A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium
Abstract
1. Introduction
2. Results
2.1. Antimicrobial Resistance
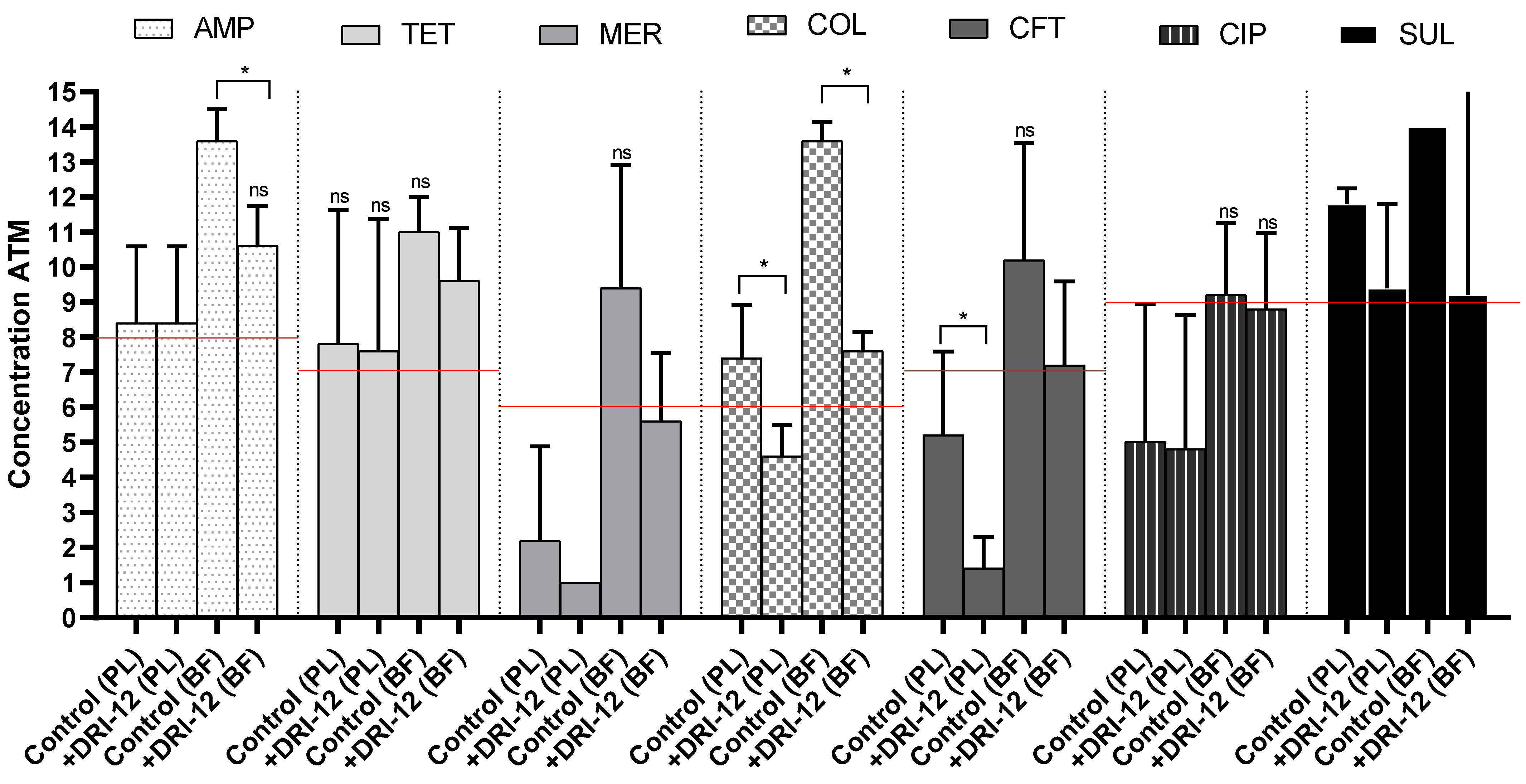
2.2. Copper Complex Synergism in Antimicrobials
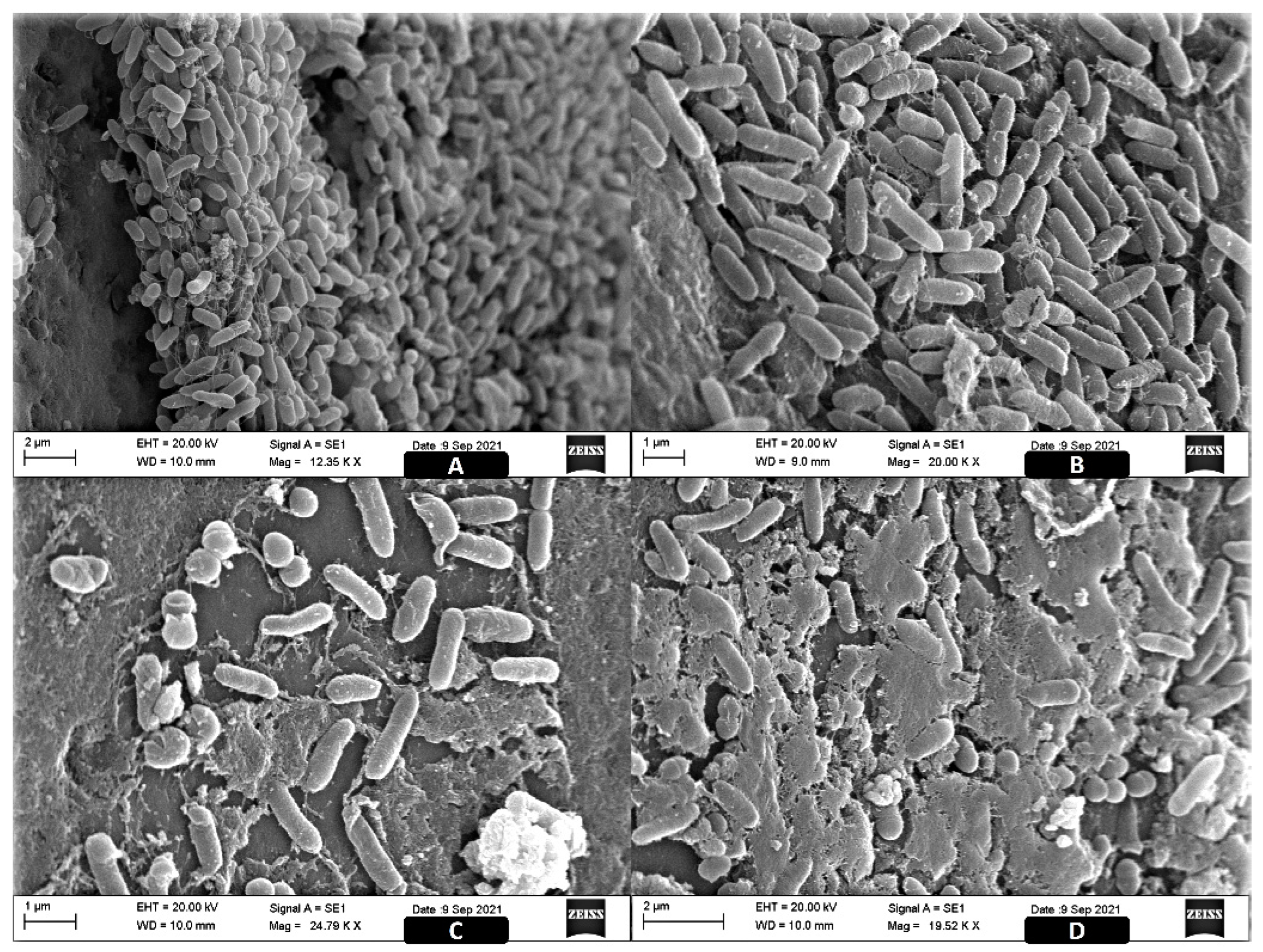
2.3. Image Analysis of the ST Biofilms
3. Discussion
3.1. Antibiotic Susceptibility
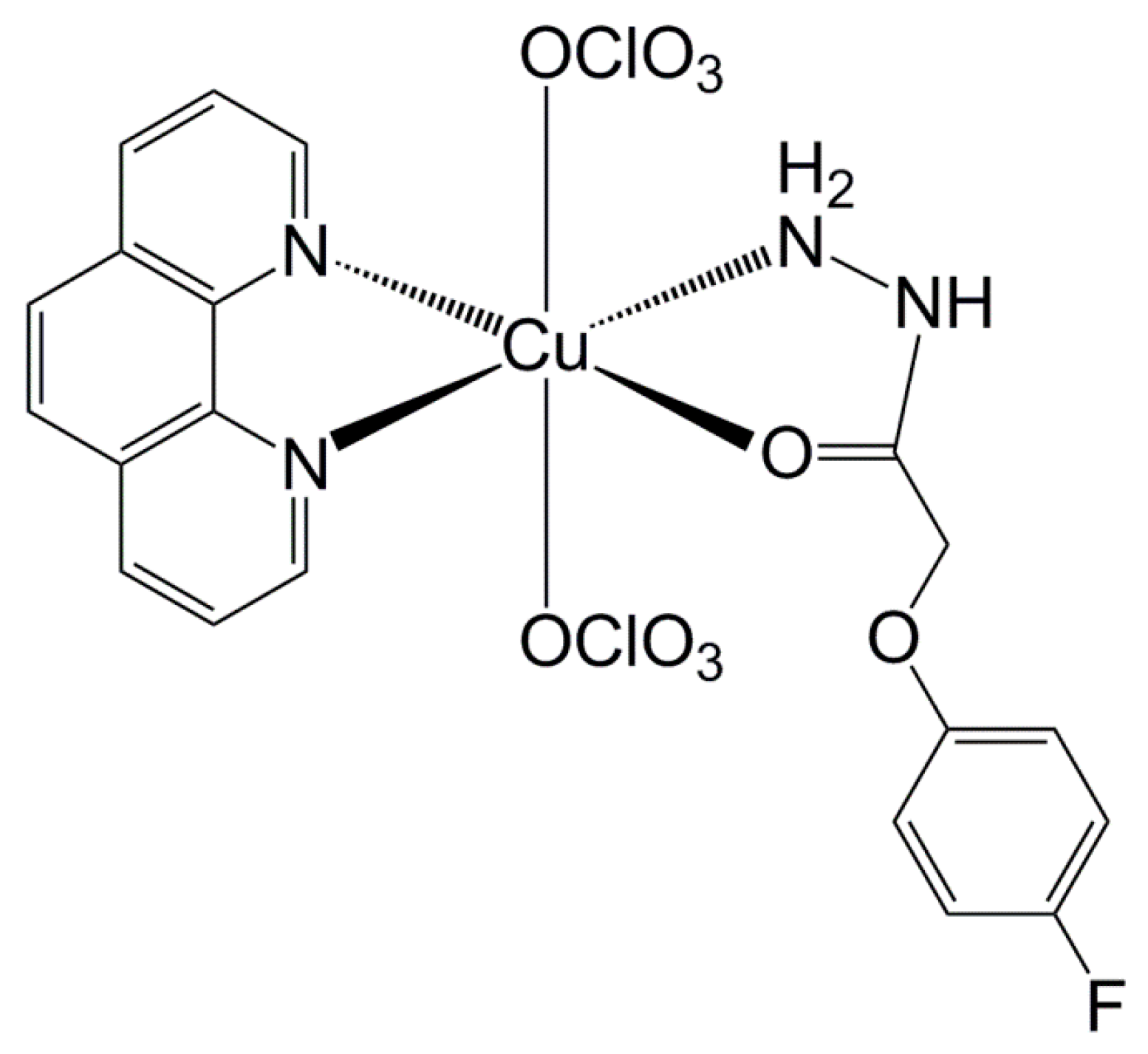
3.2. Copper Complex Effect
4. Materials and Methods
4.1. Origin of Strains and Study Design
4.2. Research of Genes Associated with Antimicrobial Resistance
4.3. Minimum Inhibitory Concentration (MIC)
4.4. The Synergistic Effect of the Copper Complex on the Planktonic and Sessile Forms of ST
4.5. Scanning Electronic Microscopy (SEM)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brasil. Ministério da Agricultura, P. e A. Anuário dos Programas de Controle de Alimentos de Origem Animal do DIPOA. Secretaria de Defesa Agropecuária, Departamento de Inspeção de Produtos de Origem Animal, Coordenação Geral de Programas Especiais 2021, 7. Available online: https://www.gov.br/agricultura/pt-br/assuntos/inspecao/produtos-animal/anuario-dos-programas-de-controle-de-alimentos-de-origem-animal-do-dipoa (accessed on 14 February 2022).
- WHO—World Health Organization. Salmonella (Non-Typhoidal). Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 14 May 2021).
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, e05926. [Google Scholar] [CrossRef]
- USDA—United States Department of Agriculture FSIS Guideline for Controlling Salmonella in Raw Poultry. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/2021-07/FSIS-GD-2021-0005.pdf (accessed on 14 January 2022).
- Brasil. Ministério da Saúde Surtos de Doenças Transmitidas por Alimentos no Brasil 2022. Available online: https://www.gov.br/saude/pt-br/assuntos/saude-de-a-a-z/d/doencas-de-transmissao-hidrica-e-alimentar-dtha/arquivos/doencas-de-transmissao-hidrica-e-alimentar-dtha/apresentacao-surtos-dtha-2022.pdf/view (accessed on 14 February 2022).
- Centers for Disease Control and Prevention. Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Contact with Dairy Calves. Available online: https://www.cdc.gov/salmonella/heidelberg-11-16/index.html (accessed on 14 January 2022).
- Grant, A.; Hashem, F.; Parveen, S. Salmonella and Campylobacter: Antimicrobial resistance and bacteriophage control in poultry. Food Microbiol. 2016, 53, 104–109. [Google Scholar] [CrossRef] [PubMed]
- dos Reis, R.O.; Souza, M.N.; Cecconi, M.C.P.; Timm, L.; Ikuta, N.; Simon, D.; Wolf, J.M.; Lunge, V.R. Increasing prevalence and dissemination of invasive nontyphoidal Salmonella serotype Typhimurium with multidrug resistance in hospitalized patients from southern Brazil. Braz. J. Infect. Dis. 2018, 22, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Helke, K.; McCrackin, M.; Galloway, A.M.; Poole, A.Z.; Salgado, C.D.; Marriott, B.P. Effects of antimicrobial use in agricultural animals on drug-resistant foodborne salmonellosis in humans: A systematic literature review. Crit. Rev. Food Sci. Nutr. 2016, 57, 472–488. [Google Scholar] [CrossRef] [PubMed]
- Wójcicki, M.; Świder, O.; Daniluk, K.; Średnicka, P.; Akimowicz, M.; Roszko, M.; Sokołowska, B.; Juszczuk-Kubiak, E. Transcriptional Regulation of the Multiple Resistance Mechanisms in Salmonella—A Review. Pathogens 2021, 10, 801. [Google Scholar] [CrossRef]
- Eran, Z.; Akçelik, M.; Yazıcı, B.C.; Özcengiz, G.; Akçelik, N. Regulation of biofilm formation by marT in Salmonella Typhimurium. Mol. Biol. Rep. 2020, 47, 5041–5050. [Google Scholar] [CrossRef]
- Frei, A.; Zuegg, J.; Elliott, A.G.; Baker, M.V.; Braese, S.; Brown, C.; Chen, F.; Dowson, C.G.; Dujardin, G.; Jung, N.; et al. Metal complexes as a promising source for new antibiotics. Chem. Sci. 2020, 11, 2627–2639. [Google Scholar] [CrossRef]
- Centola, G.; Xue, F.; Wilks, A. Metallotherapeutics development in the age of iron-clad bacteria. Metallomics 2020, 12, 1863–1877. [Google Scholar] [CrossRef]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef]
- Zalevskaya, O.A.; Gur’Eva, Y.A. Recent Studies on the Antimicrobial Activity of Copper Complexes. Russ. Russ. J. Coord. Chem. 2021, 47, 861–880. [Google Scholar] [CrossRef]
- Mendes, I.C.; Moreira, J.P.; Mangrich, A.S.; Balena, S.P.; Rodrigues, B.L.; Beraldo, H. Coordination to copper(II) strongly enhances the in vitro antimicrobial activity of pyridine-derived N(4)-tolyl thiosemicarbazones. Polyhedron 2007, 26, 3263–3270. [Google Scholar] [CrossRef]
- Iakovidis, I.; Delimaris, I.; Piperakis, S.M. Copper and Its Complexes in Medicine: A Biochemical Approach. Mol. Biol. Int. 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J. Síntese, Caracterização e Estudos Biológicos de Complexos de Cobre(II) Contendo Derivados dos Ácidos Picolínico e Nicotínico. Ph.D. Thesis, Universidade Federal de Uberlândia, Uberlândia, Brasil, 2019. [Google Scholar]
- Paixão, D.A.; de Oliveira, B.C.; Almeida, J.D.C.; Sousa, L.; Lopes, C.D.; Carneiro, Z.A.; Tezuka, D.; Clavijo, J.C.T.; Ellena, J.; Polloni, L.; et al. Crystal structure, anti-Trypanosoma cruzi and cytotoxic activities of Cu(II) complexes bearing β-diketone and α-diimine ligands. Inorg. Chim. Acta 2019, 499, 119164. [Google Scholar] [CrossRef]
- Paixão, D.A.; Marzano, I.M.; Jaimes, E.H.; Pivatto, M.; Campos, D.L.; Pavan, F.; Deflon, V.M.; Maia, P.I.D.S.; Ferreira, A.M.D.C.; Uehara, I.A.; et al. Novel copper(II) complexes with hydrazides and heterocyclic bases: Synthesis, structure and biological studies. J. Inorg. Biochem. 2017, 172, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Machado, P.H.A.; Paixão, D.A.; Lino, R.C.; de Souza, T.R.; Bontempo, N.J.D.S.; Sousa, L.M.; Azevedo, F.V.P.D.V.; Orsolin, P.C.; Lima, P.M.A.P.; Martins, I.C.; et al. A selective CuII complex with 4-fluorophenoxyacetic acid hydrazide and phenanthroline displays DNA-cleaving and pro-apoptotic properties in cancer cells. Sci. Rep. 2021, 11, 24450. [Google Scholar] [CrossRef]
- Bontempo, N.J.S.; Paixão, D.A.; Lima, P.M.A.P.; Orsolin, P.C.; Lino, R.C.; Souza, T.R.; Sousa, L.M.; Machado, P.H.A.; Martins, I.C.; Goulart, L.R.R.J.; et al. A copper(II) complex containing 4-fluorophenoxyacetic acid hydrazide and 1,10-phenanthroline modulates carcinogenic and mutagenic effects of Doxorubicin in somatic cells of Drosophila melanogaster. Sci. Rep. 2022, Submitted. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Lewis Ii, J.S.; Bobenchik, A.M.; Campeau, S.; Cullen, S.K.; Galas, M.F.; Gold, H.; Humphries, R.M.; Kirn, T.J.; Limbago, B.; et al. Performance Standards for Antimicrobial Susceptibility. Clinical and Laboratory Standards Institute (CLSI) supplement M100. CLSI 2021. Available online: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED31:2021&xormat=SPDF&src=BB (accessed on 14 January 2022).
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Antunes, P.; Machado, J.; Sousa, J.C.; Peixe, L. Dissemination of Sulfonamide Resistance Genes (sul1, sul2, and sul3) in Portuguese Salmonella enterica Strains and Relation with Integrons. Antimicrob. Agents Chemother. 2005, 49, 836–839. [Google Scholar] [CrossRef]
- Enne, I.V.; Livermore, D.M.; Stephens, P.; MC Hall, L. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 2001, 357, 1325–1328. [Google Scholar] [CrossRef]
- Perreten, V.; Boerlin, P. A New Sulfonamide Resistance Gene (sul3) in Escherichia coli Is Widespread in the Pig Population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Amajoud, N.; Bouchrif, B.; El Maadoudi, M.; Senhaji, N.S.; Karraouan, B.; El Harsal, A.; El Abrini, J. Prevalence, serotype distribution, and antimicrobial resistance of Salmonella isolated from food products in Morocco. J. Infect. Dev. Ctries. 2017, 11, 136–142. [Google Scholar] [CrossRef]
- Khallaf, M.; Ameur, N.; Terta, M.; Lakranbi, M.; Senouci, S.; Ennaji, M.M. Prevalence and antibiotic-resistance of Salmonella isolated from chicken meat marketed in Rabat, Morocco. Int. J. Innov. Appl. Stud. 2014, 6, 1123–1128. [Google Scholar]
- Proroga, Y.T.R.; Capuano, F.; Carullo, M.R.; La Tela, I.; Capparelli, R.; Barco, L.; Pasquale, V. Occurrence and antimicrobial resistance of Salmonella strains from food of animal origin in southern Italy. Folia Microbiol. 2015, 61, 21–27. [Google Scholar] [CrossRef]
- Mendonça, E.P. Características de Virulência, Resistência e Diversidade Genética de Sorovares de Salmonella com Impacto Na saúde Pública, Isolados de Frangos de Corte no Brasil. Ph.D. Thesis, Federal University of Uberlandia, Uberlândia, Brazil, 2016. [Google Scholar]
- Carattoli, A. Animal reservoirs for extended spectrum β-lactamase producers. Clin. Microbiol. Infect. 2008, 14, 117–123. [Google Scholar] [CrossRef]
- Pandini, J.A.; Pinto, F.G.D.S.; Muller, J.M.; Weber, L.D.; De Moura, A.C. Ocorrência e perfil de resistencia antimicrobiana de sorotipos de Salmonella spp. isolados de aviários do Paraná, Brasil. Arq. Inst. Biol. 2015, 82, 1–6. [Google Scholar] [CrossRef]
- Melo, R.T.; Galvão, N.N.; Guidotti-Takeuchi, M.; Peres, P.A.B.M.; Fonseca, B.B.; Profeta, R.; Azevedo, V.A.C.; Monteiro, G.P.; Brenig, B.; Rossi, D.A. Molecular Characterization and Survive Abilities of Salmonella Heidelberg Strains of Poultry Origin in Brazil. Front. Microbiol. 2021, 12, 1461. [Google Scholar] [CrossRef]
- Vinueza-Burgos, C.; Cevallos, M.; Ron, L.; Bertrand, S.; De Zutter, L. Prevalence and Diversity of Salmonella Serotypes in Ecuadorian Broilers at Slaughter Age. PLoS ONE 2016, 11, e0159567. [Google Scholar] [CrossRef]
- Rossi, F.; Girardello, R.; Cury, A.P.; Di Gioia, T.S.R.; de Almeida, J.N.; Duarte, A.J.D.S. Emergence of colistin resistance in the largest university hospital complex of São Paulo, Brazil, over five years. Braz. J. Infect. Dis. 2017, 21, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Brasil Ministério da Agricultura. Instrução Normativa No 45, De 22 De Novembro De 2016; Ministério da Agricultura: Brasília, Brazil, 2016; p. 4.
- Lin, D.; Chen, K.; Chan, E.W.-C.; Chen, S. Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 2015, 5, 14754. [Google Scholar] [CrossRef] [PubMed]
- Brasil Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa No 26, de 09 de Julho de 2009. Aprova o Regulamento Técnico para a Fabricação, o Controle de Qualidade, a Comercialização e o Emprego de Produtos Antimicrobianos de Uso Veterinário; Diário Oficial Da União; Ministério da Agricultura: Brasília, Brazil, 2009.
- Pribul, B.R.; Festivo, M.L.; Souza, M.; Rodrigues, D.D.P. Characterization of quinolone resistance in Salmonella spp. isolates from food products and human samples in Brazil. Braz. J. Microbiol. 2016, 47, 196–201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gargano, V.; Sciortino, S.; Gambino, D.; Costa, A.; Agozzino, V.; Reale, S.; Alduina, R.; Vicari, D. Antibiotic Susceptibility Profile and Tetracycline Resistance Genes Detection in Salmonella spp. Strains Isolated from Animals and Food. Antibiotics 2021, 10, 809. [Google Scholar] [CrossRef]
- Delgado-Suárez, E.J.; Palós-Guitérrez, T.; Ruíz-López, F.A.; Pérez, C.F.H.; Ballesteros-Nova, N.E.; Soberanis-Ramos, O.; Méndez-Medina, R.D.; Allard, M.W.; Rubio-Lozano, M.S. Genomic surveillance of antimicrobial resistance shows cattle and poultry are a moderate source of multi-drug resistant non-typhoidal Salmonella in Mexico. PLoS ONE 2021, 16, e0243681. [Google Scholar] [CrossRef]
- Pavelquesi, S.L.S.; Ferreira, A.C.A.D.O.; Rodrigues, A.R.M.; Silva, C.M.D.S.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef]
- Warburton, P.J.; Amodeo, N.; Roberts, A. Mosaic tetracycline resistance genes encoding ribosomal protection proteins. J. Antimicrob. Chemother. 2016, 71, 3333–3339. [Google Scholar] [CrossRef]
- McMillan, E.A.; Jackson, C.R.; Frye, J.G. Transferable Plasmids of Salmonella enterica Associated with Antibiotic Resistance Genes. Front. Microbiol. 2020, 11, 562181. [Google Scholar] [CrossRef]
- Dang, H.; Ren, J.; Song, L.; Sun, S.; An, L. Diverse Tetracycline Resistant Bacteria and Resistance Genes from Coastal Waters of Jiaozhou Bay. Microb. Ecol. 2007, 55, 237–246. [Google Scholar] [CrossRef]
- Sheykhsaran, E.; Baghi, H.B.; Soroush, M.H.; Ghotaslou, R. An overview of tetracyclines and related resistance mechanisms. Rev. Med. Microbiol. 2019, 30, 69–75. [Google Scholar] [CrossRef]
- Liao, Q.; Rong, H.; Zhao, M.; Luo, H.; Chu, Z.; Wang, R. Interaction between tetracycline and microorganisms during wastewater treatment: A review. Sci. Total Environ. 2020, 757, 143981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, M.; Sui, Q.; Tong, J.; Jiang, C.; Lu, X.; Zhang, Y.; Wei, Y. Impacts of addition of natural zeolite or a nitrification inhibitor on antibiotic resistance genes during sludge composting. Water Res. 2016, 91, 339–349. [Google Scholar] [CrossRef]
- Stefani, L.M.; Das Neves, G.B.; Brisola, M.C.; Crecencio, R.B.; Pick, E.C.; Araujo, D.N. Salmonella Heidelberg resistant to ceftiofur and disinfectants routinely used in poultry. Semin. Ciênc. Agrár. 2018, 39, 1029–1036. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Carson, C.; Li, X.-Z.; Agunos, A.; Loest, D.; Chapman, B.; Finley, R.; Mehrotra, M.; Sherk, L.M.; Gaumond, R.; Irwin, R. Ceftiofur-resistant Salmonella enterica serovar Heidelberg of poultry origin—A risk profile using the Codex framework. Epidemiol. Infect. 2019, 147, e296. [Google Scholar] [CrossRef]
- Retamal, P.; Fresno, M.; Dougnac, C.; Gutierrez, S.; Gornall, V.; Vidal, R.; Vernal, R.; Pujol, M.; Barreto, M.; González-Acuña, D.; et al. Genetic and phenotypic evidence of the Salmonella enterica serotype Enteritidis human-animal interface in Chile. Front. Microbiol. 2015, 6, 464. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Sukhnanand, S.S.; Warnick, L.D.; Su, W.-L.; McGann, P.; McDonough, P.; Wiedmann, M. Ceftiofur-Resistant Salmonella Strains Isolated from Dairy Farms Represent Multiple Widely Distributed Subtypes That Evolved by Independent Horizontal Gene Transfer. Antimicrob. Agents Chemother. 2005, 49, 4061–4067. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.-M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur Resistance in Salmonella enterica Serovar Heidelberg from Chicken Meat and Humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Perin, A.P.; Martins, B.T.F.; Barreiros, M.A.B.; Yamatogi, R.S.; Nero, L.A.; Bersot, L.D.S. Occurrence, quantification, pulse types, and antimicrobial susceptibility of Salmonella sp. isolated from chicken meat in the state of Paraná, Brazil. Braz. J. Microbiol. 2019, 51, 335–345. [Google Scholar] [CrossRef]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain through National Antimicrobial Resistance Monitoring System between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef] [PubMed]
- Tran-Dien, A.; Le Hello, S.; Bouchier, C.; Weill, F.-X. Early transmissible ampicillin resistance in zoonotic Salmonella enterica serotype Typhimurium in the late 1950s: A retrospective, whole-genome sequencing study. Lancet Infect. Dis. 2017, 18, 207–214. [Google Scholar] [CrossRef]
- FDA. Antimicrobials Sold or Distributed for Use in Food-Producing Animals; Center for Veterinary Medicine: Silver Spring, MD, USA, 2020; p. 49.
- European Medicines Agency. European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption Sales of Veterinary Antimicrobial Agents in 26 EU/EEA Countries in 2013’. (EMA/387934/2015); European Medicines Agency: Amsterdam, The Netherlands, 2015.
- Bae, D.H.; Dessie, H.K.; Baek, H.J.; Kim, S.G.; Lee, H.S.; Lee, Y.J. Prevalence and Characteristics of Salmonella spp. Isolated from Poultry Slaughterhouses in Korea. J. Vet. Med. Sci. 2013, 75, 1193–1200. [Google Scholar] [CrossRef]
- Elumalai, S.; Muthu, G.; Selvam, R.E.M.; Ramesh, S. Detection of TEM-, SHV- and CTX-M-type β-lactamase production among clinical isolates of Salmonella species. J. Med. Microbiol. 2014, 63, 962–967. [Google Scholar] [CrossRef]
- Wong, M.H.-Y.; Liu, L.; Yan, M.; Chan, E.W.-C.; Chen, S. Dissemination of IncI2 Plasmids That Harbor the blaCTX-MElement among Clinical Salmonella Isolates. Antimicrob. Agents Chemother. 2015, 59, 5026–5028. [Google Scholar] [CrossRef][Green Version]
- Hussain, M.A.; Wang, W.; Sun, C.; Gu, L.; Liu, Z.; Yu, T.; Ahmad, Y.; Jiang, Z.; Hou, J. Molecular Characterization of Pathogenic Salmonella Spp. from Raw Beef In Karachi, Pakistan. Antibiotics 2020, 9, 73. [Google Scholar] [CrossRef]
- Al-Gallas, N.; Belghouthi, K.; Barratt, N.A.; Ghedira, K.; Hotzel, H.; Tomaso, H.; El-Adawy, H.; Neubauer, H.; Laouini, D.; Zarrouk, S.; et al. Identification and characterization of multidrug-resistant ESBL-producing Salmonella enterica serovars Kentucky and Typhimurium isolated in Tunisia CTX-M-61/TEM-34, a novel cefotaxime-hydrolyzing β-lactamase of Salmonella. J. Appl. Microbiol. 2021, 132, 279–289. [Google Scholar] [CrossRef]
- Estevez, M.B.; Casaux, M.L.; Fraga, M.; Faccio, R.; Alborés, S. Biogenic Silver Nanoparticles as a Strategy in the Fight against Multi-Resistant Salmonella enterica Isolated from Dairy Calves. Front. Bioeng. Biotechnol. 2021, 9, 314. [Google Scholar] [CrossRef]
- Agência Nacional de Vigilância santária—Anvisa Monitoramento da Prevalência e do Perfil de Suscetibilidade aos Antimicrobianos em Enterococos e Salmonelas Isolados de Carcaças de Frango Congeladas Comercializadas no Brasil. 2012, 188. Available online: https://www.gov.br/anvisa/pt-br/centraisdeconteudo/publicacoes/alimentos/relatorio-prebaf-programa-nacional-de-monitoramento-da-prevalencia-e-da-resistencia-bacteriana-em-frango.pdf (accessed on 14 January 2022).
- Montero, D.A.; Arellano, C.; Pardo, M.; Vera, R.; Gálvez, R.; Cifuentes, M.; Berasain, M.A.; Gómez, M.; Ramírez, C.; Vidal, R.M. Antimicrobial properties of a novel copper-based composite coating with potential for use in healthcare facilities. Antimicrob. Resist. Infect. Control 2019, 8, 3. [Google Scholar] [CrossRef]
- Smith, J.; Tran, N.; Song, T.; Liang, D.; Qian, M. Robust bulk micro-nano hierarchical copper structures possessing exceptional bactericidal efficacy. Biomaterials 2021, 280, 121271. [Google Scholar] [CrossRef]
- Monstein, H.-J.; Östholm-Balkhed, Å.; Nilsson, M.V.; Dornbusch, K.; Nilsson, L.E. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX-M genes in Enterobacteriaceae. APMIS 2007, 115, 1400–1408. [Google Scholar] [CrossRef]
- Ude, Z.; Kavanagh, K.; Twamley, B.; Pour, M.; Gathergood, N.; Kellett, A.; Marmion, C.J. A new class of prophylactic metallo-antibiotic possessing potent anti-cancer and anti-microbial properties. Dalton Trans. 2019, 48, 8578–8593. [Google Scholar] [CrossRef]
- Reyes-Jara, A.; Cordero, N.; Aguirre, J.S.; Troncoso, M.; Figueroa, G. Antibacterial Effect of Copper on Microorganisms Isolated from Bovine Mastitis. Front. Microbiol. 2016, 7, 626. [Google Scholar] [CrossRef] [PubMed]
- Kareem, P.A.; Salh, K.K.; Ali, F.A. ZnO, TiO2 and Ag nanoparticles impact against some species of pathogenic bacteria and yeast. Cell. Mol. Biol. 2021, 67, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Randall, C.P.; Gupta, A.; Jackson, N.; Busse, D.; O’Neill, A.J. Silver resistance in Gram-negative bacteria: A dissection of endogenous and exogenous mechanisms. J. Antimicrob. Chemother. 2015, 70, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Duffy, L.L.; Osmond-McLeod, M.J.; Judy, J.; King, T. Investigation into the antibacterial activity of silver, zinc oxide and copper oxide nanoparticles against poultry-relevant isolates of Salmonella and Campylobacter. Food Control 2018, 92, 293–300. [Google Scholar] [CrossRef]
- Gudkov, S.V.; Burmistrov, D.E.; Serov, D.A.; Rebezov, M.B.; Semenova, A.A.; Lisitsyn, A.B. A Mini Review of Antibacterial Properties of ZnO Nanoparticles. Front. Phys. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Moubareck, C.A. Polymyxins and Bacterial Membranes: A Review of Antibacterial Activity and Mechanisms of Resistance. Membranes 2020, 10, 181. [Google Scholar] [CrossRef]
- Gales, A.; Jones, R.; Sader, H.; Gales, A. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: Report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin. Microbiol. Infect. 2006, 12, 315–321. [Google Scholar] [CrossRef]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.R. Current Update on Intrinsic and Acquired Colistin Resistance Mechanisms in Bacteria. Front. Med. 2021, 8, 677720. [Google Scholar] [CrossRef]
- Zaengle-Barone, J.M.; Jackson, A.C.; Besse, D.M.; Becken, B.; Arshad, M.; Seed, P.C.; Franz, K.J. Copper Influences the Antibacterial Outcomes of a β-Lactamase-Activated Prochelator against Drug-Resistant Bacteria. ACS Infect. Dis. 2018, 4, 1019–1029. [Google Scholar] [CrossRef]
- Peechakara, B.V.; Basit, H.G.M. Ampicillin; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Garza-Cervantes, J.A.; Meza-Bustillos, J.F.; Resendiz-Hernández, H.; Suárez-Cantú, I.A.; Ortega-Rivera, O.; Salinas, E.; Gonzalez, C.E.E.; Morones-Ramirez, J.R. Re-sensitizing Ampicillin and Kanamycin-Resistant E. coli and S. aureus Using Synergistic Metal Micronutrients-Antibiotic Combinations. Front. Bioeng. Biotechnol. 2020, 8, 612. [Google Scholar] [CrossRef]
- Garza-Cervantes, J.A.; Chávez-Reyes, A.; Castillo, E.C.; García-Rivas, G.; Ortega-Rivera, O.A.; Salinas, E.; Ortiz-Martínez, M.; Gómez-Flores, S.L.; Peña-Martínez, J.A.; Pepi-Molina, A.; et al. Synergistic Antimicrobial Effects of Silver/Transition-metal Combinatorial Treatments. Sci. Rep. 2017, 7, 903. [Google Scholar] [CrossRef] [PubMed]
- Rawls, S.M. Antibiotics, β-Lactam. In Encyclopedia of the Neurological Sciences; Elsevier: Amsterdam, The Netherlands, 2014; pp. 207–209. [Google Scholar]
- AbouElleef, E.M.; Mahrouka, M.M.; Salem, S.E. A Physical-Chemical Study of the Interference of Ceftriaxone Antibiotic with Copper Chloride Salt. Bioinorg. Chem. Appl. 2021, 2021, 4018843. [Google Scholar] [CrossRef] [PubMed]
- Djoko, K.Y.; Achard, M.E.S.; Phan, M.-D.; Lo, A.W.; Miraula, M.; Prombhul, S.; Hancock, S.J.; Peters, K.M.; Sidjabat, H.E.; Harris, P.N.; et al. Copper Ions and Coordination Complexes as Novel Carbapenem Adjuvants. Antimicrob. Agents Chemother. 2018, 62, e02280-17. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, R.C.A.; Rajendran, M.; Nagaiah, H.P. Re-Potentiation of β-Lactam Antibiotic by Synergistic Combination with Biogenic Copper Oxide Nanocubes against Biofilm Forming Multidrug-Resistant Bacteria. Molecules 2019, 24, 3055. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Shu, Q.; Xu, C.; Zheng, Q.; Guo, Z.; Wang, C.; Hao, Z.; Liu, X.; Wang, G.; et al. Copper Clusters: An Effective Antibacterial for Eradicating Multidrug-Resistant Bacterial Infection In Vitro and In Vivo. Adv. Funct. Mater. 2021, 31, 2008720. [Google Scholar] [CrossRef]
- Collinson, S.K.; Doig, P.C.; Doran, J.L.; Clouthier, S.; Trust, T.J.; Kay, W.W. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J. Bacteriol. 1993, 175, 12–18. [Google Scholar] [CrossRef]
- Guo, X.; Chen, J.; Beuchat, L.R.; Brackett, R.E. PCR Detection of Salmonella enterica Serotype Montevideo in and on Raw Tomatoes Using Primers Derived from hilA. Appl. Environ. Microbiol. 2000, 66, 5248–5252. [Google Scholar] [CrossRef]
- Oliveira, S.; Santos, L.; Schuch, D.; Silva, A.; Salle, C.; Canal, C. Detection and identification of salmonellas from poultry-related samples by PCR. Vet. Microbiol. 2002, 87, 25–35. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Humphries, A.D.; Weening, E.H.; de Zoete, M.R.; Winter, S.; Papaconstantinopoulou, A.; Dougan, G.; Bäumler, A.J. Molecular and Phenotypic Analysis of the CS54 Island of Salmonella enterica Serotype Typhimurium: Identification of Intestinal Colonization and Persistence Determinants. Infect. Immun. 2003, 71, 629–640. [Google Scholar] [CrossRef]
- Prager, R.; Rabsch, W.; Streckel, W.; Voigt, W.; Tietze, E.; Tschäpe, H. Molecular Properties of Salmonella enterica Serotype Paratyphi B Distinguish between Its Systemic and Its Enteric Pathovars. J. Clin. Microbiol. 2003, 41, 4270–4278. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Weill, F.-X.; Poirel, L.; Fabre, L.; Soussy, C.-J.; Nordmann, P. Prevalence of qnr genes in Salmonella in France. J. Antimicrob. Chemother. 2007, 59, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Heuzenroeder, M.W.C.J.; Murray, R.M.; Dalcin, M.B. Molecular Basis of Benign Colonisation of Salmonella Sofia in Chickens. A Report for the Rural Industries Research and Development Corporation; Rural Industries Research and Development Corporation: Canberra, Australia, 2001; Projeto no. 106.
- Kudirkienė, E.; Cohn, M.T.; Stabler, R.; Strong, P.C.R.; Šernienė, L.; Wren, B.W.; Nielsen, E.M.; Malakauskas, M.; Brøndsted, L. Phenotypic and Genotypic Characterizations of Campylobacter jejuni Isolated from the Broiler Meat Production Process. Curr. Microbiol. 2012, 65, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Brown, H.L.; Reuter, M.; Salt, L.J.; Cross, K.L.; Betts, R.P.; van Vliet, A.H.M. Chicken Juice Enhances Surface Attachment and Biofilm Formation of Campylobacter jejuni. Appl. Environ. Microbiol. 2014, 80, 7053–7060. [Google Scholar] [CrossRef]
| Concentrations | AMP | TET | MER | COL | CFT | CIP | SUL | DRI-12 |
|---|---|---|---|---|---|---|---|---|
| A | - | 24 | 5 | 4 | 1 | 16 | - | - |
| B | - | - | - | - | - | - | - | - |
| C | - | 1 | - | 1 | 1 | 4 | - | - |
| D | - | - | 1 | - | 4 | 4 | - | - |
| E | 1 | - | 2 | 3 | 8 | 1 | - | - |
| F | 14 | 1 | 18 | 22 | 19 | 11 | - | 1 |
| G | 6 | 2 | 17 | 10 | 8 | 4 | - | 2 |
| H | 1 | 1 | - | - | - | 2 | - | 17 |
| I | 2 | 12 | - | - | - | - | 1 | 21 |
| J | 19 | 2 | - | 3 | 2 | 1 | 42 | 2 |
| R (%) | 22 (51.2) | 17 (39.5) | 35 (81.4) | 35 (81.4) | 10 (23.3) | 1 (2.3) | 43 (100) | - |
| R + I (%) | 28 (65.1) | 18 (41.8) | 37 (86) | 38 (88.4) | 29 (67.4) | 18 (41.8) | 43 (100) | - |
| S | 15 (34.8) | 25 (58.1) | 6 (13.9) | 5 (11.6) | 14 (32.5) | 25 (58.1) | 0 (0) | - |
| MIC50 | 32 mg/L | <0.5 mg/L | 4 mg/L | 4 mg/L | 2 mg/L | 0.0312 mg/L | >2048 mg/L | 62.68 mg/L |
| MIC90 | >64 mg/L | 64 mg/L | 8 mg/L | 8 mg/L | 4 mg/L | 0.25 mg/L | >2048 mg/L | 62.68 mg/L |
| Gene | Primers Sequence (5′ → 3′) | Amplicon (pb) | Function | Amplification | Reference |
|---|---|---|---|---|---|
| blaTEM | CAGCGGTAAGATCCTTGAGA ACTCCCCGTCGTGTAGATAA | 643 | β-lactam resistance | 30× (94 °C, 45 s/50 °C, 45 s/72 °C, 90 s) | [96] |
| blaSHV | GGCCGCGTAGGCATGATAGA CCCGGCGATTTGCTGATTTC | 714 | β-lactam resistance | 30× (94 °C, 45 s/56 °C, 45 s/72 °C, 90 s) | [96] |
| blaCTX-M | TGGGTRAARTARGTSACCAGAAYCAGCGG CCCCGCTTATAGAGCAACAA | 593 | β-lactam resistance | 30× (94 °C, 45 s/58 °C, 60 s/72 °C, 90 s) | [72] |
| blaCMY-2 | TGGCCGTTGCCGTTATCTAC CCCGTTTTATGCACCCATGA | 870 | β-lactam resistance | 30× (94 °C, 45 s/59 °C, 53 s/72 °C, 90 s) | [96] |
| qnrA | AGAGGATTTCTCACGCCAGG TGCCAGGCACAGATCTTGAC | 580 | Fluoroquinolone resistance | 35× (95 °C, 60 s/54 °C, 60 s/72 °C, 90 s) | [97] |
| qnrS | GCAAGTTCATTGAACAGGGT TCTAAACCGTCGAGTTCGGCG | 428 | Fluoroquinolone resistance | 35× (95 °C, 60 s/54 °C, 60 s/72 °C, 90 s) | [97] |
| Carbapenamases | Β-Lactamases | AmpCs | MCR | |
|---|---|---|---|---|
| GES | Group CTX-M-1 | TEM 164C | ACC | MCR-1 |
| GIM | Subgroup CTX-M-1 | TEM 164H | ACT/MIR | MCR-2 |
| IMP | Group CTX-M-2 | TEM 164S | CMY I/MOX | |
| KPC | Subgroup CTX-M-3 | TEM 238S | CMY II | |
| NDM | Group CTX-M-8 | SHVwt | DHA | |
| OXA-23 | Group CTX-M-9 | SHV 238A | FOX | |
| OXA-24 | Subgroup CTX-M-15 | SHV 238S | ||
| OXA-48 | Group CTX-M-25 | SHV240K | ||
| OXA-58 | Group CTX-M-32 | BEL | ||
| VIM | TEM wt | GES | ||
| SPM | TEM 404K | PER | ||
| VEB | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteiro, G.P.; Melo, R.T.d.; Guidotti-Takeuchi, M.; Dumont, C.F.; Ribeiro, R.A.C.; Guerra, W.; Ramos, L.M.S.; Paixão, D.A.; Santos, F.A.L.d.; Rodrigues, D.d.P.; et al. A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium. Antibiotics 2022, 11, 388. https://doi.org/10.3390/antibiotics11030388
Monteiro GP, Melo RTd, Guidotti-Takeuchi M, Dumont CF, Ribeiro RAC, Guerra W, Ramos LMS, Paixão DA, Santos FALd, Rodrigues DdP, et al. A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium. Antibiotics. 2022; 11(3):388. https://doi.org/10.3390/antibiotics11030388
Chicago/Turabian StyleMonteiro, Guilherme Paz, Roberta Torres de Melo, Micaela Guidotti-Takeuchi, Carolyne Ferreira Dumont, Rosanne Aparecida Capanema Ribeiro, Wendell Guerra, Luana Munique Sousa Ramos, Drielly Aparecida Paixão, Fernanda Aparecida Longato dos Santos, Dália dos Prazeres Rodrigues, and et al. 2022. "A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium" Antibiotics 11, no. 3: 388. https://doi.org/10.3390/antibiotics11030388
APA StyleMonteiro, G. P., Melo, R. T. d., Guidotti-Takeuchi, M., Dumont, C. F., Ribeiro, R. A. C., Guerra, W., Ramos, L. M. S., Paixão, D. A., Santos, F. A. L. d., Rodrigues, D. d. P., Boleij, P., Hoepers, P. G., & Rossi, D. A. (2022). A Ternary Copper (II) Complex with 4-Fluorophenoxyacetic Acid Hydrazide in Combination with Antibiotics Exhibits Positive Synergistic Effect against Salmonella Typhimurium. Antibiotics, 11(3), 388. https://doi.org/10.3390/antibiotics11030388






