Prevalence and Phenotypic Antimicrobial Resistance among ESKAPE Bacteria and Enterobacterales Strains in Wild Birds
Abstract
1. Introduction
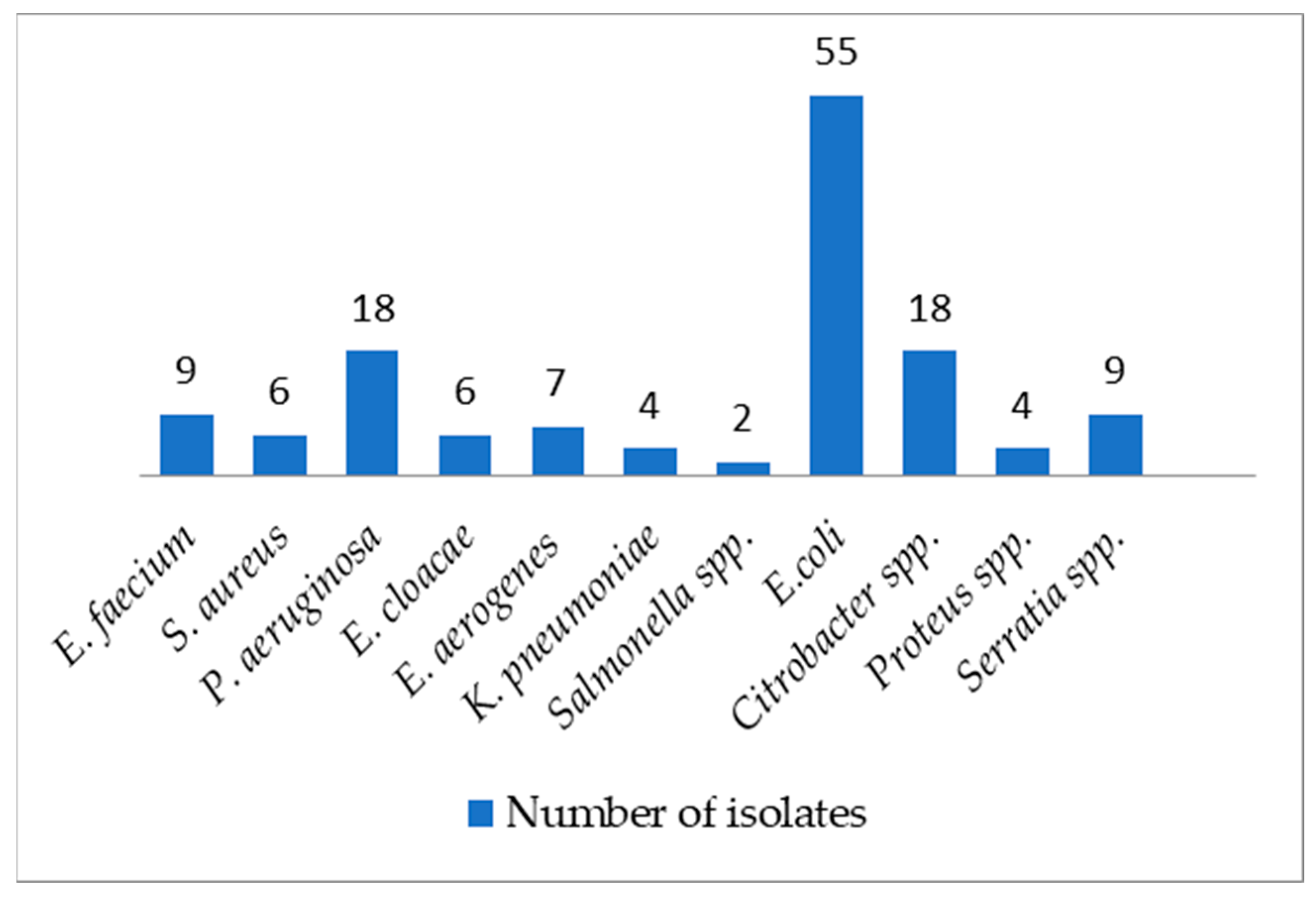
2. Results
3. Discussion
Limits
4. Materials and Methods
4.1. Sampling
4.2. Isolation and Identification of Bacterial
4.3. Determination of the AMR
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.; Parveen, S.; Schwarz, J.; Rippen, T.; Jahncke, M.; DePaola, A. Seafood Pathogens and Information on Antimicrobial Resistance: A Review. Food Microbiol. 2018, 70, 85–93. [Google Scholar] [CrossRef]
- Florez-Cuadrado, D.; Moreno, M.A.; Ugarte-Ruíz, M.; Domínguez, L. Antimicrobial Resistance in the Food Chain in the European Union. Adv. Food Nutr. Res. 2018, 86, 115–136. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control; World Health Organization. Antimicrobial Resistance Surveillance in Europe: 2022: 2020 Data; Publications Office: Luxembourg, 2022. [Google Scholar]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Navidinia, M. The Clinical Importance of Emerging ESKAPE Pathogens in Nosocomial Infections. Arch. Adv. Biosci. 2016, 7, 43–57. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Essack, S.Y. Clinical and Economic Impact of Antibiotic Resistance in Developing Countries: A Systematic Review and Meta-Analysis. PLoS ONE 2017, 12, e0189621. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef]
- Wyres, K.L.; Holt, K.E. Klebsiella Pneumoniae as a Key Trafficker of Drug Resistance Genes from Environmental to Clinically Important Bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef]
- One Health Joint Plan of Action, 2022–2026; FAO: Rome, Italy; UNEP: Nairobi, Kenya; WHO: Geneva, Switzerland; World Organisation for Animal Health (WOAH) (founded as OIE): Paris, France, 2022; ISBN 978-92-5-136957-9.
- Abulreesh, H.H.; Paget, T.A.; Goulder, R. Campylobacter in Waterfowl and Aquatic Environments: Incidence and Methods of Detection. Environ. Sci. Technol. 2006, 40, 7122–7131. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Drobni, M.; Gauthier-Clerc, M.; Hernandez, J.; Granholm, S.; Kayser, Y.; Melhus, A.; Kahlmeter, G.; Waldenström, J.; Johansson, A.; et al. Dissemination of Escherichia coli with CTX-M Type ESBL between Humans and Yellow-Legged Gulls in the South of France. PLoS ONE 2009, 4, e5958. [Google Scholar] [CrossRef] [PubMed]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.D.; Wieler, L.H.; Ewers, C. Antimicrobial Resistance Profiles of Escherichia coli from Common European Wild Bird Species. Vet. Microbiol. 2010, 144, 219–225. [Google Scholar] [CrossRef]
- Radhouani, H.; Poeta, P.; Gonçalves, A.; Pacheco, R.; Sargo, R.; Igrejas, G. Wild Birds as Biological Indicators of Environmental Pollution: Antimicrobial Resistance Patterns of Escherichia coli and Enterococci Isolated from Common Buzzards (Buteo Buteo). J. Med. Microbiol. 2012, 61, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Järhult, J.D. Antibiotic Resistance in Wild Birds. Ups. J. Med. Sci. 2014, 119, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Foti, M.; Mascetti, A.; Fisichella, V.; Fulco, E.; Orlandella, B.M.; Lo Piccolo, F. Antibiotic Resistance Assessment in Bacteria Isolated in Migratory Passeriformes Transiting through the Metaponto Territory (Basilicata, Italy). Avian Res. 2017, 8, 26. [Google Scholar] [CrossRef]
- Ahmed, Z.S.; Elshafiee, E.A.; Khalefa, H.S.; Kadry, M.; Hamza, D.A. Evidence of Colistin Resistance Genes (Mcr-1 and Mcr-2) in Wild Birds and Its Public Health Implication in Egypt. Antimicrob. Resist. Infect. Control 2019, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Giacopello, C.; Foti, M.; Mascetti, A. Antimicrobial Resistance Patterns of Enterobacteriaceae in European Wild Bird Species Admitted in a Wildlife Rescue Centre. Vet. Ital. 2016, 52, 139–144. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Cherak, Z.; Bendjama, E.; Beroual, F.; Rolain, J.-M. Detection of NDM-5 and MCR-1 Antibiotic Resistance Encoding Genes in Enterobacterales in Long-Distance Migratory Bird Species Ciconia Ciconia, Algeria. Sci. Total Environ. 2022, 814, 152861. [Google Scholar] [CrossRef]
- Chen, C.; Cui, C.-Y.; Zhang, Y.; He, Q.; Wu, X.-T.; Li, G.; Liao, X.-P.; Kreiswirth, B.N.; Liu, Y.-H.; Chen, L.; et al. Emergence of Mobile Tigecycline Resistance Mechanism in Escherichia coli Strains from Migratory Birds in China. Emerg. Microbes Infect. 2019, 8, 1219–1222. [Google Scholar] [CrossRef]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K.; et al. Clinically Relevant Antimicrobial Resistance at the Wildlife–Livestock–Human Interface in Nairobi: An Epidemiological Study. Lancet Planet. Health 2019, 3, e259–e269. [Google Scholar] [CrossRef] [PubMed]
- Oravcová, V.; Peixe, L.; Coque, T.M.; Novais, C.; Francia, M.V.; Literák, I.; Freitas, A.R. Wild Corvid Birds Colonized with Vancomycin-Resistant Enterococcus Faecium of Human Origin Harbor Epidemic VanA Plasmids. Environ. Int. 2018, 118, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Borrelli, L.; Fioretti, A.; Russo, T.P.; Barco, L.; Raia, P.; De Luca Bossa, L.M.; Sensale, M.; Menna, L.F.; Dipineto, L. First Report of Salmonella Enterica Serovar Infantis in Common Swifts (Apus apus). Avian Pathol. J. WVPA 2013, 42, 323–326. [Google Scholar] [CrossRef]
- Dipineto, L.; De Luca Bossa, L.M.; Russo, T.P.; Cutino, E.A.; Gargiulo, A.; Ciccarelli, F.; Raia, P.; Menna, L.F.; Fioretti, A. Campylobacter Spp. and Birds of Prey. Avian Dis. 2014, 58, 303–305. [Google Scholar] [CrossRef]
- Gargiulo, A.; Russo, T.P.; Schettini, R.; Mallardo, K.; Calabria, M.; Menna, L.F.; Raia, P.; Pagnini, U.; Caputo, V.; Fioretti, A.; et al. Occurrence of Enteropathogenic Bacteria in Urban Pigeons (Columba livia) in Italy. Vector-Borne Zoonotic Dis. 2014, 14, 251–255. [Google Scholar] [CrossRef]
- Russo, T.P.; Pace, A.; Varriale, L.; Borrelli, L.; Gargiulo, A.; Pompameo, M.; Fioretti, A.; Dipineto, L. Prevalence and Antimicrobial Resistance of Enteropathogenic Bacteria in Yellow-Legged Gulls (Larus michahellis) in Southern Italy. Animals 2021, 11, 275. [Google Scholar] [CrossRef]
- Hubálek, Z. An annotated checklist of pathogenic microorganisms associated with migratory birds. J. Wildl. Dis. 2004, 40, 639–659. [Google Scholar] [CrossRef] [PubMed]
- Camarda, A.; Circella, E.; Pennelli, D.; Madio, A.; Bruni, G.; Lagrasta, V.; Marzano, G.; Mallia, E.; Campagnari, E. Wild Birds As Biological Indicators Of Environmental Pollution: Biotyping And Antimicrobial Resistance Patterns Of Escherichia coli Isolated From Audouin’S Gulls (Larus Audouinii) Living In The Bay of Gallipoli (Italy). Ital. J. Anim. Sci. 2006, 5, 287–290. [Google Scholar] [CrossRef]
- Literak, I.; Dolejska, M.; Janoszowska, D.; Hrusakova, J.; Meissner, W.; Rzyska, H.; Bzoma, S.; Cizek, A. Antibiotic-Resistant Escherichia coli Bacteria, Including Strains with Genes Encoding the Extended-Spectrum Beta-Lactamase and QnrS, in Waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 2010, 76, 8126–8134. [Google Scholar] [CrossRef]
- Elmberg, J.; Berg, C.; Lerner, H.; Waldenström, J.; Hessel, R. Potential Disease Transmission from Wild Geese and Swans to Livestock, Poultry and Humans: A Review of the Scientific Literature from a One Health Perspective. Infect. Ecol. Epidemiol. 2017, 7, 1300450. [Google Scholar] [CrossRef]
- Malekian, M.; Shagholian, J.; Hosseinpour, Z. Pathogen Presence in Wild Birds Inhabiting Landfills in Central Iran. EcoHealth 2021, 18, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Matias, C.A.R.; Pereira, I.A.; Reis, E.M.F.D.; Rodrigues, D.D.P.; Siciliano, S. Frequency of Zoonotic Bacteria among Illegally Traded Wild Birds in Rio de Janeiro. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2016, 47, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Konicek, C.; Vodrážka, P.; Barták, P.; Knotek, Z.; Hess, C.; Račka, K.; Hess, M.; Troxler, S. Detection of zoonotic pathogens in wild birds in the cross-border region austria—Czech republic. J. Wildl. Dis. 2016, 52, 850–861. [Google Scholar] [CrossRef]
- Rodrigues, J.G.C.; Nair, H.P.; O’Kane, C.; Walker, C.A. Prevalence of Multidrug Resistance in Pseudomonas Spp. Isolated from Wild Bird Feces in an Urban Aquatic Environment. Ecol. Evol. 2021, 11, 14303–14311. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, R.; Yu, P.; Alvarez, P.J.J. High Levels of Antibiotic Resistance Genes and Opportunistic Pathogenic Bacteria Indicators in Urban Wild Bird Feces. Environ. Pollut. 2020, 266, 115200. [Google Scholar] [CrossRef]
- Kutkowska, J.; Turska-Szewczuk, A.; Kucharczyk, M.; Kucharczyk, H.; Zalewska, J.; Urbanik-Sypniewska, T. Methicillin-Resistant Staphylococcus Aureus and Glycopeptide-Resistant Enterococci in Fecal Samples of Birds from South-Eastern Poland. BMC Vet. Res. 2019, 15, 472. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Dec, M.; Marek, A.; Urban-Chmiel, R. Clonal Structure and Antibiotic Resistance of Enterococcus spp. from Wild Birds in Poland. Microb. Drug Resist. 2019, 25, 1227–1237. [Google Scholar] [CrossRef]
- Elsohaby, I.; Samy, A.; Elmoslemany, A.; Alorabi, M.; Alkafafy, M.; Aldoweriej, A.; Al-Marri, T.; Elbehiry, A.; Fayez, M. Migratory Wild Birds as a Potential Disseminator of Antimicrobial-Resistant Bacteria around Al-Asfar Lake, Eastern Saudi Arabia. Antibiotics 2021, 10, 260. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Lopes, A.F.; Soeiro, V.; Caniça, M.; Manageiro, V.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. Nocturnal Birds of Prey as Carriers of Staphylococcus Aureus and Other Staphylococci: Diversity, Antimicrobial Resistance and Clonal Lineages. Antibiotics 2022, 11, 240. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Gómez, P.; Alonso, C.A.; Camacho, M.C.; Ramiro, Y.; de la Puente, J.; Fernández-Fernández, R.; Quevedo, M.Á.; Blanco, J.M.; Báguena, G.; et al. Frequency and Characterization of Antimicrobial Resistance and Virulence Genes of Coagulase-Negative Staphylococci from Wild Birds in Spain. Detection of Tst-Carrying S. sciuri Isolates. Microorganisms 2020, 8, 1317. [Google Scholar] [CrossRef]
- Effah, C.Y.; Sun, T.; Liu, S.; Wu, Y. Klebsiella Pneumoniae: An Increasing Threat to Public Health. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1. [Google Scholar] [CrossRef]
- Chiaverini, A.; Cornacchia, A.; Centorotola, G.; Tieri, E.E.; Sulli, N.; Del Matto, I.; Iannitto, G.; Petrone, D.; Petrini, A.; Pomilio, F. Phenotypic and Genetic Characterization of Klebsiella Pneumoniae Isolates from Wild Animals in Central Italy. Animals 2022, 12, 1347. [Google Scholar] [CrossRef]
- Nowaczek, A.; Dec, M.; Stępień-Pyśniak, D.; Urban-Chmiel, R.; Marek, A.; Różański, P. Antibiotic Resistance and Virulence Profiles of Escherichia coli Strains Isolated from Wild Birds in Poland. Pathogens 2021, 10, 1059. [Google Scholar] [CrossRef]
- Millán, J.; Aduriz, G.; Moreno, B.; Juste, R.A.; Barral, M. Salmonella Isolates from Wild Birds and Mammals in the Basque Country (Spain). Rev. Sci. Technol. Int. Off. Epizoot. 2004, 23, 905–911. [Google Scholar] [CrossRef]
- Botti, V.; Navillod, F.V.; Domenis, L.; Orusa, R.; Pepe, E.; Robetto, S.; Guidetti, C. Salmonella Spp. and Antibiotic-Resistant Strains in Wild Mammals and Birds in North-Western Italy from 2002 to 2010. Vet. Ital. 2013, 49, 195–202. [Google Scholar]
- Refsum, T.; Handeland, K.; Baggesen, D.L.; Holstad, G.; Kapperud, G. Salmonellae in Avian Wildlife in Norway from 1969 to 2000. Appl. Environ. Microbiol. 2002, 68, 5595–5599. [Google Scholar] [CrossRef]
- Refsum, T.; Holstad, G.; Kapperud, G.; Handeland, K. An Investigation of Salmonella Bacteria in Waterfowls and Migratory Birds in Norway. Acta Vet. Scand. 2005, 46, 95–100. [Google Scholar] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019; ISBN 978-92-4-151552-8. [Google Scholar]
- Cao, J.; Hu, Y.; Liu, F.; Wang, Y.; Bi, Y.; Lv, N.; Li, J.; Zhu, B.; Gao, G.F. Metagenomic Analysis Reveals the Microbiome and Resistome in Migratory Birds. Microbiome 2020, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, H.A.; Booton, R.; Kallonen, T.; Gibbon, M.J.; Couto, N.; Passet, V.; López-Fernández, S.; Rodrigues, C.; Matthews, L.; Mitchell, S.; et al. A Large-Scale Genomic Snapshot of Klebsiella Spp. Isolates in Northern Italy Reveals Limited Transmission between Clinical and Non-Clinical Settings. Nat. Microbiol. 2022, 7, 2054–2067. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and Combating Antibiotic Resistance from One Health and Global Health Perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Pornsukarom, S.; Thakur, S. Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes Following Isolation from the Commercial Swine Operation Environment after Manure Application. Appl. Environ. Microbiol. 2017, 83, e01503-17. [Google Scholar] [CrossRef] [PubMed]
- Check List Del Paleartico Occidentale—EBN Italia. Available online: https://www.yumpu.com/it/document/read/11410474/check-list-del-paleartico-occidentale-ebn-italia (accessed on 8 December 2022).
- EUR-Lex—32010L0063—EN—EUR-Lex. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A32010L0063 (accessed on 14 November 2022).
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI: Malvern, PA, USA, 2018; Volume M100. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russo, T.P.; Minichino, A.; Gargiulo, A.; Varriale, L.; Borrelli, L.; Pace, A.; Santaniello, A.; Pompameo, M.; Fioretti, A.; Dipineto, L. Prevalence and Phenotypic Antimicrobial Resistance among ESKAPE Bacteria and Enterobacterales Strains in Wild Birds. Antibiotics 2022, 11, 1825. https://doi.org/10.3390/antibiotics11121825
Russo TP, Minichino A, Gargiulo A, Varriale L, Borrelli L, Pace A, Santaniello A, Pompameo M, Fioretti A, Dipineto L. Prevalence and Phenotypic Antimicrobial Resistance among ESKAPE Bacteria and Enterobacterales Strains in Wild Birds. Antibiotics. 2022; 11(12):1825. https://doi.org/10.3390/antibiotics11121825
Chicago/Turabian StyleRusso, Tamara Pasqualina, Adriano Minichino, Antonio Gargiulo, Lorena Varriale, Luca Borrelli, Antonino Pace, Antonio Santaniello, Marina Pompameo, Alessandro Fioretti, and Ludovico Dipineto. 2022. "Prevalence and Phenotypic Antimicrobial Resistance among ESKAPE Bacteria and Enterobacterales Strains in Wild Birds" Antibiotics 11, no. 12: 1825. https://doi.org/10.3390/antibiotics11121825
APA StyleRusso, T. P., Minichino, A., Gargiulo, A., Varriale, L., Borrelli, L., Pace, A., Santaniello, A., Pompameo, M., Fioretti, A., & Dipineto, L. (2022). Prevalence and Phenotypic Antimicrobial Resistance among ESKAPE Bacteria and Enterobacterales Strains in Wild Birds. Antibiotics, 11(12), 1825. https://doi.org/10.3390/antibiotics11121825







