrob. Agents Chemother. 2013, 57, 2902–2903. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lei, L.; Lv, Y.; Zhang, R.; Liu, X.; Li, M.; Zhang, F.; Wang, Y. blaNDM-1-Producing Multidrug-Resistant Escherichia coli Isolated from a Companion Dog in China. J. Glob. Antimicrob. Resist. 2018, 13, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Nigg, A.; Brilhante, M.; Dazio, V.; Clément, M.; Collaud, A.; Brawand, S.G.; Willi, B.; Endimiani, A.; Schuller, S.; Perreten, V. Shedding of OXA-181 Carbapenemase-Producing Escherichia coli from Companion Animals after Hospitalisation in Switzerland: An Outbreak in 2018. Eurosurveillance 2019, 24, 1900071. [Google Scholar] [CrossRef]
- Brilhante, M.; Menezes, J.; Belas, A.; Feudi, C.; Schwarz, S.; Pomba, C.; Perreten, V. OXA-181-Producing Extra-Intestinal Pathogenic Escherichia coli ST410 Isolated from a Dog in Portugal. Antimicrob. Agents Chemother. 2020, 64, e02298-19. [Google Scholar] [CrossRef]
- Pulss, S.; Stolle, I.; Stamm, I.; Leidner, U.; Heydel, C.; Semmler, T.; Prenger-Berninghoff, E.; Ewers, C. Multispecies and Clonal Dissemination of OXA-48 Carbapenemase in Enterobacteriaceae from Companion Animals in Germany, 2009–2016. Front. Microbiol. 2018, 9, 1265. [Google Scholar] [CrossRef]
- Stolle, I.; Prenger-Berninghoff, E.; Stamm, I.; Scheufen, S.; Hassdenteufel, E.; Guenther, S.; Bethe, A.; Pfeifer, Y.; Ewers, C. Emergence of OXA-48 Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae in Dogs. J. Antimicrob. Chemother. 2013, 68, 2802–2808. [Google Scholar] [CrossRef]
- Melo, L.C.; Boisson, M.N.G.; Saras, E.; Médaille, C.; Boulouis, H.J.; Madec, J.Y.; Haenni, M. OXA-48-Producing ST372 Escherichia coli in a French Dog. J. Antimicrob. Chemother. 2017, 72, 1256–1258. [Google Scholar] [CrossRef][Green Version]
- Liu, X.; Thungrat, K.; Boothe, D.M. Occurrence of OXA-48 Carbapenemase and Other β-Lactamase Genes in ESBL-Producing Multidrug Resistant: Escherichia coli from Dogs and Cats in the United States, 2009–2013. Front. Microbiol. 2016, 7, 1057. [Google Scholar] [CrossRef]
- Yousfi, M.; Touati, A.; Muggeo, A.; Mira, B.; Asma, B.; Brasme, L.; Guillard, T.; de Champs, C. Clonal Dissemination of OXA-48-Producing Enterobacter cloacae Isolates from Companion Animals in Algeria. J. Glob. Antimicrob. Resist. 2018, 12, 187–191. [Google Scholar] [CrossRef]
- Pomba, C.; Endimiani, A.; Rossano, A.; Saial, D.; Couto, N.; Perreten, V. First Report of OXA-23-Mediated Carbapenem Resistance in Sequence Type 2 Multidrug-Resistant Acinetobacter baumannii Associated with Urinary Tract Infection in a Cat. Antimicrob. Agents Chemother. 2014, 58, 1267–1268. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Klotz, P.; Leidner, U.; Stamm, I.; Prenger-Berninghoff, E.; Göttig, S.; Semmler, T.; Scheufen, S. OXA-23 and ISAba1–OXA-66 Class D β-Lactamases in Acinetobacter baumannii Isolates from Companion Animals. Int. J. Antimicrob. Agents 2017, 49, 37–44. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 16 April 2022).
- Schmiedel, J.; Falgenhauer, L.; Domann, E.; Bauerfeind, R.; Prenger-Berninghoff, E.; Imirzalioglu, C.; Chakraborty, T. Multiresistant Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae from Humans, Companion Animals and Horses in Central Hesse, Germany. BMC Microbiol. 2014, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Moreira da Silva, J.; Menezes, J.; Salas, C.; Marques, C.; Teodoro, S.; Amaral, A.J.; Pomba, C. Clinical/Research Abstracts Accepted for Presentation at the ISFM 2021 World Feline Congress. J. Feline Med. Surg. 2021, 23, 851–858. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolun, V.; Nordmann, P. Emergence of Oxacillinanse-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
- Kayama, S.; Koba, Y.; Shigemoto, N.; Kuwahara, R.; Kakuhama, T.; Kimura, K.; Hisatsune, J.; Onodera, M.; Yokozaki, M.; Ohge, H.; et al. Imipenem-Susceptible, Meropenem-Resistant Klebsiella pneumoniae Producing OXA-181 in Japan. Antimicrob. Agents Chemother. 2015, 59, 1379–1380. [Google Scholar] [CrossRef]
- Kashyap, A.; Gupta, R.; Sharma, R.; Verma, V.V.; Gupta, S.; Goyal, P. New Delhi Metallo Beta Lactamase: Menace and Its Challenges. J. Mol. Genet. Med. 2017, 11, 299. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef]
- Farhat, N.; Khan, A.U. Evolving Trends of New Delhi Metallo-Betalactamse (NDM) Variants: A Threat to Antimicrobial Resistance. Infect. Genet. Evol. 2020, 86, 104588. [Google Scholar] [CrossRef]
- Shanthi, M.; Sekar, U.; Arunagiri, K.; Goverdhandas Bramhne, H. OXA-181 Beta Lactamase Is Not a Major Mediator of Carbapenem Resistance in Enterobacteriaceae. J. Clin. Diagnostic Res. 2013, 7, 1986–1988. [Google Scholar] [CrossRef]
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.P.; Touati, A. OXA-48-like Carbapenemases Producing Enterobacteriaceae in Different Niches. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 587–604. [Google Scholar] [CrossRef]
- Bakthavatchalam, Y.D.; Anandan, S.; Veeraraghavan, B. Laboratory Detection and Clinical Implication of Oxacillinase-48 like Carbapenemase: The Hidden Threat. J. Glob. Infect. Dis. 2016, 8, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like Carbapenemases: The Phantom Menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Gülmez, D.; Woodford, N.; Palepou, M.F.I.; Mushtaq, S.; Metan, G.; Yakupogullari, Y.; Kocagoz, S.; Uzun, O.; Hascelik, G.; Livermore, D.M. Carbapenem-Resistant Escherichia Coli and Klebsiella pneumoniae Isolates from Turkey with OXA-48-like Carbapenemases and Outer Membrane Protein Loss. Int. J. Antimicrob. Agents 2008, 31, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Bonnin, R.A.; Nordmann, P. Genetic Features of the Widespread Plasmid Coding for the Carbapenemase OXA-48. Antimicrob. Agents Chemother. 2012, 56, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Potron, A.; Nordmann, P.; Lafeuille, E.; Al Maskari, Z.; Al Rashdi, F.; Poirel, L. Characterization of OXA-181, a Carbapenem-Hydrolyzing Class D β-Lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2011, 55, 4896–4899. [Google Scholar] [CrossRef]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The Role of ISAba1 in Expression of OXA Carbapenemase Genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef]
- Miriagou, V.; Cornaglia, G.; Edelstein, M.; Galani, I.; Giske, C.G.; Gniadkowski, M.; Malamou-Lada, E.; Martinez-Martinez, L.; Navarro, F.; Nordmann, P.; et al. Acquired Carbapenemases in Gram-Negative Bacterial Pathogens: Detection and Surveillance Issues. Clin. Microbiol. Infect. 2010, 16, 112–122. [Google Scholar] [CrossRef]
- Wilkinson, K.M.; Winstanley, T.G.; Lanyon, C.; Cummings, S.P.; Raza, M.W.; Perry, J.D. Comparison of Four Chromogenic Culture Media for Carbapenemase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 3102–3104. [Google Scholar] [CrossRef]
- Moran Gilad, J.; Carmeli, Y.; Schwartz, D.; Navon-Venezia, S. Laboratory Evaluation of the CHROMagar KPC Medium for Identification of Carbapenem-Nonsusceptible Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2011, 70, 565–567. [Google Scholar] [CrossRef]
- Zarakolu, P.; Day, K.M.; Sidjabat, H.E.; Kamolvit, W.; Lanyon, C.V.; Cummings, S.P.; Paterson, D.L.; Akova, M.; Perry, J.D. Evaluation of a New Chromogenic Medium, ChromID OXA-48, for Recovery of Carbapenemase-Producing Enterobacteriaceae from Patients at a University Hospital in Turkey. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Comparison of the SUPERCARBA, CHROMagar KPC, and Brilliance CRE Screening Media for Detection of Enterobacteriaceae with Reduced Susceptibility to Carbapenems. Diagn. Microbiol. Infect. Dis. 2013, 75, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid Detection of Producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Pauly, N.; Hammerl, J.A.; Grobbel, M.; Tenhagen, B.A.; Käsbohrer, A.; Bisenius, S.; Fuchs, J.; Horlacher, S.; Lingstädt, H.; Mauermann, U.; et al. ChromID® CARBA Agar Fails to Detect Carbapenem-Resistant Enterobacteriaceae with Slightly Reduced Susceptibility to Carbapenems. Front. Microbiol. 2020, 11, 1678. [Google Scholar] [CrossRef] [PubMed]
- Panagea, T.; Galani, I.; Souli, M.; Adamou, P.; Antoniadou, A.; Giamarellou, H. Evaluation of CHROMagarTM KPC for the Detection of Carbapenemase-Producing Enterobacteriaceae in Rectal Surveillance Cultures. Int. J. Antimicrob. Agents 2011, 37, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Dortet, L.; Bréchard, L.; Poirel, L.; Nordmann, P. Rapid Detection of Carbapenemase-Producing Enterobacteriaceae from Blood Cultures. Clin. Microbiol. Infect. 2014, 20, 340–344. [Google Scholar] [CrossRef]
- Vasoo, S.; Cunningham, S.A.; Kohner, P.C.; Simner, P.J.; Mandrekar, J.N.; Lolans, K.; Hayden, M.K.; Patel, R. Comparison of a Novel, Rapid Chromogenic Biochemical Assay, the Carba NP Test, with the Modified Hodge Test for Detection of Carbapenemase-Producing Gram-Negative Bacilli. J. Clin. Microbiol. 2013, 51, 3097–3101. [Google Scholar] [CrossRef]
- Tijet, N.; Boyd, D.; Patel, S.N.; Mulvey, M.R.; Melano, R.G. Evaluation of the Carba NP Test for Rapid Detection of Carbapenemase—Producing Enterobacteriaceae and Pseudomonas Aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 4578–4580. [Google Scholar] [CrossRef]
- Skov, R.; Skov, G. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance; EUCAST: Växjö, Sweden, 2012; pp. 1–47. [Google Scholar]
- Van Der Zwaluw, K.; De Haan, A.; Pluister, G.N.; Bootsma, H.J.; De Neeling, A.J.; Schouls, L.M. The Carbapenem Inactivation Method (CIM), a Simple and Low-Cost Alternative for the Carba NP Test to Assess Phenotypic Carbapenemase Activity in Gram-Negative Rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef]
- Pires, J.; Novais, A.; Peixe, L. Blue-Carba, an Easy Biochemical Test for Detection of Diverse Carbapenemase Producers Directly from Bacterial Cultures. J. Clin. Microbiol. 2013, 51, 4281–4283. [Google Scholar] [CrossRef] [PubMed]
- Pasteran, F.; Veliz, O.; Ceriana, P.; Lucero, C.; Rapoport, M.; Albornoz, E.; Gomez, S.; Corso, A. Evaluation of the Blue-Carba Test for Rapid Detection of Carbapenemases in Gram-Negative Bacilli. J. Clin. Microbiol. 2015, 53, 1996–1998. [Google Scholar] [CrossRef] [PubMed]
- Bernabeu, S.; Dortet, L.; Naas, T. Evaluation of the β-CARBATM Test, a Colorimetric Test for the Rapid Detection of Carbapenemase Activity in Gram-Negative Bacilli. J. Antimicrob. Chemother. 2017, 72, 1646–1658. [Google Scholar] [CrossRef] [PubMed]
- Giske, C.G.; Gezelius, L.; Samuelsen, Ø.; Warner, M.; Sundsfjord, A.; Woodford, N. A Sensitive and Specific Phenotypic Assay for Detection of Metallo-β-Lactamases and KPC in Klebsiella Pneumoniae with the Use of Meropenem Disks Supplemented with Aminophenylboronic Acid, Dipicolinic Acid and Cloxacillin. Clin. Microbiol. Infect. 2011, 17, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Leverstein-Van Hall, M.A.; Cohen Stuart, J.W.T. A Disc Diffusion Assay for Detection of Class A, B and OXA-48 Carbapenemases in Enterobacteriaceae Using Phenyl Boronic Acid, Dipicolinic Acid and Temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Giske, C.G. Contemporary Resistance Trends and Mechanisms for the Old Antibiotics Colistin, Temocillin, Fosfomycin, Mecillinam and Nitrofurantoin. Clin. Microbiol. Infect. 2015, 21, 899–905. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). CLSI Archived Methods. In Performance Standards for Antimicrobial Susceptibility testing, 28th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Wareham, D.W.; Abdul Momin, M.H.F. Rapid Detection of Carbapenemases in Enterobacteriaceae: Evaluation of the Resist-3 O.K.N. (OXA-48, KPC, NDM) Lateral Flow Multiplexed Assay. J. Clin. Microbiol. 2017, 55, 1223–1225. [Google Scholar] [CrossRef]
- Glupczynski, Y.; Jousset, A.; Evrard, S.; Bonnin, R.A.; Huang, T.D.; Dortet, L.; Bogaerts, P.; Naas, T. Prospective Evaluation of the OKN K-SeT Assay, a New Multiplex Immunochromatographic Test for the Rapid Detection of OXA-48-like, KPC and NDM Carbapenemases. J. Antimicrob. Chemother. 2017, 72, 1955–1960. [Google Scholar] [CrossRef]
- Meunier, D.; Vickers, A.; Pike, R.; Hill, R.L.; Woodford, N.; Hopkins, K.L. Evaluation of the K-SeT R.E.S.I.S.T. Immunochromatographic Assay for the Rapid Detection of KPC and OXA-48-like Carbapenemases. J. Antimicrob. Chemother. 2016, 71, 2357–2359. [Google Scholar] [CrossRef]
- Wareham, D.W.; Phee, L.M.; Momin, M.H.F.A. Direct Detection of Carbapenem Resistance Determinants in Clinical Specimens Using Immunochromatographic Lateral Flow Devices. J. Antimicrob. Chemother. 2018, 73, 1997–1998. [Google Scholar] [CrossRef]
- Kolenda, C.; Benoit, R.; Carricajo, A.; Bonnet, R.; Dauwalder, O.; Laurent, F. Evaluation of the New Multiplex Immunochromatographic O.K.N.V. K -SeT Assay for Rapid Detection of OXA-48-like, KPC, NDM, and VIM Carbapenemases. J. Clin. Microbiol. 2018, 56, e01247-18. [Google Scholar] [CrossRef] [PubMed]
- Rösner, S.; Kamalanabhaiah, S.; Küsters, U.; Kolbert, M.; Pfennigwerth, N.; Mack, D. Evaluation of a Novel Immunochromatographic Lateral Flow Assay for Rapid Detection of OXA-48, NDM, KPC and VIM Carbapenemases in Multidrug-Resistant Enterobacteriaceae. J. Med. Microbiol. 2019, 68, 379–381. [Google Scholar] [CrossRef] [PubMed]
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A Multiplex Lateral Flow Immunoassay for the Rapid Identification of NDM-, KPC-, IMP- and VIM-Type and OXA-48-like Carbapenemase-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar] [CrossRef]
- Hong, J.; Kang, D.; Kim, D. Performance Evaluation of the Newly Developed In Vitro Rapid Diagnostic Test for Detecting OXA-48-Like, KPC-, NDM-, VIM- and IMP-Type Carbapenemases: The RESIST-5 O.K.N.V.I. Multiplex Lateral Flow Assay. Antibiotics 2021, 10, 460. [Google Scholar] [CrossRef]
- Cohen Stuart, J.; Leverstein-Van Hall, M.A. Guideline for Phenotypic Screening and Confirmation of Carbapenemases in Enterobacteriaceae. Int. J. Antimicrob. Agents 2010, 36, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Multiplex Real-Time PCR Detection of Klebsiella pneumoniae Carbapenemase (KPC) and New Delhi Metallo-β-lactamase (NDM-1). Available online: https://www.cdc.gov/hai/pdfs/labsettings/KPC-NDM-protocol-2011.pdf (accessed on 11 April 2022).
- Girlich, D.; Bogaerts, P.; Bouchahrouf, W.; Bernabeu, S.; Langlois, I.; Begasse, C.; Arangia, N.; Dortet, L.; Huang, T.D.; Glupczynski, Y.; et al. Evaluation of the Novodiag CarbaR+, a Novel Integrated Sample to Result Platform for the Multiplex Qualitative Detection of Carbapenem and Colistin Resistance Markers. Microb. Drug Resist. 2021, 27, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, S.A.; Vasoo, S.; Patel, R. Evaluation of the Check-Points Check MDR CT103 and CT103 XL Microarray Kits by Use of Preparatory Rapid Cell Lysis. J. Clin. Microbiol. 2016, 54, 1368–1371. [Google Scholar] [CrossRef]
- Sękowska, A.; Bogiel, T. The Evaluation of Eazyplex® SuperBug CRE Assay Usefulness for the Detection of ESBLs and Carbapenemases Genes Directly from Urine Samples and Positive Blood Cultures. Antibiotics 2022, 11, 138. [Google Scholar] [CrossRef]
- Bilozor, A.; Balode, A.; Chakhunashvili, G.; Chumachenko, T.; Egorova, S.; Ivanova, M.; Kaftyreva, L.; Kõljalg, S.; Kõressaar, T.; Lysenko, O.; et al. Application of Molecular Methods for Carbapenemase Detection. Front. Microbiol. 2019, 10, 1755. [Google Scholar] [CrossRef]
- Rossen, J.W.A.; Friedrich, A.W.; Moran-Gilad, J. Practical Issues in Implementing Whole-Genome-Sequencing in Routine Diagnostic Microbiology. Clin. Microbiol. Infect. 2018, 24, 355–360. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical Metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I.; Zimmermann, S. Using Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry to Detect Carbapenem Resistance within 1 to 2.5 Hours. J. Clin. Microbiol. 2011, 49, 3321–3324. [Google Scholar] [CrossRef] [PubMed]
- Mirande, C.; Canard, I.; Buffet Croix Blanche, S.; Charrier, J.P.; van Belkum, A.; Welker, M.; Chatellier, S. Rapid Detection of Carbapenemase Activity: Benefits and Weaknesses of MALDI-TOF MS. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Pomba, C.; Belas, A.; Menezes, J.; Marques, C. The Public Health Risk of Companion Animal to Human Transmission of Antimicrobial Resistance during Different Types of Animal Infection. In Advances in Animal Health, Medicine and Production; Springer Nature: Cham, Switzerland, 2020; pp. 265–278. [Google Scholar] [CrossRef]
- Marques, C.; Belas, A.; Aboim, C.; Cavaco-Silva, P.; Trigueiro, G.; Gama, L.T.; Pomba, C. Evidence of Sharing of Klebsiella Pneumoniae Strains between Healthy Companion Animals and Cohabiting Humans. J. Clin. Microbiol. 2019, 57, e01537-18. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bunt, G.; Fluit, A.C.; Spaninks, M.P.; Timmerman, A.J.; Geurts, Y.; Kant, A.; Scharringa, J.; Mevius, D.; Wagenaar, J.A.; Bonten, M.J.M.; et al. Faecal Carriage, Risk Factors, Acquisition and Persistence of ESBL-Producing Enterobacteriaceae in Dogs and Cats and Co-Carriage with Humans Belonging to the Same Household. J. Antimicrob. Chemother. 2020, 75, 342–350. [Google Scholar] [CrossRef]
- Belas, A.; Menezes, J.; Gama, L.T.; Pomba, C. Sharing of Clinically Important Antimicrobial Resistance Genes by Companion Animals and Their Human Household Members. Microb. Drug Resist. 2020, 26, 1174–1185. [Google Scholar] [CrossRef]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in Antimicrobial Resistance and Emergence of Major International High-Risk Clonal Lineages in Dogs and Cats with Urinary Tract Infection: 16 Year Retrospective Study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Carbapenem-Resistant Enterobacteriaceae, Second Update; ECDC Stock; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2019; pp. 1–17.
- Marques, C.; Menezes, J.; Belas, A.; Aboim, C.; Cavaco-Silva, P.; Trigueiro, G.; Gama, L.T.; Pomba, C. Klebsiella Pneumoniae Causing Urinary Tract Infections in Companion Animals and Humans: Population Structure, Antimicrobial Resistance and Virulence Genes. J. Antimicrob. Chemother. 2019, 74, 594–602. [Google Scholar] [CrossRef]
- Marques, C.; Gama, L.T.; Belas, A.; Bergström, K.; Beurlet, S.; Briend-Marchal, A.; Broens, E.M.; Costa, M.; Criel, D.; Damborg, P.; et al. European Multicenter Study on Antimicrobial Resistance in Bacteria Isolated from Companion Animal Urinary Tract Infections. BMC Vet. Res. 2016, 12, 213. [Google Scholar] [CrossRef]
- Abraham, S.; Wong, H.S.; Turnidge, J.; Johnson, J.R.; Trott, D.J. Carbapenemase-Producing Bacteria in Companion Animals: A Public Health Concern on the Horizon. J. Antimicrob. Chemother. 2014, 69, 1155–1157. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine; 6th Revision; Licence CC BY-NC-SA 3.0 IGO; World Health Organization: Geneve, Switzerland, 2019. [Google Scholar]
- European Centre for Disease Prevention and Control. Rapid Risk Assessment: Carbapenem-Resistant Enterobacteriaceae—First Update 4 June 2018; ECDC: Stockholm, Sweden, 2018.
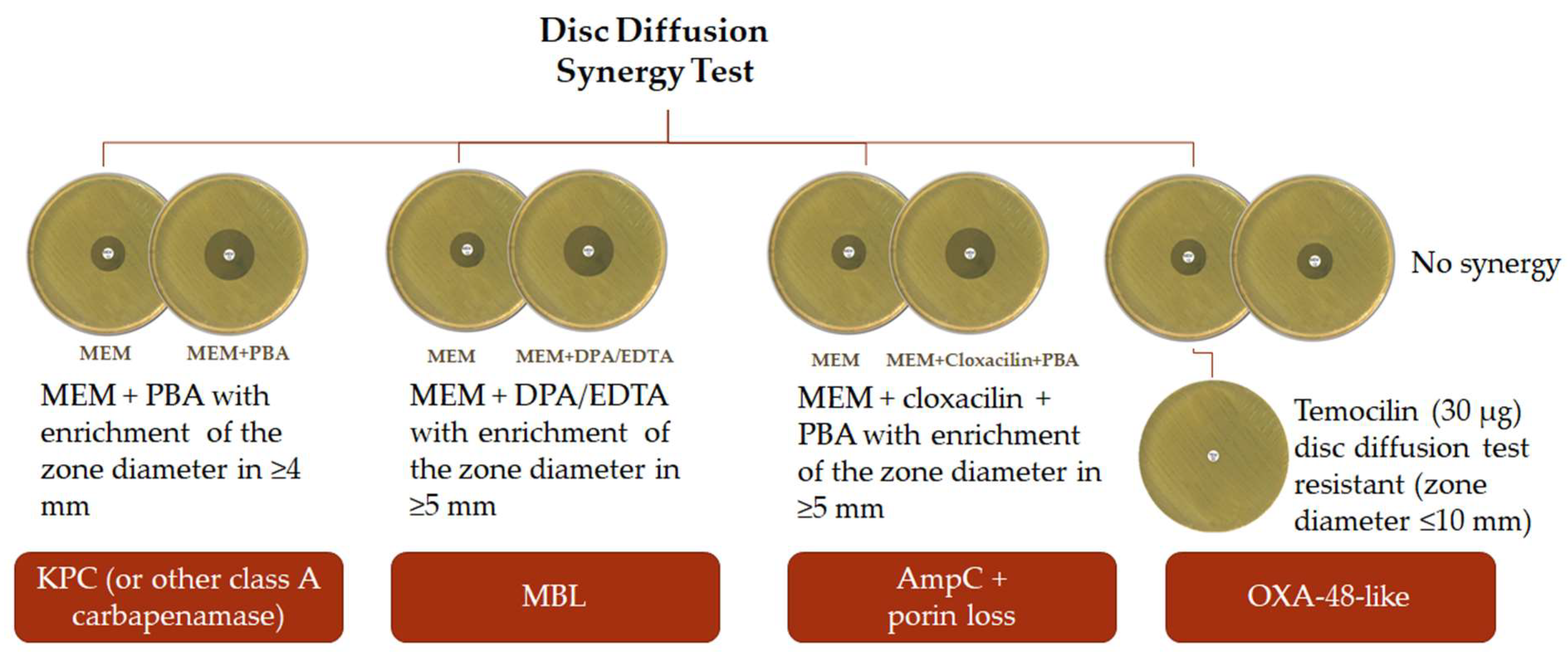
| Amber Class | Representative Carbapenemase Type | Hydrolysis Profile | Refs. | |||
|---|---|---|---|---|---|---|
| Narrow Spectrum Cephalosporins | Extended Spectrum Cephalosporins | Imipenem * | Meropenem * | |||
| Class A | KPC | + | + | + | + | [2,9] |
| Class B | IMP, VIM, NDM, | + | + | + | + | [3] |
| Class D | OXA-48-like | + | - | Variable 1 | - | [4,38,39] |
| OXA-23-like | + | + | + | + | [4] | |
| Technique | Sensitivity (%) | Specificity (%) | Turnaround Time (h) | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Selective Culture Medium | |||||
| SUPERCARBA | 95.6–96.5 | 60.7 | 18–24 | Colour identification of bacterial species | Extensive turnaround time; possible growth of non-carbapenemase producing bacteria; positive control needed. |
| CRE Agar | 78 | 60–66 | |||
| ChromID CARBA Smart | 91 | 76–89 | |||
| CHROMagar™ KPC | 100 | NDA | Only detects KPC-producing bacteria | ||
| CHROMagar™ OXA-48 | 75.8 | 99.3 | Only detects OXA-48-producing bacteria | ||
| Biochemical Tests | |||||
| Rapidec® CarbaNP | 100 | 100 | 2 | Rapid Detection of carbapenemase-producing bacteria | Non-specific detection; colour interpretation; expensive |
| CIM | NDA | NDA | 8 | Affordable; no commercial kit necessary | Non-specific detection; negative control strain needed; non-standardized |
| BlueCarba | 100 | 100 | 2 | Rapid Detection of carbapenemase-producing bacteria | Non-specific detection; positive control needed; expensive |
| β CARBA Test™ | 84.9 | 95.6 | 0.5 | Rapid Detection of carbapenemase-producing bacteria | Non-specific detection; expensive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, J.M.d.; Menezes, J.; Marques, C.; Pomba, C.F. Companion Animals—An Overlooked and Misdiagnosed Reservoir of Carbapenem Resistance. Antibiotics 2022, 11, 533. https://doi.org/10.3390/antibiotics11040533
Silva JMd, Menezes J, Marques C, Pomba CF. Companion Animals—An Overlooked and Misdiagnosed Reservoir of Carbapenem Resistance. Antibiotics. 2022; 11(4):533. https://doi.org/10.3390/antibiotics11040533
Chicago/Turabian StyleSilva, Joana Moreira da, Juliana Menezes, Cátia Marques, and Constança Ferreira Pomba. 2022. "Companion Animals—An Overlooked and Misdiagnosed Reservoir of Carbapenem Resistance" Antibiotics 11, no. 4: 533. https://doi.org/10.3390/antibiotics11040533
APA StyleSilva, J. M. d., Menezes, J., Marques, C., & Pomba, C. F. (2022). Companion Animals—An Overlooked and Misdiagnosed Reservoir of Carbapenem Resistance. Antibiotics, 11(4), 533. https://doi.org/10.3390/antibiotics11040533







