Antibiotic Susceptibility Testing with Raman Biosensing
Abstract
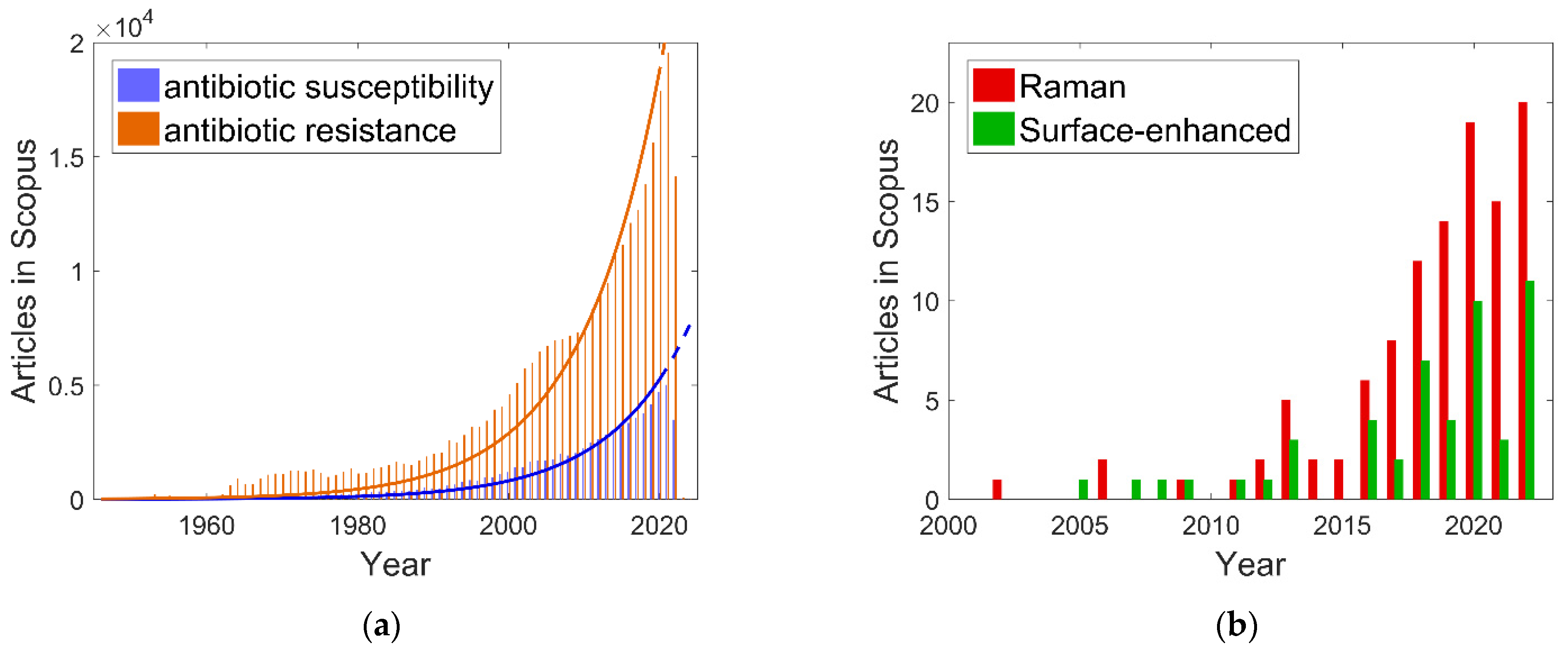
1. Introduction
2. Antibiotic Susceptibility
2.1. Antibiotics and Raman Sensing
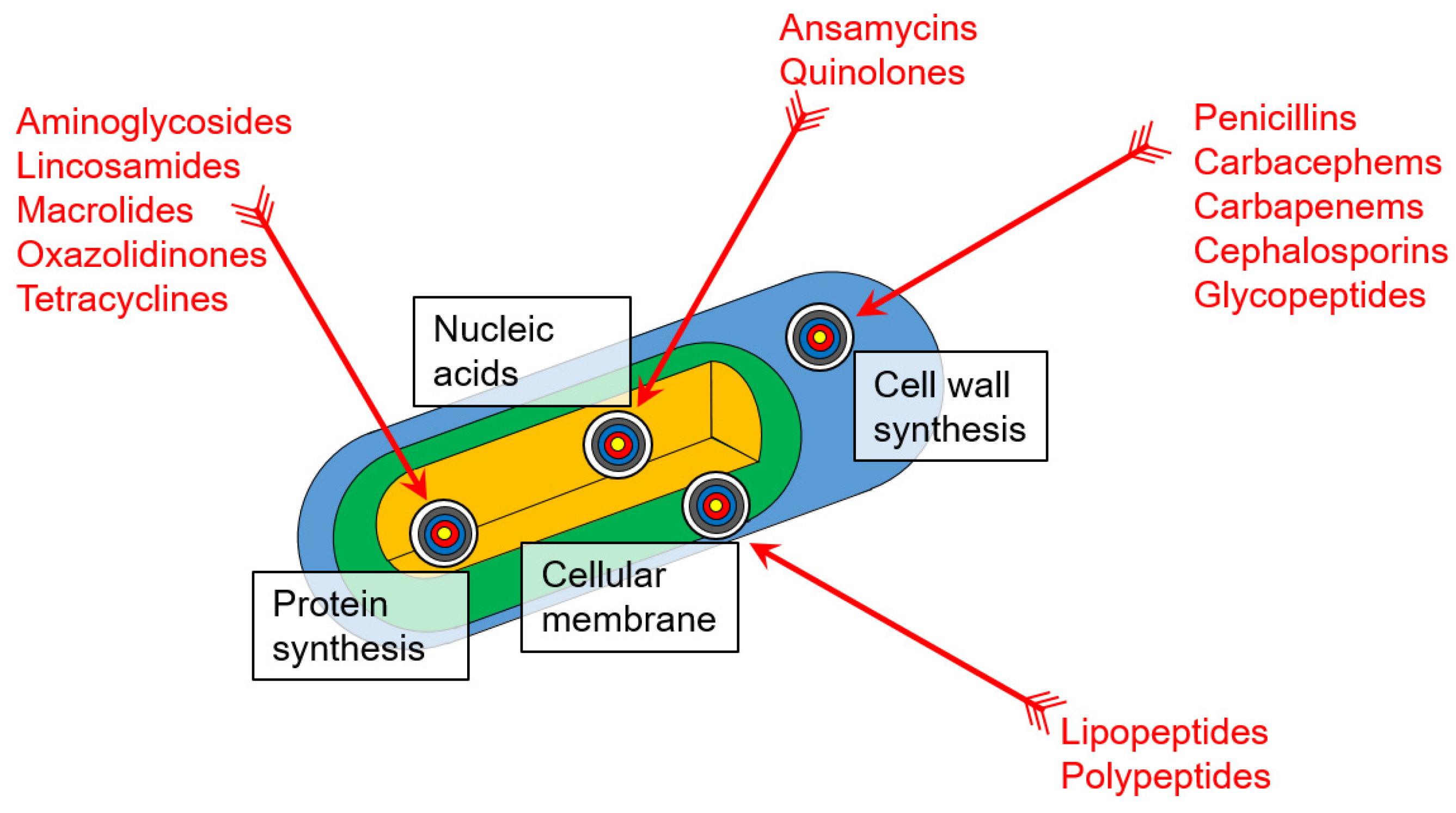
2.2. Antibiotic Action on Bacteria and Mechanisms of Their Resistance
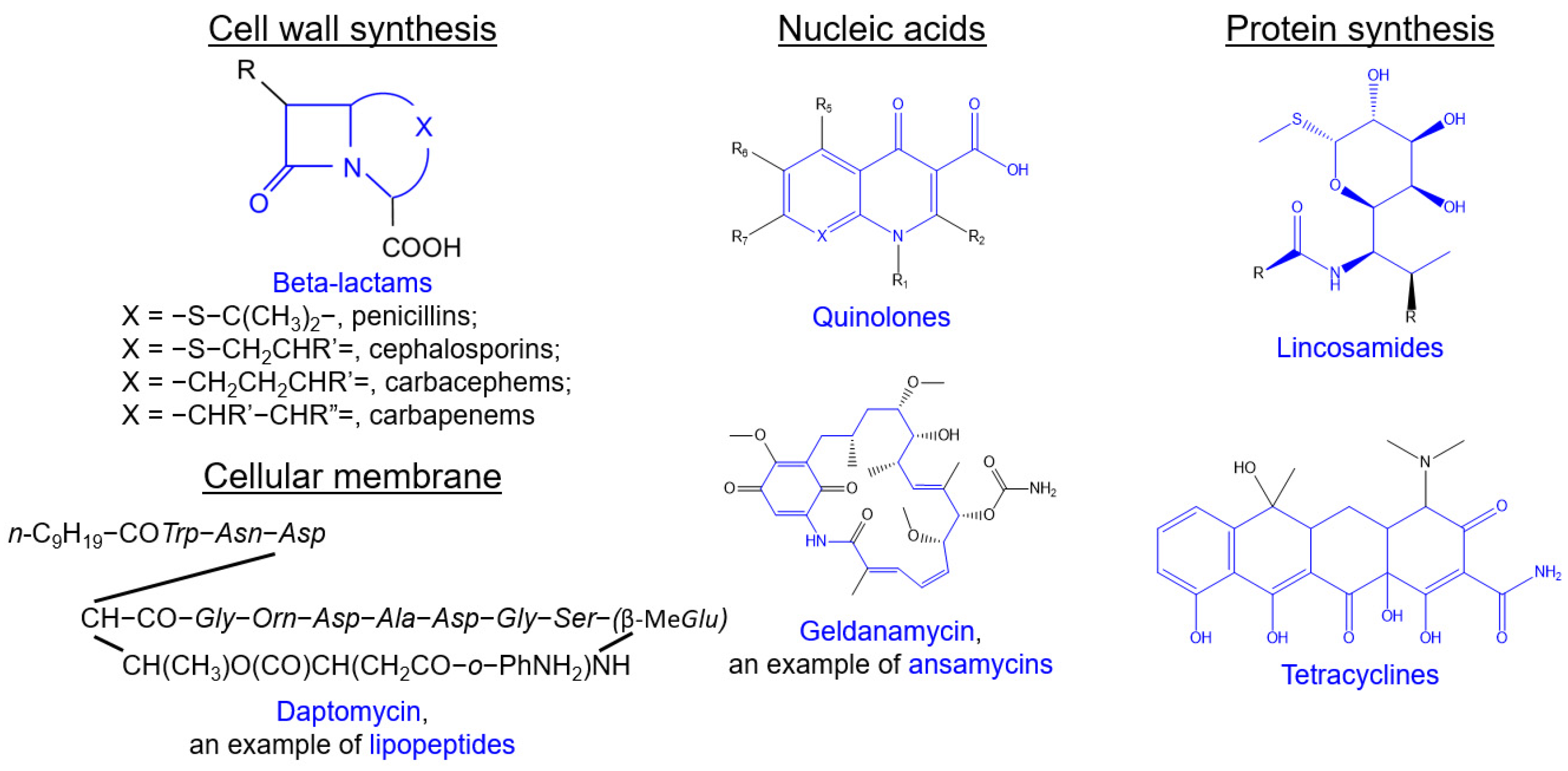
2.2.1. Beta-Lactams
2.2.2. Glycopeptides
2.2.3. Polypeptides
2.2.4. Lipopeptides
2.2.5. Quinolones
2.2.6. Aminoglycosides
2.2.7. Macrolides
2.2.8. Tetracyclines
2.3. Raman Spectroscopy and Raman Spectra of Bacteria
2.4. Features in the Raman Spectra of Bacteria Affected by Antibiotics
2.5. Antibiotic Susceptibility Testing Using Raman Spectroscopy of Bacteria
2.5.1. Laser Tweezer–Assisted Normal Raman Spectroscopy
2.5.2. Resonance Raman Spectroscopy
2.5.3. Stimulated Raman Spectroscopy
2.5.4. Surface-Enhanced Raman Spectroscopy
2.5.5. DEP-Raman Spectroscopy
3. Prospects: Whole-Cell Biosensing of Antibiotics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nauclér, P.; Huttner, A.; van Werkhoven, C.H.; Singer, M.; Tattevin, P.; Einav, S.; Tängdén, T. Impact of Time to Antibiotic Therapy on Clinical Outcome in Patients with Bacterial Infections in the Emergency Department: Implications for Antimicrobial Stewardship. Clin. Microbiol. Infect. 2021, 27, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hutchings, M.; Truman, A.; Wilkinson, B. Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Paulus, G.K.; Hornstra, L.M.; Alygizakis, N.; Slobodnik, J.; Thomaidis, N.; Medema, G. The Impact of On-Site Hospital Wastewater Treatment on the Downstream Communal Wastewater System in Terms of Antibiotics and Antibiotic Resistance Genes. Int. J. Hyg. Environ. Health 2019, 222, 635–644. [Google Scholar] [CrossRef]
- Bielen, A.; Šimatović, A.; Kosić-Vukšić, J.; Senta, I.; Ahel, M.; Babić, S.; Jurina, T.; González Plaza, J.J.; Milaković, M.; Udiković-Kolić, N. Negative Environmental Impacts of Antibiotic-Contaminated Effluents from Pharmaceutical Industries. Water Res. 2017, 126, 79–87. [Google Scholar] [CrossRef]
- Chng, K.R.; Li, C.; Bertrand, D.; Ng, A.H.Q.; Kwah, J.S.; Low, H.M.; Tong, C.; Natrajan, M.; Zhang, M.H.; Xu, L.; et al. Cartography of Opportunistic Pathogens and Antibiotic Resistance Genes in a Tertiary Hospital Environment. Nat. Med. 2020, 26, 941–951. [Google Scholar] [CrossRef]
- Thoma, R.; Seneghini, M.; Seiffert, S.N.; Vuichard Gysin, D.; Scanferla, G.; Haller, S.; Flury, D.; Boggian, K.; Kleger, G.R.; Filipovic, M.; et al. The Challenge of Preventing and Containing Outbreaks of Multidrug-Resistant Organisms and Candida Auris during the Coronavirus Disease 2019 Pandemic: Report of a Carbapenem-Resistant Acinetobacter Baumannii Outbreak and a Systematic Review of the Literature. Antimicrob. Resist. Infect. Control 2022, 11, 12. [Google Scholar] [CrossRef]
- Meschiari, M.; Onorato, L.; Bacca, E.; Orlando, G.; Menozzi, M.; Franceschini, E.; Bedini, A.; Cervo, A.; Santoro, A.; Sarti, M.; et al. Long-Term Impact of the COVID-19 Pandemic on In-Hospital Antibiotic Consumption and Antibiotic Resistance: A Time Series Analysis (2015–2021). Antibiotics 2022, 11, 826. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Li, J.; Wuethrich, A.; Sina, A.A.I.; Cheng, H.H.; Wang, Y.; Behren, A.; Mainwaring, P.N.; Trau, M. A Digital Single-Molecule Nanopillar SERS Platform for Predicting and Monitoring Immune Toxicities in Immunotherapy. Nat. Commun. 2021, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Matricardi, C.; Hanske, C.; Garcia-Pomar, J.L.; Langer, J.; Mihi, A.; Liz-Marzán, L.M. Gold Nanoparticle Plasmonic Superlattices as Surface-Enhanced Raman Spectroscopy Substrates. ACS Nano 2018, 12, 8531–8539. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Zamaleeva, A.I.; Fakhrullin, R.F.; Culha, M. Layer-by-Layer Coating of Bacteria with Noble Metal Nanoparticles for Surface-Enhanced Raman Scattering. Anal. Bioanal. Chem. 2009, 395, 2559–2567. [Google Scholar] [CrossRef] [PubMed]
- Gorbachevskii, M.V.; Kopitsyn, D.S.; Kotelev, M.S.; Ivanov, E.V.; Vinokurov, V.A.; Novikov, A.A. Amplification of Surface-Enhanced Raman Scattering by the Oxidation of Capping Agents on Gold Nanoparticles. RSC Adv. 2018, 8, 19051–19057. [Google Scholar] [CrossRef]
- Wonner, K.; Murke, S.; Alfarano, S.R.; Hosseini, P.; Havenith, M.; Tschulik, K. Operando Electrochemical SERS Monitors Nanoparticle Reactions by Capping Agent Fingerprints. Nano Res. 2022, 15, 4517–4524. [Google Scholar] [CrossRef]
- Lindquist, N.C.; de Albuquerque, C.D.L.; Sobral-Filho, R.G.; Paci, I.; Brolo, A.G. High-Speed Imaging of Surface-Enhanced Raman Scattering Fluctuations from Individual Nanoparticles. Nat. Nanotechnol. 2019, 14, 981–987. [Google Scholar] [CrossRef]
- Wolf, S.; Popp, J.; Frosch, T. Multifocal Hyperspectral Raman Imaging Setup for Multi-Well Plates. Sens. Actuators B Chem. 2023, 375, 132949. [Google Scholar] [CrossRef]
- Kawagoe, H.; Ando, J.; Asanuma, M.; Dodo, K.; Miyano, T.; Ueda, H.; Sodeoka, M.; Fujita, K. Multiwell Raman Plate Reader for High-Throughput Biochemical Screening. Sci. Rep. 2021, 11, 15742. [Google Scholar] [CrossRef]
- Yu, H.; Lyu, Q.; Chen, X.; Guo, D.; He, D.; Jia, X.; Han, L.; Xiao, W. Nylon Membranes Modified by Gold Nanoparticles as Surface-Enhanced Raman Spectroscopy Substrates for Several Pesticides Detection. RSC Adv. 2021, 11, 24183–24189. [Google Scholar] [CrossRef]
- Logan, N.; Haughey, S.A.; Liu, L.; Burns, D.T.; Quinn, B.; Cao, C.; Elliott, C.T. Handheld SERS Coupled with QuEChERs for the Sensitive Analysis of Multiple Pesticides in Basmati Rice. NPJ Sci. Food 2022, 6, 3. [Google Scholar] [CrossRef]
- Adomavičiūtė, S.; Velička, M.; Šablinskas, V. Detection of Aspirin Traces in Blood by Means of Surface-Enhanced Raman Scattering Spectroscopy. J. Raman Spectrosc. 2020, 51, 919–931. [Google Scholar] [CrossRef]
- Turzhitsky, V.; Zhang, L.; Horowitz, G.L.; Vitkin, E.; Khan, U.; Zakharov, Y.; Qiu, L.; Itzkan, I.; Perelman, L.T. Picoanalysis of Drugs in Biofluids with Quantitative Label-Free Surface-Enhanced Raman Spectroscopy. Small 2018, 14, 1802392. [Google Scholar] [CrossRef] [PubMed]
- Markina, N.E.; Goryacheva, I.Y.; Markin, A.V. Surface-Enhanced Raman Spectroscopy for the Determination of Medical and Narcotic Drugs in Human Biofluids. J. Anal. Chem. 2022, 77, 930–947. [Google Scholar] [CrossRef]
- Lin, X.; Lin, D.; Chen, Y.; Lin, J.; Weng, S.; Song, J.; Feng, S. High Throughput Blood Analysis Based on Deep Learning Algorithm and Self-Positioning Super-Hydrophobic SERS Platform for Non-Invasive Multi-Disease Screening. Adv. Funct. Mater. 2021, 31, 2103382. [Google Scholar] [CrossRef]
- Zhou, C.; Zou, H.; Sun, C.; Li, Y. Recent Advances in Biosensors for Antibiotic Detection: Selectivity and Signal Amplification with Nanomaterials. Food Chem. 2021, 361, 130109. [Google Scholar] [CrossRef]
- Girmatsion, M.; Mahmud, A.; Abraha, B.; Xie, Y.; Cheng, Y.; Yu, H.; Yao, W.; Guo, Y.; Qian, H. Rapid Detection of Antibiotic Residues in Animal Products Using Surface-Enhanced Raman Spectroscopy: A Review. Food Control 2021, 126, 108019. [Google Scholar] [CrossRef]
- Huang, J.; Wen, J.; Zhou, M.; Ni, S.; Le, W.; Chen, G.; Wei, L.; Zeng, Y.; Qi, D.; Pan, M.; et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations. Anal. Chem. 2021, 93, 9174–9182. [Google Scholar] [CrossRef]
- Yu, H.; Xiao, M.; Lai, W.; Alam, M.F.; Zhang, W.; Pei, H.; Wan, Y.; Li, L. A Self-Calibrating Surface-Enhanced Raman Scattering-Active System for Bacterial Phenotype Detection. Anal. Chem. 2020, 92, 4491–4497. [Google Scholar] [CrossRef]
- Kopitsyn, D.S.; Gorbachevskii, M.V.; Botchkova, E.A.; Bychenko, M.A.; Novikov, A.A. Identification of Bacteria by Surface-Enhanced Raman Spectra after Peroxide Treatment. Appl. Biochem. Microbiol. 2019, 55, 78–82. [Google Scholar] [CrossRef]
- Prakash, O.; Sil, S.; Verma, T.; Umapathy, S. Direct Detection of Bacteria Using Positively Charged Ag/Au Bimetallic Nanoparticles: A Label-Free Surface-Enhanced Raman Scattering Study Coupled with Multivariate Analysis. J. Phys. Chem. C 2020, 124, 861–869. [Google Scholar] [CrossRef]
- Chiu, S.W.Y.; Cheng, H.W.; Chen, Z.X.; Wang, H.H.; Lai, M.Y.; Wang, J.K.; Wang, Y.L. Quantification of Biomolecules Responsible for Biomarkers in the Surface-Enhanced Raman Spectra of Bacteria Using Liquid Chromatography-Mass Spectrometry. Phys. Chem. Chem. Phys. 2018, 20, 8032–8041. [Google Scholar] [CrossRef] [PubMed]
- Gorbachevskii, M.V.; Filatova, S.V.; Filimonova, A.V.; Kopitsyn, D.S.; Panchenko, A.A.; Vinokurov, V.A.; Novikov, A.A. Detection of Bacterial Colonization by the Spectral Changes of Surface-Enhanced Raman Reporters. Biochem. Biophys. Res. Commun. 2021, 546, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Kuku, G.; Saricam, M.; Akhatova, F.; Danilushkina, A.; Fakhrullin, R.; Culha, M. Surface-Enhanced Raman Scattering to Evaluate Nanomaterial Cytotoxicity on Living Cells. Anal. Chem. 2016, 88, 9813–9820. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qiao, Y.; Zhang, H.; Zhao, J.; Li, W.; Xie, T.; Zhong, D.; Wei, Q.; Hua, S.; Yu, Y.; et al. Gold–Silver Nanoshells Promote Wound Healing from Drug-Resistant Bacteria Infection and Enable Monitoring via Surface-Enhanced Raman Scattering Imaging. Biomaterials 2020, 234, 119763. [Google Scholar] [CrossRef] [PubMed]
- Neter, E. Relative Susceptibility of Staphylococci to the Bacteriostatic Action of Antibiotics. Proc. Soc. Exp. Biol. Med. 1945, 58, 126–128. [Google Scholar] [CrossRef]
- Webb, S.J.; Stoneham, M.E. Resonance between 1011 and 1012 Hz in Active Bacterial Cells as Seen by Laser Raman Spectroscopy. Phys. Lett. A 1977, 60, 267–268. [Google Scholar] [CrossRef]
- Howard, W.F.; Nelson, W.H.; Sperry, J.F. Resonance Raman Method for the Rapid Detection and Identification of Bacteria in Water. Appl. Spectrosc. 1980, 34, 72–75. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman Spectra of Pyridine Adsorbed at a Silver Electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Efrima, S.; Bronk, B.V. Silver Colloids Impregnating or Coating Bacteria. J. Phys. Chem. B 1998, 102, 5947–5950. [Google Scholar] [CrossRef]
- Zeiri, L.; Bronk, B.V.; Shabtai, Y.; Czégé, J.; Efrima, S. Silver Metal Induced Surface Enhanced Raman of Bacteria. Colloids Surf. A Physicochem. Eng. Asp. 2002, 208, 357–362. [Google Scholar] [CrossRef]
- Zeiri, L.; Bronk, B.V.; Shabtai, Y.; Eichler, J.; Efrima, S. Surface-Enhanced Raman Spectroscopy as a Tool for Probing Specific Biochemical Components in Bacteria. Appl. Spectrosc. 2004, 58, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, R.M.; Goodacre, R. Discrimination of Bacteria Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2004, 76, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, R.M.; Brooker, A.; Goodacre, R. Surface-Enhanced Raman Spectroscopy for Bacterial Discrimination Utilizing a Scanning Electron Microscope with a Raman Spectroscopy Interface. Anal. Chem. 2004, 76, 5198–5202. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot Spots of Horizontal Gene Transfer (HGT) in Aquatic Environments, with a Focus on a New HGT Mechanism. FEMS Microbiol. Ecol. 2021, 96, fiaa031. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Ma, J.; Li, G.; Rillig, M.C.; Zhu, Y.G. Soil Plastispheres as Hotpots of Antibiotic Resistance Genes and Potential Pathogens. ISME J. 2022, 16, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.M.; da Silva, B.N.M.; Barbosa, G.; Barreiro, E.J. β-Lactam Antibiotics: An Overview from a Medicinal Chemistry Perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Deja, E.N. Novel SS-Lactamase Inhibitors: New Weapons in the Arms Race against Antimicrobial Resistance. Clin. Microbiol. Newsl. 2021, 43, 119–125. [Google Scholar] [CrossRef]
- Tan, C.T.; Xu, X.; Qiao, Y.; Wang, Y. A Peptidoglycan Storm Caused by β-Lactam Antibiotic’s Action on Host Microbiota Drives Candida Albicans Infection. Nat. Commun. 2021, 12, 2560. [Google Scholar] [CrossRef]
- Rao, J.; Lahiri, J.; Isaacs, L.; Weis, R.M.; Whitesides, G.M. A Trivalent System from Vancomycin-D-Ala-D-Ala with Higher Affinity than Avidin-Biotin. Science 1998, 280, 708–711. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin Resistance in Gram-Positive Cocci. Clin. Infect. Dis. 2006, 42, S25–S34. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin Resistant Staphylococcus Aureus Infections: A Review of Case Updating and Clinical Features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Okano, A.; Isley, N.A.; Boger, D.L. Peripheral Modifications of [Ψ[CH2NH]Tpg4]Vancomycin with Added Synergistic Mechanisms of Action Provide Durable and Potent Antibiotics. Proc. Natl. Acad. Sci. USA 2017, 114, E5052–E5061. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.C.; Isley, N.A.; Okano, A.; Weiss, W.J.; Boger, D.L. C1-CBP-Vancomycin: Impact of a Vancomycin C-Terminus Trimethylammonium Cation on Pharmacological Properties and Insights into Its Newly Introduced Mechanism of Action. J. Org. Chem. 2020, 85, 1365–1375. [Google Scholar] [CrossRef]
- Roberts, K.D.; Azad, M.A.K.; Wang, J.; Horne, A.S.; Thompson, P.E.; Nation, R.L.; Velkov, T.; Li, J. Antimicrobial Activity and Toxicity of the Major Lipopeptide Components of Polymyxin B and Colistin: Last-Line Antibiotics against Multidrug-Resistant Gram-Negative Bacteria. ACS Infect. Dis. 2015, 1, 568–575. [Google Scholar] [CrossRef]
- Takada, Y.; Itoh, H.; Paudel, A.; Panthee, S.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Discovery of Gramicidin A Analogues with Altered Activities by Multidimensional Screening of a One-Bead-One-Compound Library. Nat. Commun. 2020, 11, 4935. [Google Scholar] [CrossRef]
- Ketchem, R.R.; Hu, W.; Cross, T.A. High-Resolution Conformation of Gramicidin A in a Lipid Bilayer by Solid-State NMR. Science 1993, 261, 1457–1460. [Google Scholar] [CrossRef]
- Chistyulin, D.K.; Rokitskaya, T.I.; Kovalchuk, S.I.; Sorochkina, A.I.; Firsov, A.M.; Kotova, E.A.; Antonenko, Y.N. PH-Dependent Properties of Ion Channels Formed by N-Terminally Glutamate Substituted Gramicidin A in Planar Lipid Bilayers. Biochim. Biophys. Acta Biomembr. 2017, 1859, 896–902. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Brown, P.; Dawson, M.J. Development of New Polymyxin Derivatives for Multi-Drug Resistant Gram-Negative Infections. J. Antibiot. 2017, 70, 386–394. [Google Scholar] [CrossRef]
- Jadhav, K.B.; Stein, C.; Makarewicz, O.; Pradel, G.; Lichtenecker, R.J.; Sack, H.; Heinemann, S.H.; Arndt, H.D. Bioactivity of Topologically Confined Gramicidin A Dimers. Bioorganic Med. Chem. 2017, 25, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Wenzel, M.; Strahl, H.; Grein, F.; Saaki, T.N.V.; Kohl, B.; Siersma, T.; Bandow, J.E.; Sahl, H.G.; Schneider, T.; et al. Daptomycin Inhibits Cell Envelope Synthesis by Interfering with Fluid Membrane Microdomains. Proc. Natl. Acad. Sci. USA 2016, 113, E7077–E7086. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Palmer, M. The Action Mechanism of Daptomycin. Bioorganic Med. Chem. 2016, 24, 6253–6268. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Munita, J.M.; Arias, C.A. Mechanisms of Drug Resistance: Daptomycin Resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 32–53. [Google Scholar] [CrossRef]
- Roch, M.; Gagetti, P.; Davis, J.; Ceriana, P.; Errecalde, L.; Corso, A.; Rosato, A.E. Daptomycin Resistance in Clinical MRSA Strains Is Associated with a High Biological Fitness Cost. Front. Microbiol. 2017, 8, 2303. [Google Scholar] [CrossRef]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA Topoisomerase Inhibition by a New Class of Antibacterial Agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of Quinolone Action and Resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Millanao, A.R.; Mora, A.Y.; Villagra, N.A.; Bucarey, S.A.; Hidalgo, A.A. Biological Effects of Quinolones: A Family of Broad-Spectrum Antimicrobial Agents. Molecules 2021, 26, 7153. [Google Scholar] [CrossRef]
- Kolarič, A.; Anderluh, M.; Minovski, N. Two Decades of Successful SAR-Grounded Stories of the Novel Bacterial Topoisomerase Inhibitors (NBTIs). J. Med. Chem. 2020, 63, 5664–5674. [Google Scholar] [CrossRef]
- Bellucci, M.C.; Volonterio, A. Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles. Antibiotics 2020, 9, 504. [Google Scholar] [CrossRef]
- Becker, B.; Cooper, M.A. Aminoglycoside Antibiotics in the 21st Century. ACS Chem. Biol. 2013, 8, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Thamban Chandrika, N.; Garneau-Tsodikova, S. Comprehensive Review of Chemical Strategies for the Preparation of New Aminoglycosides and Their Biological Activities. Chem. Soc. Rev. 2018, 47, 1189–1249. [Google Scholar] [CrossRef] [PubMed]
- Tenson, T.; Lovmar, M.; Ehrenberg, M. The Mechanism of Action of Macrolides, Lincosamides and Streptogramin B Reveals the Nascent Peptide Exit Path in the Ribosome. J. Mol. Biol. 2003, 330, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C. Resistance to Macrolide, Lincosamide, Streptogramin, Ketolide, and Oxazolidinone Antibiotics. Appl. Biochem. Biotechnol. Part B Mol. Biotechnol. 2004, 28, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Chukwudi, C.U. RRNA Binding Sites and the Molecular Mechanism of Action of the Tetracyclines. Antimicrob. Agents Chemother. 2016, 60, 4433–4441. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Gasparrini, A.J.; Reck, M.R.; Symister, C.T.; Elliott, J.L.; Vogel, J.P.; Wencewicz, T.A.; Dantas, G.; Tolia, N.H. Plasticity, Dynamics, and Inhibition of Emerging Tetracycline Resistance Enzymes. Nat. Chem. Biol. 2017, 13, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, C.; Schwarz, S.; Liu, W.; Yang, Q.; Luan, T.; Wang, L.; Liu, S.; Zhang, W. Identification of a Novel Tetracycline Resistance Gene, Tet(63), Located on a Multiresistance Plasmid from Staphylococcus Aureus. J. Antimicrob. Chemother. 2021, 76, 576–581. [Google Scholar] [CrossRef]
- Smekal, A. Zur Quantentheorie Der Dispersion. Naturwissenschaften 1923, 11, 873–875. [Google Scholar] [CrossRef]
- Raman, C.V.; Krishnan, K.S. A New Type of Secondary Radiation. Nature 1928, 121, 501–502. [Google Scholar] [CrossRef]
- Landsberg, G.; Mandelstam, L. Eine Neue Erscheinung Bei Der Lichtzerstreuung in Krystallen. Naturwissenschaften 1928, 16, 557–558. [Google Scholar] [CrossRef]
- Cardona, M.; Merlin, R. Light Scattering in Solids IX; Springer: Berlin/Heidelberg, Germany, 2006; Volume 108, ISBN 3540344357. [Google Scholar]
- Shorygin, P. Resonance Combinatorial Light Scattering. Dokl. Akad. Nauk. SSSR 1952, 87, 201–204. [Google Scholar]
- Jeanmaire, D.L.; Van Duyne, R.P. Surface Raman Spectroelectrochemistry. Part I. Heterocyclic, Aromatic, and Aliphatic Amines Adsorbed on the Anodized Silver Electrode. J. Electroanal. Chem. 1977, 84, 1–20. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously Intense Raman Spectra of Pyridine at a Silver Electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Gill, D.; Kilponen, R.G.; Rimai, L. Resonance Raman Scattering of Laser Radiation by Vibrational Modes of Carotenoid Pigment Molecules in Intact Plant Tissues. Nature 1970, 227, 743–744. [Google Scholar] [CrossRef]
- Webb, S.J. Laser-Raman Spectroscopy of Living Cells. Phys. Rep. 1980, 60, 201–224. [Google Scholar] [CrossRef]
- Gukowsky, J.C.; Yang, T.; He, L. Assessment of Three SERS Approaches for Studying E. Coli O157:H7 Susceptibility to Ampicillin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120239. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.W.; Cheng, H.W.; Shiue, J.; Wang, J.K.; Wang, Y.L.; Huang, N.T. Antibiotic Susceptibility Test with Surface-Enhanced Raman Scattering in a Microfluidic System. Anal. Chem. 2019, 91, 10988–10995. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, X.; Wang, T.; Li, Z.; Han, D.; Yu, C.; Yang, C.; Qu, H.; Chi, H.; Wang, Y.; et al. A Sensitive and Rapid Bacterial Antibiotic Susceptibility Test Method by Surface Enhanced Raman Spectroscopy. Braz. J. Microbiol. 2020, 51, 875–881. [Google Scholar] [CrossRef]
- Liao, C.C.; Chen, Y.Z.; Lin, S.J.; Cheng, H.W.; Wang, J.K.; Wang, Y.L.; Han, Y.Y.; Huang, N.T. A Microfluidic Microwell Device Operated by the Automated Microfluidic Control System for Surface-Enhanced Raman Scattering-Based Antimicrobial Susceptibility Testing. Biosens. Bioelectron. 2021, 191, 113483. [Google Scholar] [CrossRef]
- Liu, T.T.; Lin, Y.H.; Hung, C.S.; Liu, T.J.; Chen, Y.; Huang, Y.C.; Tsai, T.H.; Wang, H.H.; Wang, D.W.; Wang, J.K.; et al. A High Speed Detection Platform Based on Surface-Enhanced Raman Scattering for Monitoring Antibiotic-Induced Chemical Changes in Bacteria Cell Wall. PLoS ONE 2009, 4, e5470. [Google Scholar] [CrossRef]
- López-Díez, E.C.; Winder, C.L.; Ashton, L.; Currie, F.; Goodacre, R. Monitoring the Mode of Action of Antibiotics Using Raman Spectroscopy: Investigating Subinhibitory Effects of Amikacin on Pseudomonas Aeruginosa. Anal. Chem. 2005, 77, 2901–2906. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Han, Y.Y.; Shih, P.H.; Lian, W.N.; Wang, H.H.; Lin, C.H.; Hsueh, P.R.; Wang, J.K.; Wang, Y.L. Rapid Bacterial Antibiotic Susceptibility Test Based on Simple Surface-Enhanced Raman Spectroscopic Biomarkers. Sci. Rep. 2016, 6, 23375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Fu, Y.; Zhao, H.; Liu, X.; Wu, X.; Lin, T.; Wang, H.; Song, L.; Fang, Y.; Lu, W.; et al. Dynamic Insights into Increasing Antibiotic Resistance in Staphylococcus Aureus by Label-Free SERS Using a Portable Raman Spectrometer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 273, 121070. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, X.H.; Su, L.; Wang, H.Q.; Lin, T.F.; Fang, Y.P.; Zhao, H.M.; Lu, W.J.; Liu, M.J.; Liu, W.B.; et al. Rapid, Label-Free Prediction of Antibiotic Resistance in Salmonella Typhimurium by Surface-Enhanced Raman Spectroscopy. Int. J. Mol. Sci. 2022, 23, 1356. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.W.; Liu, R.; Matthews, D.L. Application of Laser Tweezers Raman Spectroscopy Techniques to the Monitoring of Single Cell Response to Stimuli. In Proceedings of the Biophotonics: Photonic Solutions for Better Health Care III; Popp, J., Drexler, W., Tuchin, V.V., Matthews, D.L., Eds.; SPIE: Bellingham, DC, USA, 2012; Volume 8427, p. 84270M. [Google Scholar]
- Wang, P.; Pang, S.; Zhang, H.; Fan, M.; He, L. Characterization of Lactococcus Lactis Response to Ampicillin and Ciprofloxacin Using Surface-Enhanced Raman Spectroscopy. Anal. Bioanal. Chem. 2016, 408, 933–941. [Google Scholar] [CrossRef]
- Hadjigeorgiou, K.; Kastanos, E.; Pitris, C.; Andreou, C. Surface Enhanced Raman Spectroscopy as a Sensitive Method for UTI Diagnosis. IEEE Sens. J. 2022, 22, 10063–10074. [Google Scholar] [CrossRef]
- Schröder, U.C.; Kirchhoff, J.; Hübner, U.; Mayer, G.; Glaser, U.; Henkel, T.; Pfister, W.; Fritzsche, W.; Popp, J.; Neugebauer, U. On-Chip Spectroscopic Assessment of Microbial Susceptibility to Antibiotics within 3.5 Hours. J. Biophotonics 2017, 10, 1547–1557. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Karanja, C.W.; Abutaleb, N.S.; Younis, W.; Zhang, X.; Seleem, M.N.; Cheng, J.X. Antibiotic Susceptibility Determination within One Cell Cycle at Single-Bacterium Level by Stimulated Raman Metabolic Imaging. Anal. Chem. 2018, 90, 3737–3743. [Google Scholar] [CrossRef]
- Kirchhoff, J.; Glaser, U.; Bohnert, J.A.; Pletz, M.W.; Popp, J.; Neugebauer, U. Simple Ciprofloxacin Resistance Test and Determination of Minimal Inhibitory Concentration within 2 h Using Raman Spectroscopy. Anal. Chem. 2018, 90, 1811–1818. [Google Scholar] [CrossRef]
- Schröder, U.C.; Beleites, C.; Assmann, C.; Glaser, U.; Hübner, U.; Pfister, W.; Fritzsche, W.; Popp, J.; Neugebauer, U. Detection of Vancomycin Resistances in Enterococci within 3 1/2; Hours. Sci. Rep. 2015, 5, 8217. [Google Scholar] [CrossRef] [PubMed]
- Novelli-Rousseau, A.; Espagnon, I.; Filiputti, D.; Gal, O.; Douet, A.; Mallard, F.; Josso, Q. Culture-Free Antibiotic-Susceptibility Determination from Single-Bacterium Raman Spectra. Sci. Rep. 2018, 8, 3957. [Google Scholar] [CrossRef]
- Tao, Y.; Wang, Y.; Huang, S.; Zhu, P.; Huang, W.E.; Ling, J.; Xu, J. Metabolic-Activity-Based Assessment of Antimicrobial Effects by D2O-Labeled Single-Cell Raman Microspectroscopy. Anal. Chem. 2017, 89, 4108–4115. [Google Scholar] [CrossRef] [PubMed]
- Moritz, T.J.; Taylor, D.S.; Polage, C.R.; Krol, D.M.; Lane, S.M.; Chan, J.W. Effect of Cefazolin Treatment on the Nonresonant Raman Signatures of the Metabolic State of Individual Escherichia Coli Cells. Anal. Chem. 2010, 82, 2703–2710. [Google Scholar] [CrossRef]
- Moritz, T.J.; Polage, C.R.; Taylor, D.S.; Krol, D.M.; Lane, S.M.; Chan, J.W. Evaluation of Escherichia Coli Cell Response to Antibiotic Treatment by Use of Raman Spectroscopy with Laser Tweezers. J. Clin. Microbiol. 2010, 48, 4287–4290. [Google Scholar] [CrossRef] [PubMed]
- Pilát, Z.; Bernatová, S.; Ježek, J.; Kirchhoff, J.; Tannert, A.; Neugebauer, U.; Samek, O.; Zemánek, P. Microfluidic Cultivation and Laser Tweezers Raman Spectroscopy of E. Coli under Antibiotic Stress. Sensors 2018, 18, 1623. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Shi, L.; Shao, L.; Fu, P.; Wang, K.; Xiao, R.; Wang, S.; Gu, B. Rapid Identification and Antibiotic Susceptibility Test of Pathogens in Blood Based on Magnetic Separation and Surface-Enhanced Raman Scattering. Microchim. Acta 2019, 186, 475. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.Z.; Zhu, X.; Su, J.Q.; Ren, B.; Zhu, Y.G.; Cui, L. Rapid Antibiotic Susceptibility Testing of Pathogenic Bacteria Using Heavy-Water-Labeled Single-Cell Raman Spectroscopy in Clinical Samples. Anal. Chem. 2019, 91, 6296–6303. [Google Scholar] [CrossRef]
- Zhang, W.; Li, Y.; Yao, L.; Wu, F.; Chen, C.; Zhou, L.; Zheng, B.; Wang, P.; Hong, W. Rapid Antimicrobial Susceptibility Testing by Stimulated Raman Scattering Metabolic Imaging and Morphological Deformation of Bacteria. Anal. Chim. Acta 2021, 1168, 338622. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.K.; Huang, N.T. A Microfluidic Microwell Device Integrating Surface-Enhanced Raman Scattering for Rapid Antibiotic Susceptibility Test of Blood-Borne Pathogen. In Proceedings of the 14th Annual IEEE International Conference on Nano/Micro Engineered and Molecular Systems, NEMS 2019, Bangkok, Thailand, 11–14 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 277–281. [Google Scholar]
- Götz, T.; Dahms, M.; Kirchhoff, J.; Beleites, C.; Glaser, U.; Bohnert, J.A.; Pletz, M.W.; Popp, J.; Schlattmann, P.; Neugebauer, U. Automated and Rapid Identification of Multidrug Resistant Escherichia Coli against the Lead Drugs of Acylureidopenicillins, Cephalosporins, and Fluoroquinolones Using Specific Raman Marker Bands. J. Biophotonics 2020, 13, e202000149. [Google Scholar] [CrossRef]
- Neugebauer, U.; Schmid, U.; Baumann, K.; Holzgrabe, U.; Ziebuhr, W.; Kozitskaya, S.; Kiefer, W.; Schmitt, M.; Popp, J. Characterization of Bacterial Growth and the Influence of Antibiotics by Means of UV Resonance Raman Spectroscopy. Biopolymers 2006, 82, 306–311. [Google Scholar] [CrossRef]
- Barzan, G.; Sacco, A.; Mandrile, L.; Giovannozzi, A.M.; Brown, J.; Portesi, C.; Alexander, M.R.; Williams, P.; Hardie, K.R.; Rossi, A.M. New Frontiers against Antibiotic Resistance: A Raman-Based Approach for Rapid Detection of Bacterial Susceptibility and Biocide-Induced Antibiotic Cross-Tolerance. Sens. Actuators B Chem. 2020, 309, 127774. [Google Scholar] [CrossRef]
- Bi, L.; Wang, X.; Cao, X.; Liu, L.; Bai, C.; Zheng, Q.; Choo, J.; Chen, L. SERS-Active Au@Ag Core-Shell Nanorod (Au@AgNR) Tags for Ultrasensitive Bacteria Detection and Antibiotic-Susceptibility Testing. Talanta 2020, 220, 121397. [Google Scholar] [CrossRef]
- Yi, X.; Song, Y.; Xu, X.; Peng, D.; Wang, J.; Qie, X.; Lin, K.; Yu, M.; Ge, M.; Wang, Y.; et al. Development of a Fast Raman-Assisted Antibiotic Susceptibility Test (FRAST) for the Antibiotic Resistance Analysis of Clinical Urine and Blood Samples. Anal. Chem. 2021, 93, 5098–5106. [Google Scholar] [CrossRef]
- Gukowsky, J.C.; He, L. Development of a Portable SERS Method for Testing the Antibiotic Sensitivity of Foodborne Bacteria. J. Microbiol. Methods 2022, 198, 106496. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chen, Y.; Lin, K.; Zou, L.; Lu, X.; He, N.; Liu, R.; Zhang, S.; Shen, D.; Song, Z.; et al. Single Cell Raman Spectroscopy Deuterium Isotope Probing for Rapid Antimicrobial Susceptibility Test of Elizabethkingia spp. Front. Microbiol. 2022, 13, 876925. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.Y.; Lin, Y.C.; Cheng, W.C.; Lin, Y.T.; Teng, L.J.; Wang, J.K.; Wang, Y.L. Rapid Antibiotic Susceptibility Testing of Bacteria from Patients’ Blood via Assaying Bacterial Metabolic Response with Surface-Enhanced Raman Spectroscopy. Sci. Rep. 2020, 10, 12538. [Google Scholar] [CrossRef]
- Jayan, H.; Pu, H.; Sun, D.W. Recent Developments in Raman Spectral Analysis of Microbial Single Cells: Techniques and Applications. Crit. Rev. Food Sci. Nutr. 2022, 62, 4294–4308. [Google Scholar] [CrossRef] [PubMed]
- Korpela, M.T.; Kurittu, J.S.; Karvinen, J.T.; Karp, M.T. A Recombinant Escherichia Coli Sensor Strain for the Detection of Tetracyclines. Anal. Chem. 1998, 70, 4457–4462. [Google Scholar] [CrossRef]
- Melamed, S.; Lalush, C.; Elad, T.; Yagur-Kroll, S.; Belkin, S.; Pedahzur, R. A Bacterial Reporter Panel for the Detection and Classification of Antibiotic Substances. Microb. Biotechnol. 2012, 5, 536–548. [Google Scholar] [CrossRef]
- Bianchi, A.A.; Baneyx, F. Stress Responses as a Tool to Detect and Characterize the Mode of Action of Antibacterial Agents. Appl. Environ. Microbiol. 1999, 65, 5023–5027. [Google Scholar] [CrossRef] [PubMed]
- Urban, A.; Eckermann, S.; Fast, B.; Metzger, S.; Gehling, M.; Ziegelbauer, K.; Rübsamen-Waigmann, H.; Freiberg, C. Novel Whole-Cell Antibiotic Biosensors for Compound Discovery. Appl. Environ. Microbiol. 2007, 73, 6436–6443. [Google Scholar] [CrossRef] [PubMed]
- Ngo, H.T.; Vo-Dinh, T. SERS Biosensors for Point-of-Care Infectious Disease Diagnostics. In SERS for Point-of-Care and Clinical Applications; Elsevier: Amsterdam, The Netherlands, 2022; pp. 115–134. [Google Scholar]

| Organism | Antibiotic Exposure | Raman Features * | Raman Technique | Reference |
|---|---|---|---|---|
| E. coli | Kanamycin 16–64 µg·mL−1 2 h | r740 of susceptible bacteria decreases; r740 of resistant bacteria varies slightly | SERS | [89] |
| E. coli | Amikacin, Ciprofloxacin, Polymyxin, Tigecycline 2−2–2−5 µg·mL−1 2 h | I7342MIC < I734MIC < I734control < I7340.5MIC < I7340.25MIC | SERS | [90] |
| S. aureus | Ciprofloxacin, Chloramphenicol, Erythromycin, Vancomycin 2−2–2−5 µg·mL−1 2 h | |||
| E. coli | Ampicillin 4–32 µg·mL−1 3 h | I733, r733 of susceptible bacteria increase until the MIC is reached, and then decrease; I733, r733 of resistant bacteria vary slightly; | SERS | [91] |
| E. coli | Ampicillin 20 µg·mL−1 (5MIC) Up to 3 h; first features revealed after 20 min of antibiotic exposure | I725, I1095 decrease, which is accompanied by new SERS peak appearance over 90 min | SERS | [92] |
| S. aureus | Oxacillin 5 µg·mL−1 (5MIC) Up to 3 h; first features revealed after 50 min of antibiotic exposure | Appearance of new peaks at 50 min, sharp decline of I732, 10–20 min later, newly formed peaks disappeared, I732 recovery, 732 peak disappearance after 120–180 min | ||
| Gentamicin, Tetracycline Up to 12 h | Characteristic SERS response I732 was not noted until after 9–13 h of treatment | |||
| P. aeruginosa | Amikacin 0.25–6 µg·mL−1 Overnight incubation | I1607 decreases | UV resonance Raman | [93] |
| S. aureus | Oxacillin, Vancomycin 0.5–2 µg·mL−1 Up to 6 h for Oxacillin; Up to 2 h for Vancomycin; first features revealed within one hour | r730 decreases; r730 of resistant bacteria varies slightly | SERS | [94] |
| E. coli | Imipenem 0.03–012 µg·mL−1 Up to 6 h; first features revealed within one hour | r654, r724 decrease; r654, r724 of resistant bacteria vary slightly | SERS | |
| E. coli | Ampicillin 1 mg·mL−1 (excess) 3 h of incubation | I733 of susceptible bacteria increases; I733 of resistant bacteria varies slightly | SERS | [88] |
| S. aureus | Oxacillin, Cefazolin 0.125–32 µg·mL−1 21 days of exposure | I734/I867 decreases, I1372/I1349, I1163/I959 increase as antibiotic resistance develops | SERS | [95] |
| Salmonella typhimurium | Cefotaxime 0.5–4 µg·mL−1 50 days of exposure | I990/I1348 increases, I1165/I1205, I958/I1017 decrease as antibiotic resistance develops | SERS | [96] |
| Antibiotic | Target Organism | Raman Protocol | Reference |
|---|---|---|---|
| Ampicillin, Vancomycin, Cephotaxim, Oxacillin, Gentamicin, Tetracycline | S. aureus, E. coli | SERS; Raman microscope; Excitation at 632.8 nm; 105 W × cm−2; Ag/AAO substrate | [92] |
| Amikacin | P. aeruginosa | RRS; Raman spectrometer; Excitation at 244 nm; 0.5 mW; CaF2 substrate | [93] |
| Penicillin/streptomycin | E. coli | NR; laser tweezer Raman microscope; Excitation at 785 nm; 28 mW; Cell chamber with a fused-silica microscope coverslip | [97] |
| Ampicillin, Ciprofloxacin | L. lactis | SERS; Raman microscope; Excitation at 780 nm; 1 mW; 50 nm citrate-capped AgNPs on a gold-stained glass slide substrate | [98] |
| Oxacillin, Imipenem, Vancomycin | S. aureus, E. coli, A. baumannii, K. pneumoniae | SERS; Raman microscope; Excitation at 632.8 nm; 105 mW × cm−2; two-dimensional hexagonally packed AgNPs embedded in nanochannels of anodic aluminum oxide substrate | [94] |
| Augmentin, Amoxicillin, Cefaclor, Cefuroxime, Cefazolin, Ceftriaxone, Ciprofloxacin | Proteus sp., K. pneumoniae, E. coli | SERS; portable Raman spectrometer; Excitation at 532 nm; 50 mW; 100 μm AgNPs substrate | [99] |
| Ciprofloxacin | E. coli | NR; Raman microscope; Excitation at 532 nm; 36 mW; DEP (dielectrophoresis)-based microfluidic device | [100] |
| Vancomycin, Linezolid, Daptomycin, Gentamicin, Erythromycin | E. faecalis, E. coli, K. pneumoniae, S. aureus | SRS; custom-built dual-laser Raman microscope; Excitation at 847 + 1040 nm; Agar gel pad on coverglass | [101] |
| Ciprofloxacin | E. coli | NR; confocal Raman microscope; Excitation at 532 nm; 15 mW; DEP (dielectrophoresis) chip | [102] |
| Vancomycin | E. faecalis, E. faecium | NR; confocal Raman microscope; Excitation at 532 nm; 15 mW DEP (dielectrophoresis) chip | [103] |
| Gentamicin, Ciprofloxacin, Amoxicillin | E. coli | NR; confocal Raman microscope; Excitation at 532 nm; 9 mW; Coverslip substrate | [104] |
| Ampicillin | S. mutans | NR; Raman microscope; Excitation at 532 nm; 3–5 mW; CaF2 substrate | [105] |
| Cefazolin | E. coli | NR; laser tweezer Raman microscope; Excitation at 785 nm; 28 mW; Cell chamber with a fused-silica microscope coverslip | [106] |
| Penicillin, G-streptomycin, Cefazolin | E. coli | NR; laser tweezer Raman microscope; Excitation at 785 nm; 28 mW; Cell chamber with a fused-silica microscope coverslip | [107] |
| Cefotaxime | E. coli | NR; laser tweezer Raman microscope; Excitation at 785 nm, 532 nm; 150 mW; Microfluidic chip | [108] |
| Oxacillin, Imipenem, Methicillin | S. aureus, A. baumannii, P. aeruginosa | SERS; portable Raman spectrometer; Excitation at 785 nm; 25 mW; AgNPs SERS substrate | [109] |
| Kanamycin | E. coli | SERS; Raman microscope; Excitation at 632.8 nm; 5 mW; Silver island film sputtered substrate in a microfluidic system | [89] |
| Ampicillin, Chloramphenicol, Kanamycin, Meropenem | E. coli, P. vulgaris, S. entérica, S. flexneri,K. variicola, E. fergusonii, P. rettgeri | NR; confocal Raman microscope; Excitation at 532 nm; D2O-labeling | [110] |
| Cefotaxime | E. coli | SRS; custom-built dual-laser Raman microscope; Excitation at 852 nm (~20 mW) + 1045 nm (~300 mW); Coverslip substrate | [111] |
| Kanamycin | E. coli | SERS; Raman microscope; Excitation at 632.8 nm; 50 mW; Microfluidic microwell device AgNP@AAO substrate | [112] |
| Ciprofloxacin, Cefotaxime, Piperacillin | E. coli | NR; confocal Raman microscope; Excitation at 532 nm; DEP setup | [113] |
| Ciprofloxacin | B. pumilus | RRS; Raman microscope; Excitation at 244 nm; 32 mW | [114] |
| Amikacin, Ciprofloxacin, Polymyxin B, Tigecycline, Ciprofloxacin, Chloramphenicol, Erythromycin, Vancomycin | E. coli, S. aureus | SERS; Raman microscope; Excitation at 532 nm; 14 mW; Bacteria-aptamer@AgNPs substrate | [90] |
| Ciprofloxacin | E. coli | NR; confocal Raman microscope; Excitation at 532 nm; 10 mW; DEP microfluidic device | [115] |
| Ampicillin | E. coli | SERS; confocal Raman microscope; Excitation at 785 nm; 20 mW; Au@AgNR tag substrate | [116] |
| Ampicillin | E. coli | SERS; Raman microscope; Excitation at 632.8 nm; 5 mW; Microfluidic microwell device substrate | [91] |
| Amikacin, Azithromycin, Aztreonam, Cefazolin, Cefepime, Cefmetazole Na, Cefoperazone/sulbactam, Cefoxitin, Ceftazidime, Ceftazidime/avibactam, Ceftolozane/tazobactam, Ceftriaxone, Cefuroxime, Ciprofloxacin, Clindamycin, Doxycycline, Ertapenem, Erythromycin, Gentamicin, Imipenem, Levofloxacin, Linezolid, Meropenem, Minocycline, Moxifloxacin, Nitrofurantoin, Nitrofurantoin, Oxacillin, Penicillin, Piperacillin, Piperacillin/tazobactam, Polymyxin, Rifampicin, Teicoplanin, Tetracycline, Ticarcillin/clavulanic acid, Tigecycline, Tobramycin, Tobramycin, Trimethoprim−sulfamethoxazole, Vancomycin | E. coli, P. aeruginosa, K. pneumoniae, E. faecium, S. aureus, S. epidermidis, S. hominis | NR; confocal Raman microscope; Excitation at 532 nm; D2O-labeling; Aluminum-coated slide substrate | [117] |
| Ampicillin | E. coli | SERS; confocal Raman microscope; Excitation at 780 nm; 4 mW; 55 nm AuNPs substrate | [88] |
| Oxacillin, Cefazolin | S. aureus | SERS; portable Raman spectrometer; Excitation at 785 nm; 200 mW; 50 nm AuNPs substrate | [95] |
| Cefotaxime | S. typhimurium | SERS; portable Raman spectrometer; Excitation at 785 nm; 200 mW; 40–60 nm AuNPs substrate | [96] |
| Ampicillin, Neomycin, Chlortetracycline | Escherichia coli, Bacillus cereus, Salmonella enterica | SERS; portable Raman spectrometer; Excitation wavelength not reported; 55 nm AuNPs substrate | [118] |
| Minocycline, Levofloxacin | Elizabethkingia spp. | NR; confocal Raman microscope; Excitation wavelength not reported; 5 mW; Aluminum-coated slide substrate | [119] |
| Oxacillin, Cefotaxime | S. aureus E. coli | SERS; Raman microscope; Excitation at 632.8 nm 105 Mw × cm−2; AgNPs array embedded in nanochannels of anodic aluminum oxide substrate | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikov, A.; Sayfutdinova, A.; Botchkova, E.; Kopitsyn, D.; Fakhrullin, R. Antibiotic Susceptibility Testing with Raman Biosensing. Antibiotics 2022, 11, 1812. https://doi.org/10.3390/antibiotics11121812
Novikov A, Sayfutdinova A, Botchkova E, Kopitsyn D, Fakhrullin R. Antibiotic Susceptibility Testing with Raman Biosensing. Antibiotics. 2022; 11(12):1812. https://doi.org/10.3390/antibiotics11121812
Chicago/Turabian StyleNovikov, Andrei, Adeliya Sayfutdinova, Ekaterina Botchkova, Dmitry Kopitsyn, and Rawil Fakhrullin. 2022. "Antibiotic Susceptibility Testing with Raman Biosensing" Antibiotics 11, no. 12: 1812. https://doi.org/10.3390/antibiotics11121812
APA StyleNovikov, A., Sayfutdinova, A., Botchkova, E., Kopitsyn, D., & Fakhrullin, R. (2022). Antibiotic Susceptibility Testing with Raman Biosensing. Antibiotics, 11(12), 1812. https://doi.org/10.3390/antibiotics11121812









