Bovine Respiratory Disease: Conventional to Culture-Independent Approaches to Studying Antimicrobial Resistance in North America
Abstract
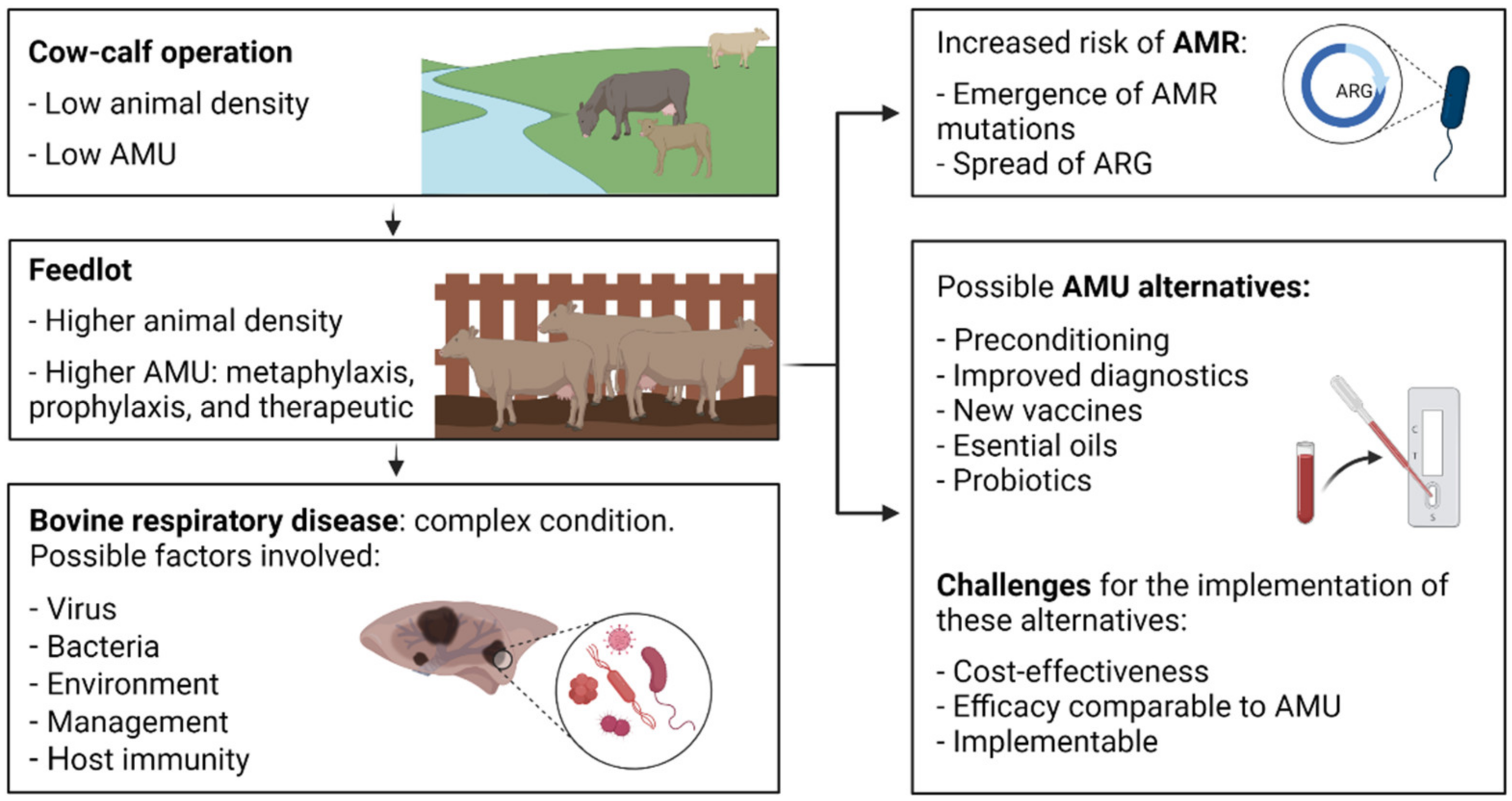
1. Introduction
2. Surveillance Studies Search Strategy
3. Culture-Dependent Surveillance Studies
3.1. Pasteurellaceae
3.2. Mycoplasma Bovis
3.3. Occasional BRD Opportunistic Pathogens
4. Integrative and Conjugative Elements
4.1. Pasteurellaceae
4.2. Mycoplasma Bovis
5. Culture-Independent Surveillance Studies
6. Novel Approaches for the Rapid Detection of Antimicrobial-Resistant BRD Bacteria
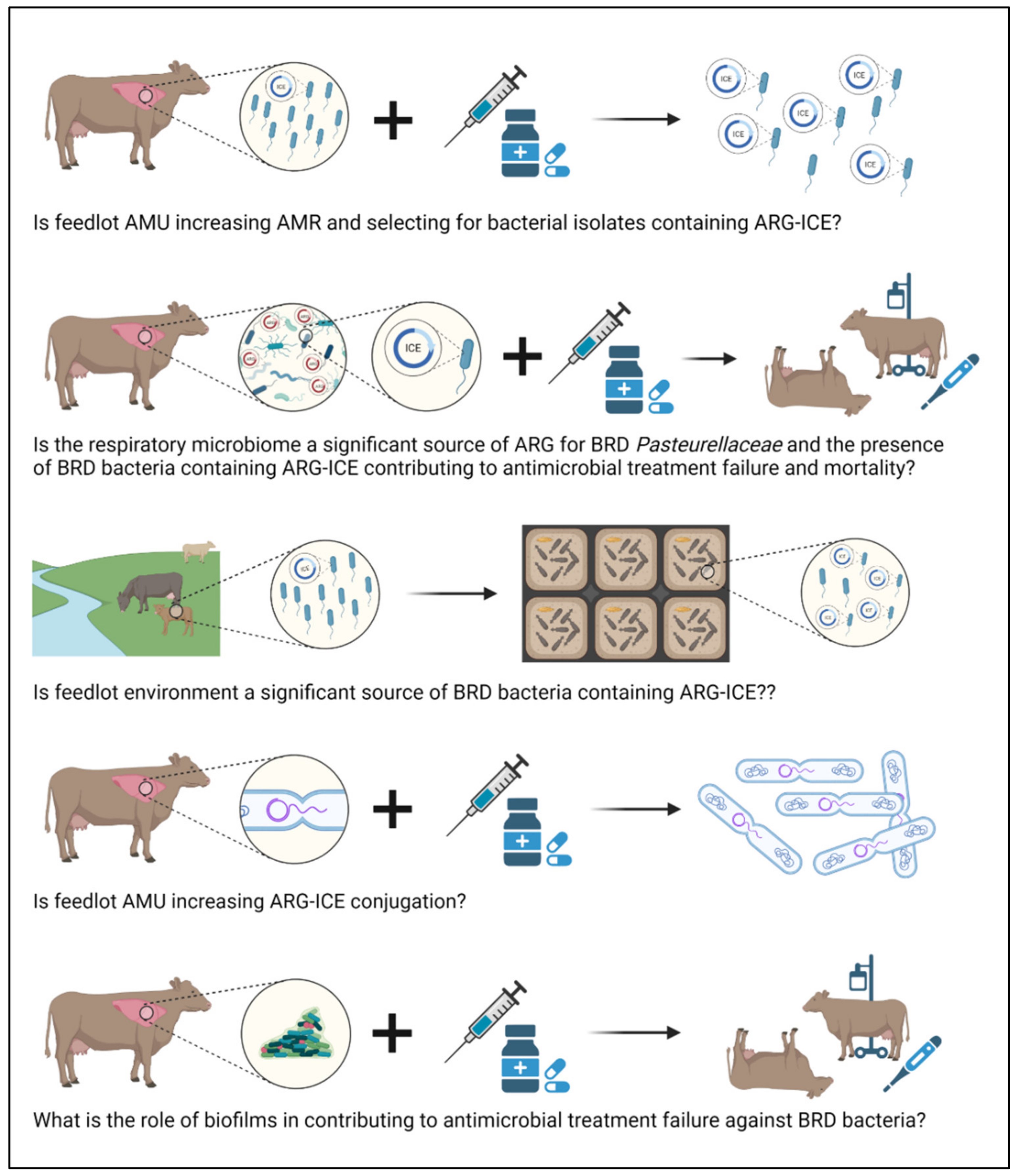
7. Further Considerations
7.1. Understanding the Genomic Epidemiology of ICE
7.2. Strategies to Mitigate ICE
7.3. Defining the Fitness of BRD Pathogens in the Environment
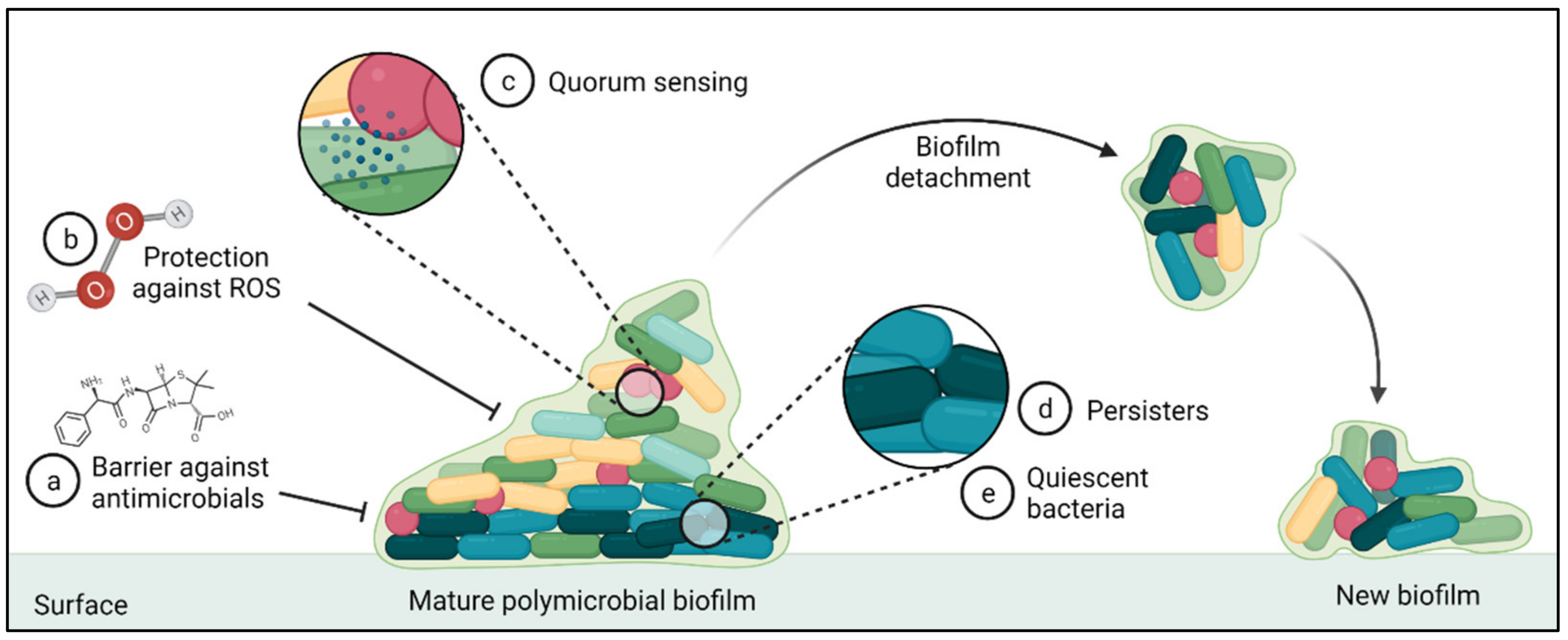
7.4. Role of Biofilms in AMR within BRD Pathogens
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Waldner, C.L.; Parker, S.; Gow, S.; Wilson, D.J.; Campbell, J.R. Antimicrobial usage in western Canadian cow-calf herds. Can. Vet. J. 2019, 60, 255–267. [Google Scholar] [PubMed]
- Taylor, J.D.; Fulton, R.W.; Lehenbauer, T.W.; Step, D.L.; Confer, A.W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can. Vet. J. 2010, 51, 1095–1102. [Google Scholar]
- Stroebel, C.; Alexander, T.; Workentine, M.L.; Timsit, E. Effects of transportation to and co-mingling at an auction market on nasopharyngeal and tracheal bacterial communities of recently weaned beef cattle. Vet. Microbiol. 2018, 223, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.; Chengappa, M.M.; Kuszak, J.; McVey, D.S. Bacterial pathogens of the bovine respiratory disease complex. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Hilton, W.M. BRD in 2014: Where have we been, where are we now, and where do we want to go? Anim. Health Res. Rev. 2014, 15, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.L.; Sweeney, M.T. Antimicrobial resistance in bovine respiratory disease pathogens: Measures, trends, and impact on efficacy. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Step, D.L.; Woolums, A.R. Bovine respiratory disease: Looking back and looking forward, what do we see? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef]
- Brault, S.A.; Hannon, S.J.; Gow, S.P.; Warr, B.N.; Withell, J.; Song, J.; Williams, C.M.; Otto, S.J.G.; Booker, C.W.; Morley, P.S. Antimicrobial use on 36 beef feedlots in western Canada: 2008–2012. Front. Vet. Sci. 2019, 6, 329. [Google Scholar] [CrossRef]
- Baptiste, K.E.; Kyvsgaard, N.C. Do antimicrobial mass medications work? A systematic review and meta-analysis of randomised clinical trials investigating antimicrobial prophylaxis or metaphylaxis against naturally occurring bovine respiratory disease. Pathog. Dis. 2017, 75, 7. [Google Scholar] [CrossRef]
- U.S. Food and Drug Agency: Timeline of FDA Action on Antimicrobial Resistance. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/timeline-fda-action-antimicrobial-resistance (accessed on 14 June 2021).
- Public Health Agency of Canada: Responsible Use of Medically Important Antimicrobials in Animals. Available online: https://www.canada.ca/en/public-health/services/antibiotic-antimicrobial-resistance/animals/actions/responsible-use-antimicrobials.html (accessed on 14 September 2021).
- Federation of Veterinarians of Europe: FVE Guidelines for Responsible Use of Antibiotics. Available online: https://fve.org/publications/fve-guidelines-responsible-use-of-antibiotics/ (accessed on 14 February 2022).
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Pardon, B.; Buczinski, S. Bovine respiratory disease diagnosis: What progress has been made in infectious diagnosis? Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Quintais, L.; Garcillan-Barcia, M.P.; de la Cruz, F.; Rocha, E.P. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011, 7, e1002222. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, R.A.; Waldor, M.K. Integrative and conjugative elements: Mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 2010, 8, 552–563. [Google Scholar] [CrossRef]
- Stanford, K.; Zaheer, R.; Klima, C.; McAllister, T.; Peters, D.; Niu, Y.D.; Ralston, B. Antimicrobial resistance in members of the bacterial bovine respiratory disease complex isolated from lung tissue of cattle mortalities managed with or without the use of antimicrobials. Microorganisms 2020, 8, 288. [Google Scholar] [CrossRef] [PubMed]
- Klima, C.L.; Cook, S.R.; Zaheer, R.; Laing, C.; Gannon, V.P.; Xu, Y.; Rasmussen, J.; Potter, A.; Hendrick, S.; Alexander, T.W.; et al. Comparative genomic analysis of Mannh. Haemolytica from bovine sources. PLoS ONE 2016, 11, e0149520. [Google Scholar] [CrossRef] [PubMed]
- Eidam, C.; Poehlein, A.; Leimbach, A.; Michael, G.B.; Kadlec, K.; Liesegang, H.; Daniel, R.; Sweeney, M.T.; Murray, R.W.; Watts, J.L.; et al. Analysis and comparative genomics of ICEMh1, a novel integrative and conjugative element (ICE) of Mannheimia haemolytica. J. Antimicrob. Chemother. 2015, 70, 93–97. [Google Scholar] [CrossRef]
- Citti, C.; Dordet-Frisoni, E.; Nouvel, L.X.; Kuo, C.H.; Baranowski, E. Horizontal gene transfers in Mycoplasmas (Mollicutes). Curr. Issues Mol. Biol. 2018, 29, 3–22. [Google Scholar] [CrossRef]
- Dordet-Frisoni, E.; Sagne, E.; Baranowski, E.; Breton, M.; Nouvel, L.X.; Blanchard, A.; Marenda, M.S.; Tardy, F.; Sirand-Pugnet, P.; Citti, C. Chromosomal transfers in mycoplasmas: When minimal genomes go mobile. mBio 2014, 5, e01958. [Google Scholar] [CrossRef]
- Klima, C.L.; Alexander, T.W.; Read, R.R.; Gow, S.P.; Booker, C.W.; Hannon, S.; Sheedy, C.; McAllister, T.A.; Selinger, L.B. Genetic characterization and antimicrobial susceptibility of Mannheimia haemolytica isolated from the nasopharynx of feedlot cattle. Vet. Microbiol. 2011, 149, 390–398. [Google Scholar] [CrossRef]
- Alexander, T.W.; Cook, S.; Klima, C.L.; Topp, E.; McAllister, T.A. Susceptibility to tulathromycin in Mannheimia haemolytica isolated from feedlot cattle over a 3-year period. Front. Microbiol. 2013, 4, 297. [Google Scholar] [CrossRef][Green Version]
- Klima, C.L.; Alexander, T.W.; Hendrick, S.; McAllister, T.A. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can. J. Vet. Res. 2014, 78, 38–45. [Google Scholar] [PubMed]
- Noyes, N.R.; Benedict, K.M.; Gow, S.P.; Booker, C.W.; Hannon, S.J.; McAllister, T.A.; Morley, P.S. Mannheimia haemolytica in feedlot cattle: Prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J. Vet. Int. Med. 2015, 29, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.; Credille, B.; Berghaus, R.; Giguere, S. Prevalence of multi drug antimicrobial resistance in Mannhemia haemolytica isolated from high-risk stocker cattle at arrival and two weeks after processing. J. Anim. Sci. Biotechnol. 2017, 95, 1124–1131. [Google Scholar]
- Woolums, A.R.; Karisch, B.B.; Frye, J.G.; Epperson, W.; Smith, D.R.; Blanton, J., Jr.; Austin, F.; Kaplan, R.; Hiott, L.; Woodley, T.; et al. Multidrug resistant Mannheimia haemolytica isolated from high-risk beef stocker cattle after antimicrobial metaphylaxis and treatment for bovine respiratory disease. Vet. Microbiol. 2018, 221, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; McMullen, C.; Timsit, E.; Hallewell, J.; Orsel, K.; van der Meer, F.; Yan, S.; Alexander, T.W. Genetic relatedness and antimicrobial resistance in respiratory bacteria from beef calves sampled from spring processing to 40 days after feedlot entry. Vet. Microbiol. 2020, 240, 108478. [Google Scholar] [CrossRef] [PubMed]
- Andres-Lasheras, S.; Ha, R.; Zaheer, R.; Lee, C.; Booker, C.W.; Dorin, C.; Van Donkersgoed, J.; Deardon, R.; Gow, S.; Hannon, S.J.; et al. Prevalence and risk factors associated with antimicrobial resistance in bacteria related to bovine respiratory disease-A broad cross-sectional study of beef cattle at entry into Canadian feedlots. Front. Vet. Sci. 2021, 8, 692646. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, D.; Andres-Lasheras, S.; Zaheer, R.; McAllister, T.; Homerosky, E.; Anholt, R.M.; Dorin, C. Prevalence, risk factors, and antimicrobial resistance profile of respiratory pathogens isolated from suckling beef calves to reprocessing at the feedlot: A longitudinal study. Front. Vet. Sci. 2021, 8, 764701. [Google Scholar] [CrossRef]
- Welsh, R.D.; Dye, L.B.; Payton, M.E.; Confer, A.W. Isolation and antimicrobial susceptibilities of bacterial pathogens from bovine pneumonia: 1994–2002. J. Vet. Diagn. Investig. 2004, 16, 426–431. [Google Scholar] [CrossRef]
- Portis, E.; Lindeman, C.; Johansen, L.; Stoltman, G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex--Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni—In the United States and Canada. J. Vet. Diagn. Investig. 2012, 24, 932–944. [Google Scholar] [CrossRef]
- Lubbers, B.V.; Hanzlicek, G.A. Antimicrobial multidrug resistance and coresistance patterns of Mannheimia haemolytica isolated from bovine respiratory disease cases—A three-year (2009–2011) retrospective analysis. J. Vet. Diagn. Investig. 2013, 25, 413–417. [Google Scholar] [CrossRef]
- Klima, C.L.; Zaheer, R.; Cook, S.R.; Booker, C.W.; Hendrick, S.; Alexander, T.W.; McAllister, T.A. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J. Clin. Microbiol. 2014, 52, 438–448. [Google Scholar] [CrossRef] [PubMed]
- Anholt, R.M.; Klima, C.; Allan, N.; Matheson-Bird, H.; Schatz, C.; Ajitkumar, P.; Otto, S.J.; Peters, D.; Schmid, K.; Olson, M.; et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front. Vet. Sci. 2017, 4, 207. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Hallewell, J.; Booker, C.; Tison, N.; Amat, S.; Alexander, T.W. Prevalence and antimicrobial susceptibility of Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni isolated from the lower respiratory tract of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2017, 208, 118–125. [Google Scholar] [CrossRef]
- Rosenbusch, R.F.; Kinyon, J.M.; Apley, M.; Funk, N.D.; Smith, S.; Hoffman, L.J. In vitro antimicrobial inhibition profiles of Mycoplasma bovis isolates recovered from various regions of the United States from 2002 to 2003. J. Vet. Diagn. Investig. 2005, 17, 436–441. [Google Scholar] [CrossRef]
- Hendrick, S.H.; Bateman, K.G.; Rosengren, L.B. The effect of antimicrobial treatment and preventive strategies on bovine respiratory disease and genetic relatedness and antimicrobial resistance of Mycoplasma bovis isolates in a western Canadian feedlot. Can. Vet. J. 2013, 54, 1146–1156. [Google Scholar] [PubMed]
- Cai, H.Y.; McDowall, R.; Parker, L.; Kaufman, E.I.; Caswell, J.L. Changes in antimicrobial susceptibility profiles of Mycoplasma bovis over time. Can. J. Vet. Res. 2019, 83, 34–41. [Google Scholar] [PubMed]
- Jelinski, M.; Kinnear, A.; Gesy, K.; Andres-Lasheras, S.; Zaheer, R.; Weese, S.; McAllister, T.A. Antimicrobial sensitivity testing of Mycoplasma bovis isolates derived from Western Canadian feedlot cattle. Microorganisms 2020, 8, 124. [Google Scholar] [CrossRef]
- Hannan, P.C. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. International Research Programme on Comparative Mycoplasmology. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef]
- CLSI Document VET01S: Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020.
- CLSI Supplement VET08: Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- CLSI Report VET09: Understanding Susceptibility Test Data as a Component of Antimicrobial Stewardship in Veterinary Settings, 1st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019.
- Saini, V.; McClure, J.T.; Leger, D.; Dufour, S.; Sheldon, A.G.; Scholl, D.T.; Barkema, H.W. Antimicrobial use on Canadian dairy farms. J. Dairy Sci. 2012, 95, 1209–1221. [Google Scholar] [CrossRef]
- Andres-Lasheras, S.; Zaheer, R.; Ha, R.; Lee, C.; Jelinski, M.; McAllister, T.A. A direct qPCR screening approach to improve the efficiency of Mycoplasma bovis isolation in the frame of a broad surveillance study. J. Microbiol. Methods 2020, 169, 105805. [Google Scholar] [CrossRef]
- Kinnear, A.; McAllister, T.A.; Zaheer, R.; Waldner, M.; Ruzzini, A.C.; Andres-Lasheras, S.; Parker, S.; Hill, J.E.; Jelinski, M.D. Investigation of macrolide resistance genotypes in Mycoplasma bovis isolates from Canadian feedlot cattle. Pathogens 2020, 9, 622. [Google Scholar] [CrossRef] [PubMed]
- Ledger, L.; Eidt, J.; Cai, H.Y. Identification of antimicrobial resistance-associated genes through whole genome sequencing of Mycoplasma bovis isolates with different antimicrobial resistances. Pathogens 2020, 9, 588. [Google Scholar] [CrossRef] [PubMed]
- CLSI Document VET06: Methods for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria Isolated from Animals, 1st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016.
- Gautier-Bouchardon, A.V. Antimicrobial Resistance in Mycoplasma spp. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Soehnlen, M.K.; Kunze, M.E.; Karunathilake, K.E.; Henwood, B.M.; Kariyawasam, S.; Wolfgang, D.R.; Jayarao, B.M. In vitro antimicrobial inhibition of Mycoplasma bovis isolates submitted to the Pennsylvania Animal Diagnostic Laboratory using flow cytometry and a broth microdilution method. J. Vet. Diagn. Investig. 2011, 23, 547–551. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gagea, M.I.; Bateman, K.G.; Shanahan, R.A.; van Dreumel, T.; McEwen, B.J.; Carman, S.; Archambault, M.; Caswell, J.L. Naturally occurring Mycoplasma bovis-associated pneumonia and polyarthritis in feedlot beef calves. J. Vet. Diagn. Investig. 2006, 18, 29–40. [Google Scholar] [CrossRef]
- Lysnyansky, I.; Ayling, R.D. Mycoplasma bovis: Mechanisms of resistance and trends in antimicrobial susceptibility. Front. Microbiol. 2016, 7, 595. [Google Scholar] [CrossRef] [PubMed]
- Heuvelink, A.; Reugebrink, C.; Mars, J. Antimicrobial susceptibility of Mycoplasma bovis isolates from veal calves and dairy cattle in the Netherlands. Vet. Microbiol. 2016, 189, 1–7. [Google Scholar] [CrossRef]
- Ayling, R.D.; Rosales, R.S.; Barden, G.; Gosney, F.L. Changes in antimicrobial susceptibility of Mycoplasma bovis isolates from Great Britain. Vet. Rec. 2014, 175, 486. [Google Scholar] [CrossRef]
- Holman, D.B.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 86. [Google Scholar] [CrossRef]
- Blackall, P.J.; Bojesen, A.M.; Christensen, H.; Bisgaard, M. Reclassification of [Pasteurella] trehalosi as Bibersteinia trehalosi gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 666–674. [Google Scholar] [CrossRef]
- Dyer, N.W.; Ward, A.C.; Weiser, G.C.; White, D.G. Seasonal incidence and antibiotic susceptibility patterns of Pasteurellaceae isolated from American bison (Bison bison). Can. J. Vet. Res. 2001, 65, 7–14. [Google Scholar] [PubMed]
- Hanthorn, C.J.; Dewell, R.D.; Cooper, V.L.; Frana, T.S.; Plummer, P.J.; Wang, C.; Dewell, G.A. Randomized clinical trial to evaluate the pathogenicity of Bibersteinia trehalosi in respiratory disease among calves. BMC Vet. Res. 2014, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Cortese, V.S.; Braun, D.A.; Crouch, D.; Townsend, C.; Zukowski, B. Case report—Peracute to acute fatal pneuinonia in cattle. caused by Bibersteinia trehalosi. Bov. Pract. 2012, 46, 138–142. [Google Scholar]
- APHA Disease Surveillance Report: Bibersteinia trehalosi Infection in Cattle and Sheep; Animal and Plant Health Agency: Bristol, UK, 2014.
- Watson, P.J.; Scholes, S.F. Bibersteinia trehalosi necrotising hepatitis associated with sudden death in an adult cow. Vet. Rec. 2010, 167, 100–102. [Google Scholar] [CrossRef]
- Collins, R.L. Bibersteinia trehalosi in cattle—Another component in the bovine respiratory disease complex? Cattle Pract. 2011, 19, 9–12. [Google Scholar]
- Narayanan, S.; Bates, H.; Confer, A.; Couger, B.; Ramachandran, A. Whole-genome sequence of multidrug-resistant Bibersteinia trehalosi strain OADDL-BT1. Microbiol. Resour. Announc. 2019, 8, e01690-18. [Google Scholar] [CrossRef]
- Confer, A.W. Update on bacterial pathogenesis in BRD. Anim. Health Res. Rev. 2009, 10, 145–148. [Google Scholar] [CrossRef]
- Hall, J.A.; Isaiah, A.; Estill, C.T.; Pirelli, G.J.; Suchodolski, J.S. Weaned beef calves fed selenium-biofortified alfalfa hay have an enriched nasal microbiota compared with healthy controls. PLoS ONE 2017, 12, e0179215. [Google Scholar] [CrossRef]
- Holman, D.B.; Timsit, E.; Booker, C.W.; Alexander, T.W. Injectable antimicrobials in commercial feedlot cattle and their effect on the nasopharyngeal microbiota and antimicrobial resistance. Vet. Microbiol. 2018, 214, 140–147. [Google Scholar] [CrossRef]
- Klima, C.L.; Holman, D.B.; Ralston, B.J.; Stanford, K.; Zaheer, R.; Alexander, T.W.; McAllister, T.A. Lower respiratory tract microbiome and resistome of bovine respiratory disease mortalities. Microb. Ecol. 2019, 78, 446–456. [Google Scholar] [CrossRef]
- McMullen, C.; Orsel, K.; Alexander, T.W.; van der Meer, F.; Plastow, G.; Timsit, E. Evolution of the nasopharyngeal bacterial microbiota of beef calves from spring processing to 40 days after feedlot arrival. Vet. Microbiol. 2018, 225, 139–148. [Google Scholar] [CrossRef] [PubMed]
- McMullen, C.; Orsel, K.; Alexander, T.W.; van der Meer, F.; Plastow, G.; Timsit, E. Comparison of the nasopharyngeal bacterial microbiota of beef calves raised without the use of antimicrobials between healthy calves and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2019, 231, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Lowe, J.; de Godoy, M.; Maradiaga, N.; Ramirez, C.; Ghanem, M.; Abd El-Raof, Y.; Aldridge, B. Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet. Microbiol. 2017, 208, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.M.; Lowe, J.F.; Grimmer, E.D.; de Godoy, M.R.C.; Ghanem, M.M.; Abd El-Raof, Y.M.; Aldridge, B.M. Relationship between nasopharyngeal and bronchoalveolar microbial communities in clinically healthy feedlot cattle. BMC Microbiol. 2017, 17, 138. [Google Scholar] [CrossRef]
- Beef Magazine: Bibersteinia Trehalosi on the Rise in Beef Cattle. Available online: https://www.beefmagazine.com/animal-health/bibersteinia-trehalosi-rise-beef-cattle (accessed on 25 November 2021).
- Iowa State University. Iowa State University Veterinary Diagnostic Laboratory: 2012 Annual Report; Iowa State University: Iowa, IA, USA, 2012. [Google Scholar]
- Holschbach, C.L.; Aulik, N.; Poulsen, K.; Ollivett, T.L. Prevalence and temporal trends in antimicrobial resistance of bovine respiratory disease pathogen isolates submitted to the Wisconsin Veterinary Diagnostic Laboratory: 2008–2017. J. Dairy Sci. 2020, 103, 9464–9472. [Google Scholar] [CrossRef]
- Michael, G.B.; Bosse, J.T.; Schwarz, S. Antimicrobial resistance in Pasteurellaceae of veterinary origin. Microbiol. Spectr. 2018, 6, 3. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Meunier, D.; Targant, H.; Cloeckaert, A.; Schwarz, S.; Madec, J.Y. Plasmid-mediated florfenicol resistance in Pasteurella trehalosi. J. Antimicrob. Chemother. 2006, 58, 13–17. [Google Scholar] [CrossRef]
- Quinn, P.J.; Markey, B.K.; Leonard, F.C.; FitzPatrick, E.S.; Hartigan, P.J. Veterinary Microbiology and Microbial Disease, 2nd ed.; Wiley-Blackwell: Oxford, UK, 2011. [Google Scholar]
- Fessler, A.T.; Schwarz, S. Antimicrobial Resistance in Corynebacterium spp., Arcanobacterium spp. and Trueperella pyogenes. Microbiol. Spectr. 2017, 5, 6. [Google Scholar] [CrossRef]
- Trinh, H.T.; Billington, S.J.; Field, A.C.; Songer, J.G.; Jost, B.H. Susceptibility of Arcanobacterium pyogenes from different sources to tetracycline, macrolide and lincosamide antimicrobial agents. Vet. Microbiol. 2002, 85, 353–359. [Google Scholar] [CrossRef]
- Michael, G.B.; Kadlec, K.; Sweeney, M.T.; Brzuszkiewicz, E.; Liesegang, H.; Daniel, R.; Murray, R.W.; Watts, J.L.; Schwarz, S. ICEPmu1, an integrative conjugative element (ICE) of Pasteurella multocida: Analysis of the regions that comprise 12 antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 84–90. [Google Scholar] [CrossRef]
- Beker, M.; Rose, S.; Lykkebo, C.A.; Douthwaite, S. Integrative and Conjugative Elements (ICEs) in Pasteurellaceae species and their detection by multiplex PCR. Front. Microbiol. 2018, 9, 1329. [Google Scholar] [CrossRef] [PubMed]
- Mohd-Zain, Z.; Turner, S.L.; Cerdeno-Tarraga, A.M.; Lilley, A.K.; Inzana, T.J.; Duncan, A.J.; Harding, R.M.; Hood, D.W.; Peto, T.E.; Crook, D.W. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 2004, 186, 8114–8122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bhatt, K.; Timsit, E.; Rawlyk, N.; Potter, A.; Liljebjelke, K. Integrative conjugative element ICEHs1 encodes for antimicrobial resistance and metal tolerance in Histophilus somni. Front. Vet. Sci. 2018, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Clawson, M.L.; Murray, R.W.; Sweeney, M.T.; Apley, M.D.; DeDonder, K.D.; Capik, S.F.; Larson, R.L.; Lubbers, B.V.; White, B.J.; Kalbfleisch, T.S.; et al. Genomic signatures of Mannheimia haemolytica that associate with the lungs of cattle with respiratory disease, an integrative conjugative element, and antibiotic resistance genes. BMC Genom. 2016, 17, 982. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.C.; Daher, R.K.; Stanford, K.; Amoako, K.K.; Boissinot, M.; Bergeron, M.G.; Alexander, T.; Cook, S.; Ralston, B.; Zaheer, R.; et al. A sensitive and accurate recombinase polymerase amplification assay for detection of the primary bacterial pathogens causing bovine respiratory disease. Front. Vet. Sci. 2020, 7, 208. [Google Scholar] [CrossRef]
- Tardy, F.; Mick, V.; Dordet-Frisoni, E.; Marenda, M.S.; Sirand-Pugnet, P.; Blanchard, A.; Citti, C. Integrative conjugative elements are widespread in field isolates of Mycoplasma species pathogenic for ruminants. Appl. Environ. Microbiol. 2015, 81, 1634–1643. [Google Scholar] [CrossRef] [PubMed]
- Faucher, M.; Nouvel, L.X.; Dordet-Frisoni, E.; Sagne, E.; Baranowski, E.; Hygonenq, M.C.; Marenda, M.S.; Tardy, F.; Citti, C. Mycoplasmas under experimental antimicrobial selection: The unpredicted contribution of horizontal chromosomal transfer. PLoS Genet. 2019, 15, e1007910. [Google Scholar] [CrossRef]
- Francoz, D.; Fortin, M.; Fecteau, G.; Messier, S. Determination of Mycoplasma bovis susceptibilities against six antimicrobial agents using the E test method. Vet. Microbiol. 2005, 105, 57–64. [Google Scholar] [CrossRef]
- Murray, G.L.; Bradshaw, C.S.; Bissessor, M.; Danielewski, J.; Garland, S.M.; Jensen, J.S.; Fairley, C.K.; Tabrizi, S.N. Increasing macrolide and fluoroquinolone resistance in Mycoplasma genitalium. Emerg. Infect. Dis. 2017, 23, 809. [Google Scholar] [CrossRef]
- Delaney, N.F.; Balenger, S.; Bonneaud, C.; Marx, C.J.; Hill, G.E.; Ferguson-Noel, N.; Tsai, P.; Rodrigo, A.; Edwards, S.V. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 2012, 8, e1002511. [Google Scholar] [CrossRef]
- Davis, M.A.; Hancock, D.D.; Besser, T.E.; Daniels, J.B.; Baker, K.N.; Call, D.R. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet. Microbiol. 2007, 119, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Merck Veterinary Manual: Cephalosporins and Cephamycins. Available online: https://www.merckvetmanual.com/pharmacology/antibacterial-agents/cephalosporins-and-cephamycins (accessed on 14 December 2021).
- Lamm, C.G.; Love, B.C.; Krehbiel, C.R.; Johnson, N.J.; Step, D.L. Comparison of antemortem antimicrobial treatment regimens to antimicrobial susceptibility patterns of postmortem lung isolates from feedlot cattle with bronchopneumonia. J. Vet. Diagn. Investig. 2012, 24, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.C.; Van Bibber-Krueger, C.L.; Ogunrinu, O.J.; Amachawadi, R.G.; Scott, H.M.; Drouillard, J.S. Effects of intermittent feeding of tylosin phosphate during the finishing period on feedlot performance, carcass characteristics, antimicrobial resistance, and incidence and severity of liver abscesses in steers. J. Anim. Sci. Biotechnol. 2018, 96, 2877–2885. [Google Scholar] [CrossRef] [PubMed]
- Agga, G.E.; Schmidt, J.W.; Arthur, T.M. Effects of in-feed chlortetracycline prophylaxis in beef cattle on animal health and antimicrobial-resistant Escherichia coli. Appl. Environ. Microbiol. 2016, 82, 7197–7204. [Google Scholar] [CrossRef]
- Agga, G.E.; Cook, K.L.; Netthisinghe, A.M.P.; Gilfillen, R.A.; Woosley, P.B.; Sistani, K.R. Persistence of antibiotic resistance genes in beef cattle backgrounding environment over two years after cessation of operation. PLoS ONE 2019, 14, e0212510. [Google Scholar] [CrossRef]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase polymerase amplification for diagnostic applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Latorre-Perez, A.; Pascual, J.; Porcar, M.; Vilanova, C. A lab in the field: Applications of real-time, in situ metagenomic sequencing. Biol. Methods Protoc. 2020, 5, bpaa016. [Google Scholar] [CrossRef]
- Torres-Sangiao, E.; Leal Rodriguez, C.; Garcia-Riestra, C. Application and perspectives of MALDI-TOF mass spectrometry in clinical microbiology laboratories. Microorganisms 2021, 9, 1539. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Higa, Y.; Uemura, R.; Yamazaki, W.; Goto, S.; Goto, Y.; Sueyoshi, M. An improved loop-mediated isothermal amplification assay for the detection of Mycoplasma bovis. J. Med. Sci. 2016, 78, 1343–1346. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Garrigos, A.; Maruthamuthu, M.K.; Ault, A.; Davidson, J.L.; Rudakov, G.; Pillai, D.; Koziol, J.; Schoonmaker, J.P.; Johnson, T.; Verma, M.S. On-farm colorimetric detection of Pasteurella multocida, Mannheimia haemolytica and Histophilus somni in crude bovine nasal samples. Vet. Res. 2021, 52, 126. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Wang, Y.; Zhang, L.; Xu, J.; Ye, C. Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of Enterococcus faecalis and Staphylococcus aureus. Front. Microbiol. 2017, 8, 192. [Google Scholar] [CrossRef]
- Sun, D.; Wang, J.; Wu, R.; Wang, C.; He, X.; Zheng, J.; Yang, H. Development of a novel LAMP diagnostic method for visible detection of swine Pasteurella multocida. Vet. Res. Commun. 2010, 34, 649–657. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, G.; Sun, B.; Liu, S.; Wang, Y.; Gao, M.; Fan, Y.; Zhang, G.; Shi, G.; Kang, X. Rapid detection of mecA and femA genes by loop-mediated isothermal amplification in a microfluidic system for discrimination of different Staphylococcal species and prediction of methicillin resistance. Front. Microbiol. 2020, 11, 1487. [Google Scholar] [CrossRef]
- Sakai, J.; Maeda, T.; Tarumoto, N.; Misawa, K.; Tamura, S.; Imai, K.; Yamaguchi, T.; Iwata, S.; Murakami, T.; Maesaki, S. A novel detection procedure for mutations in the 23S rRNA gene of Mycoplasma pneumoniae with peptide nucleic acid-mediated loop-mediated isothermal amplification assay. J. Microbiol. Methods 2017, 141, 90–96. [Google Scholar] [CrossRef]
- Zhou, B.; Liang, T.; Zhan, Z.; Liu, R.; Li, F.; Xu, H. Rapid and simultaneous quantification of viable Escherichia coli O157:H7 and Salmonella spp. in milk through multiplex real-time PCR. J. Dairy Sci. 2017, 100, 8804–8813. [Google Scholar] [CrossRef]
- Han, L.; Wang, K.; Ma, L.; Delaquis, P.; Bach, S.; Feng, J.; Lu, X. Viable but nonculturable Escherichia coli O157:H7 and Salmonella enterica in fresh produce: Rapid determination by loop-mediated isothermal amplification coupled with a propidium monoazide treatment. Appl. Environ. Microbiol. 2020, 86, e02566-19. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Liu, X.; Chen, G.; Chen, X.; Chen, J.; Liu, Z.; Gong, J.; Yang, G.; Lan, Q. Development of propidium monoazide-recombinase polymerase amplification (PMA-RPA) assay for rapid detection of Streptococcus pyogenes and Streptococcus agalactiae. Mol. Cell. Probes 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Andermann, T.; Torres Jimenez, M.F.; Matos-Maravi, P.; Batista, R.; Blanco-Pastor, J.L.; Gustafsson, A.L.S.; Kistler, L.; Liberal, I.M.; Oxelman, B.; Bacon, C.D.; et al. A guide to carrying out a phylogenomic target sequence capture project. Front. Genet. 2019, 10, 1407. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Wu, B.; Yan, G.; Li, G.; Sun, L.; Lu, G.; Zhou, W. Combined nanopore adaptive sequencing and enzyme-based host depletion efficiently enriched microbial sequences and identified missing respiratory pathogens. BMC Genom. 2021, 22, 732. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Bokma, J.; Haesebrouck, F.; Nauwynck, H.; Boyen, F.; Pardon, B.; Theuns, S. High quality genome assemblies of Mycoplasma bovis using a taxon-specific Bonito basecaller for MinION and Flongle long-read nanopore sequencing. BMC Bioinform. 2020, 21, 517. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.; Naidenov, B.; Bates, H.; Willyerd, K.; Snider, T.; Couger, M.B.; Chen, C.; Ramachandran, A. Nanopore ultra-long read sequencing technology for antimicrobial resistance detection in Mannheimia haemolytica. J. Microbiol. Methods 2019, 159, 138–147. [Google Scholar] [CrossRef]
- Benkova, M.; Soukup, O.; Marek, J. Antimicrobial susceptibility testing: Currently used methods and devices and the near future in clinical practice. J. Appl. Microbiol. 2020, 129, 806–822. [Google Scholar] [CrossRef]
- Burrer, A.; Findeisen, P.; Jager, E.; Ghebremedhin, B.; Grundt, A.; Ahmad-Nejad, P.; Miethke, T.; Neumaier, M. Rapid detection of cefotaxime-resistant Escherichia coli by LC-MS. Int. J. Med. Microbiol. 2015, 305, 860–864. [Google Scholar] [CrossRef]
- Griffin, P.M.; Price, G.R.; Schooneveldt, J.M.; Schlebusch, S.; Tilse, M.H.; Urbanski, T.; Hamilton, B.; Venter, D. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J. Clin. Microbiol. 2012, 50, 2918–2931. [Google Scholar] [CrossRef]
- Van Driessche, L.; Bokma, J.; Gille, L.; Ceyssens, P.J.; Sparbier, K.; Haesebrouck, F.; Deprez, P.; Boyen, F.; Pardon, B. Rapid detection of tetracycline resistance in bovine Pasteurella multocida isolates by MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA). Sci. Rep. 2018, 8, 13599. [Google Scholar] [CrossRef]
- Ludwig, A.; Berthiaume, P.; Boerlin, P.; Gow, S.; Leger, D.; Lewis, F.I. Identifying associations in Escherichia coli antimicrobial resistance patterns using additive Bayesian networks. Prev. Vet. Med. 2013, 110, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Hartnack, S.; Odoch, T.; Kratzer, G.; Furrer, R.; Wasteson, Y.; L’Abée-Lund, T.M.; Skjerve, E. Additive Bayesian networks for antimicrobial resistance and potential risk factors in non-typhoidal Salmonella isolates from layer hens in Uganda. BMC Vet. Res. 2019, 15, 212. [Google Scholar] [CrossRef] [PubMed]
- In Ha, R.; Zaheer, R.; Sargeant, C.; Klima, C.; McAllister, T. Intra- and inter-species horizontal transfer potential of integrative and conjugative elements carrying antimicrobial resistance genes. In Proceedings of the Canadian Society of Microbiologists, Sherbrooke, QC, Canada, 10–13 June 2019. [Google Scholar]
- Timsit, E.; McMullen, C.; Amat, S.; Alexander, T.W. Respiratory bacterial microbiota in cattle: From development to modulation to enhance respiratory health. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Cameron, A.; Zaheer, R.; McAllister, T.A. Emerging variants of the integrative and conjugant element ICEMh1 in livestock pathogens: Structural insights, potential host range, and implications for bacterial fitness and antimicrobial therapy. Front. Microbiol. 2019, 10, 2608. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Huang, S.; Smith, R.P.; Srimani, J.K.; Sysoeva, T.A.; Bewick, S.; Karig, D.K.; You, L. Antibiotics as a selective driver for conjugation dynamics. Nat. Biotechnol. 2016, 1, 16044. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Burriel, A.R. Isolation of Pasteurella haemolytica from grass, drinking water, and straw bedding used by sheep. Curr. Microbiol. 1997, 35, 316–318. [Google Scholar] [CrossRef]
- Neupane, S.; Nayduch, D.; Zurek, L. House flies (Musca domestica) pose a risk of carriage and transmission of bacterial pathogens associated with bovine respiratory disease (BRD). Insects 2019, 10, 358. [Google Scholar] [CrossRef]
- Romling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef]
- Seneviratne, C.J.; Wang, Y.; Jin, L.; Wong, S.S.; Herath, T.D.; Samaranayake, L.P. Unraveling the resistance of microbial biofilms: Has proteomics been helpful? Proteomics 2012, 12, 651–665. [Google Scholar] [CrossRef]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E.; Ceri, H.; Morck, D.W.; Buret, A.G.; Read, R.R. Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002, 66, 86–92. [Google Scholar] [PubMed]
- Boukahil, I.; Czuprynski, C.J. Characterization of Mannheimia haemolytica biofilm formation in vitro. Vet. Microbiol. 2015, 175, 114–122. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, L.; Ellis, R.J.; Miles, K.; Ayling, R.D.; Nicholas, R.A.J. Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology 2006, 152, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Hannon, S.J.; Brault, S.A.; Otto, S.J.G.; Morley, P.S.; McAllister, T.A.; Booker, C.W.; Gow, S.P. Feedlot cattle antimicrobial use surveillance network: A Canadian journey. Front. Vet. Sci. 2020, 7, 596042. [Google Scholar] [CrossRef] [PubMed]



| Geographical Area | Sampling Year (s) | Targeted Bacteria | Authors |
|---|---|---|---|
| Pasteurellaceae AMR active surveillance studies | |||
| Southern Alberta, Canada | 2008–2009 | Mh | Klima et al. 2011 [23] |
| Southern Alberta, Canada | 2007–2010 | Mh | Alexander et al., 2013 [24] |
| Southern Alberta, Canada | 2008–2009 | Mh | Klima et al., 2014 [25] |
| Southern and central Alberta, Canada | 2007–2010 | Mh | Noyes et al., 2015 [26] |
| Central Georgia, USA | 2016 | Mh | Snyder et al., 2017 [27] |
| Mississippi, USA | 2016 | Mh, Pm, Hs | Woolums et al., 2018 [28] |
| Southern Alberta, Canada | 2016 | Mh, Pm, Hs | Guo et al., 2020 [29] |
| Alberta, Canada | 2017–2019 | Mh, Pm, Hs | Andres-Lasheras et al., 2021 [30] |
| Alberta, Canada | 2017–2017 | Mh, Pm, Hs | Nobrega et al., 2021 [31] |
| Pasteurellaceae AMR passive surveillance studies | |||
| Oklahoma Animal Disease Diagnostic Laboratory, USA | 1994–2002 | Mh, Pm, Hs | Welsh et al., 2004 [32] |
| Veterinary diagnostic laboratories across USA and Canada | 2000–2009 | Mh, Pm, Hs | Portis et al., 2012 [33] |
| Kansas State Veterinary Diagnostic Laboratory, USA | 2009–2011 | Mh | Lubbers and Hanzlicek. 2013 [34] |
| Alberta (Canada), Texas, and Nebraska (USA) | NS | Mh, Pm, Hs | Klima et al., 2014 [35] |
| Southern Alberta, Canada | 2014–2015 | Mh, Pm, Hs | Anholt et al., 2017 [36] |
| Southern Alberta, Canada | 2015–2016 | Mh, Pm, Hs | Timsit et al., 2017 [37] |
| Mycoplasma bovis AMR active and passive surveillance studies | |||
| Different regions across USA | 2002–2003 | Mb | Rosenbusch et al., 2005 [38] |
| Saskatchewan, Canada | 2007–2008 | Mb | Hendrick et al., 2013 [39] |
| Southern Alberta, Canada | 2015–2016 | Mb | Anholt et al., 2017 [36] |
| Animal Health Laboratory at the University of Guelph, Canada | 1978–2019 | Mb | Cai et al., 2019 [40] |
| Western Canada and Idaho, USA | 2006–2018 | Mb | Jelinski et al., 2020 [41] |
| Alberta, Canada | 2017–2019 | Mb | Andres-Lasheras et al., 2021 [30] |
| Alberta, Canada | 2017 | Mb | Nobrega et al., 2021 [31] |
| Antimicrobial | Registration | Common Use (s) |
|---|---|---|
| Tildipirosin | 2012 | Treatment of BRD |
| Gamithromycin | 2010 | Treatment of BRD |
| Tulathromycin | 2005 | Prevention/treatment of BRD |
| Enrofloxacin | 2004 | Treatment of BRD |
| Danofloxacin | 2002 | Treatment of BRD |
| Tylosin | 1997 | Prevention/treatment of liver abscess |
| Florfenicol | 1996 | Treatment of BRD and footrot |
| Spectinomycin | 1996 | Treatment of BRD |
| Chlortetracycline | 1995 | Prevention of footrot and BRD; prevention/treatment of liver abscess; treatment of enteritis |
| Ceftiofur | 1994 | Treatment of BRD |
| Oxytetracycline | 1994 | Prevention/treatment of liver abscess; prevention/treatment of BRD; prevention of bloat |
| Trimethoprim | 1994 | Treatment of BRD |
| Tilmicosin | 1992 | Prevention/treatment of BRD |
| Geographical Area | Sampling Year (s) | Bacterial Species | Number of ARGs Reported Linked to ICE | Authors |
|---|---|---|---|---|
| USA | NS | Hs | 1 | Mohd-Zain et al., 2004 [84] |
| Nebraska, USA | 2005 | Pm | 12 | Michael et al., 2012 [82] |
| Texas and Nebraska, USA | NS | Mh | Up to 14 | Klima et al., 2014 [35] |
| Pennsylvania, USA | 2007 | Mh | 5 | Eidam et al., 2015 [20] |
| Canada and USA | 2002–2013 | Mh | Up to 12 | Clawson et al., 2016 [86] |
| Alberta, Canada | 2012–2016 | Hs | 2 (including metal-tolerance) | Bhatt et al., 2018 [85] |
| Alberta, Canada | NS | Mh, Pm, HS | Up to 13 | Stanford et al., 2020 [18] |
| Alberta, Canada | 2017–2019 | Mh, Pm, Hs | Up to 7 * | Conrad et al., 2020 [87] |
| Antimicrobial | Gene | Escherichia coli K-12 Numbering | Mycoplasma bovis PG45 Numbering | Effect on MIC Values 1 |
|---|---|---|---|---|
| Enrofloxacin | gyrA | Ser83-Phe | Ser150-Phe | 32-fold increase |
| gyrB | Asp362-Asn | Asp382-Asn | Up to 2-fold increase (slight increase) | |
| parC | Ser80-Ile | Ser91-Ile | 2 to 8-fold increase | |
| Tetracyclines | rrs | A965T | A956T | Resistance to oxytetracycline. Mutation present in both rrs alleles of the 16S rRNA gene: rrs3 and rrs4 |
| A967T | A958T | |||
| Macrolides | rrl | G748A | G788A | Up to 64 and 256-fold increase for TYLT and TIL, respectively. Mutation present in both rrl alleles of the 23S rRNA gene: rrl3 and rrl4 |
| A2059G | A2057G | Up to 32-fold increase for TYLT and TIL. Mutation present in one or both rrl alleles of the 23S rRNA gene: rrl3 and rrl4 | ||
| rplV (protein L22) | Q93H | Q93H | Up to 8 and 16-fold increase for TYLT and TIL, respectively |
| Geographical Area | Sampling Year (s) | Approach | Authors |
|---|---|---|---|
| Alberta, Canada | 2008–2010 | qPCR | Holman et al., 2018 [68] |
| Alberta, Canada | 2007–2010 | qPCR | Holman et al., 2019 [57] |
| Alberta, Canada | 2016 | Metagenomic sequencing | Klima et al., 2019 [69] |
| Alberta, Canada | 2016 | qPCR | Guo et al., 2020 [29] |
| Technique | Chute-Side Potential | Turnaround Time | Advantages 1 | Disadvantages | Reference |
|---|---|---|---|---|---|
| RPA | Yes | <30 min | Simple; High sensitivity and specificity; Stable when PCR inhibitors present; Fast; Efficient; Offers multiplexing; Cost-effective | Sample processing; Update emerging resistance genes/mutations | [87,99] |
| LAMP | Yes | 30–60 min | Simple; High sensitivity and specificity; Stable when PCR inhibitors present; Fast; Some sample types do not need processing; Cost-effective | High number of primers per target; Limited multiplexing; Update emerging resistance genes/mutations | [100,101] |
| Metagenomics–Nanopore Minion | Yes | 6–8 h | Detection of every pathogen present; Detection of ARG; High sensitivity and specificity; Fast; Cost-effective | Sample processing; Detection of non-pathogenic bacteria genes; Enrichment of low copy number genes by PCR; Host DNA sample contamination | [102] |
| MALDI-TOF | No | 30–180 min | Low-cost consumables; Simple; High sensitivity and specificity; Fast; Various samples in a single run; Cost-effective | Up-front high cost; Requires bacterial isolation or sample processing; Update emerging resistance genes/mutations | [103] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrés-Lasheras, S.; Jelinski, M.; Zaheer, R.; McAllister, T.A. Bovine Respiratory Disease: Conventional to Culture-Independent Approaches to Studying Antimicrobial Resistance in North America. Antibiotics 2022, 11, 487. https://doi.org/10.3390/antibiotics11040487
Andrés-Lasheras S, Jelinski M, Zaheer R, McAllister TA. Bovine Respiratory Disease: Conventional to Culture-Independent Approaches to Studying Antimicrobial Resistance in North America. Antibiotics. 2022; 11(4):487. https://doi.org/10.3390/antibiotics11040487
Chicago/Turabian StyleAndrés-Lasheras, Sara, Murray Jelinski, Rahat Zaheer, and Tim A. McAllister. 2022. "Bovine Respiratory Disease: Conventional to Culture-Independent Approaches to Studying Antimicrobial Resistance in North America" Antibiotics 11, no. 4: 487. https://doi.org/10.3390/antibiotics11040487
APA StyleAndrés-Lasheras, S., Jelinski, M., Zaheer, R., & McAllister, T. A. (2022). Bovine Respiratory Disease: Conventional to Culture-Independent Approaches to Studying Antimicrobial Resistance in North America. Antibiotics, 11(4), 487. https://doi.org/10.3390/antibiotics11040487







