Exposure of Pseudomonas aeruginosa to Cinnamaldehyde Selects Multidrug Resistant Mutants
Abstract
1. Introduction
2. Results
2.1. In Vitro Selection of Mutants Showing an Increase Resistance to CNA
2.2. CNA Can Select Multidrug Resistance Mutants
2.3. Genetic Traits of CNA Resistant Mutants
2.4. Role of MutS/L in Emergence of CNA-Resistant Mutants
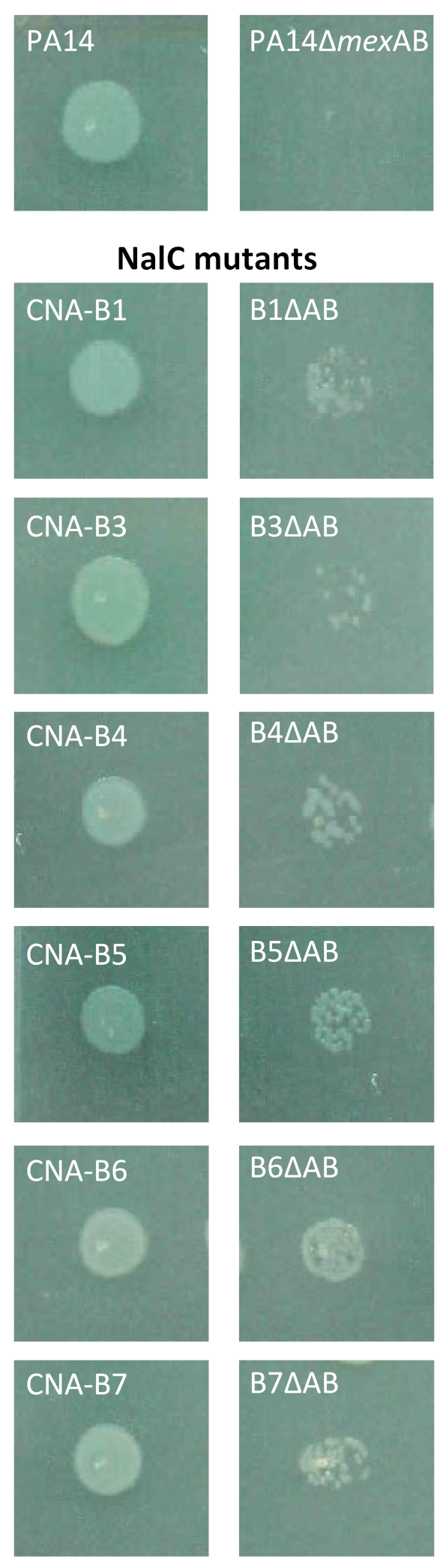
2.5. Role of NalC in Bacterial Adaptation to CAN
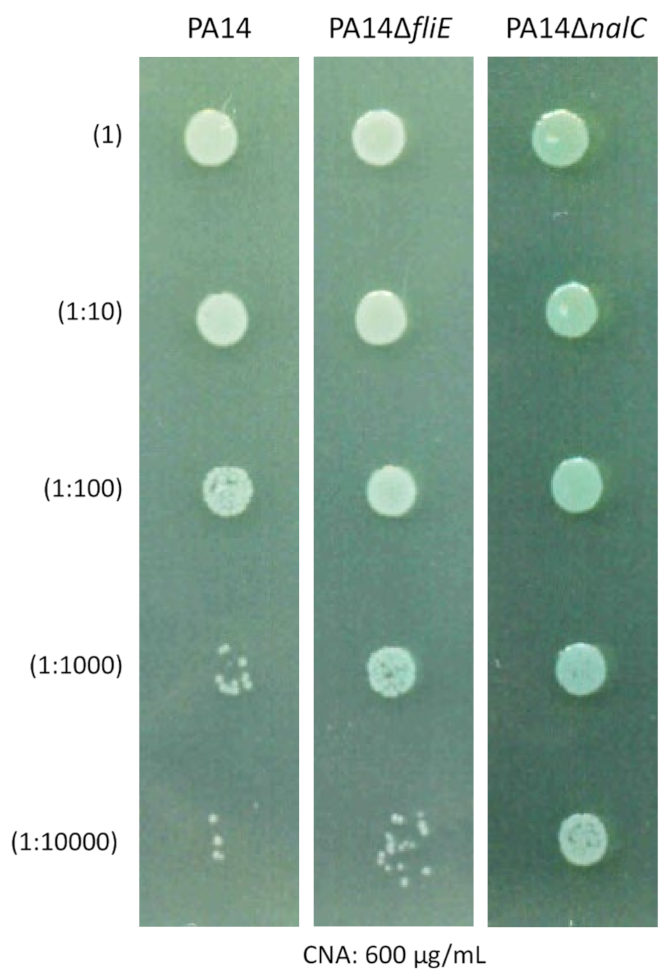
2.6. Role of Bacterial Motility in CNA Adaptation
3. Discussion
4. Material and Methods
4.1. Bacterial Strains, Plasmids and Growth Conditions
4.2. Mutagenesis Frequency Evaluation
4.3. Selection of PA14 CNA Adaptation Mutants
4.4. Drug Susceptibility Testing
4.5. RT-qPCR Experiments
4.6. Construction of PA14 fliE-Defective Mutant
4.7. Genome Sequencing and in Silico Analysis
4.8. Motility and Cell Adhesion Assay
4.8.1. Swarming
4.8.2. Detection of Biofilm Formation by Adherence Test
4.9. Metabolite Extraction and Detection Using Thin Layer Chromatography (TLC)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef]
- Inouye, S.; Takizawa, T.; Yamaguchi, H. Antibacterial activity of essential oils and their major constituents against respiratory tract pathogens by gaseous contact. J. Antimicrob. Chemother. 2001, 47, 565–573. [Google Scholar] [CrossRef]
- Gadisa, E.; Weldearegay, G.; Desta, K.; Tsegaye, G.; Hailu, S.; Jote, K.; Takele, A. Combined antibacterial effect of essential oils from three most commonly used Ethiopian traditional medicinal plants on multidrug resistant bacteria. BMC Complement. Altern. Med. 2019, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Castaneda-Correa, A.; Corral-Luna, A.; Hume, M.E.; Anderson, R.C.; Ruiz-Barrera, O.; Castillo-Castillo, Y.; Rodriguez-Almeida, F.; Salinas-Chavira, J.; Arzola-Alvarez, C. Effects of thymol and carvacrol, alone or in combination, on fermentation and microbial diversity during in vitro culture of bovine rumen microbes. J. Environ. Sci. Health B 2019, 54, 170–175. [Google Scholar] [CrossRef]
- Langeveld, W.T.; Veldhuizen, E.J.; Burt, S.A. Synergy between essential oil components and antibiotics: A review. Crit. Rev. Microbiol. 2014, 40, 76–94. [Google Scholar] [CrossRef]
- Lv, F.; Liang, H.; Yuan, Q.P.; Li, C.F. In vitro antimicrobial effects and mechanism of action of selected plant essential oil combinations against four food-related microorganisms. Food Res. Int. 2011, 44, 3057–3064. [Google Scholar] [CrossRef]
- Lorenzi, V.; Muselli, A.; Bernardini, A.F.; Berti, L.; Pages, J.M.; Amaral, L.; Bolla, J.M. Geraniol restores antibiotic activities against multidrug-resistant isolates from Gram-negative species. Antimicrob. Agents Chemother. 2009, 53, 2209–2211. [Google Scholar] [CrossRef]
- Qiu, J.; Feng, H.; Lu, J.; Xiang, H.; Wang, D.; Dong, J.; Wang, J.; Wang, X.; Liu, J.; Deng, X. Eugenol reduces the expression of virulence-related exoproteins in Staphylococcus aureus. Appl. Environ. Microbiol. 2010, 76, 5846–5851. [Google Scholar] [CrossRef]
- Wendakoon, C.N.; Sakaguchi, M. Inhibition of amino acid decarboxylase activity of Enterobacter aerogenes by active components in spices. J. Food Prot. 1995, 58, 280–283. [Google Scholar] [CrossRef]
- Sibanda, T.; Okoh, A.I. The challenges of overcoming antibiotic resistance: Plant extracts as potential sources of antimicrobial and resistance modifying agents. Afr. J. Biotechnol. 2007, 6, 2886–2896. [Google Scholar]
- Mann, C.M.; Cox, S.D.; Markham, J.L. The outer membrane of Pseudomonas aeruginosa NCTC 6749 contributes to its tolerance to the essential oil of Melaleuca alternifolia (tea tree oil). Lett. Appl. Microbiol. 2000, 30, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, C.J.; Carson, C.F.; Chang, B.J.; Riley, T.V. Role of the MexAB-OprM efflux pump of Pseudomonas aeruginosa in tolerance to tea tree (Melaleuca alternifolia) oil and its monoterpene components Terpinen-4-ol, 1,8-Cineole, and α-Terpineol. Appl. Environ. Microbiol. 2008, 74, 1932–1935. [Google Scholar] [CrossRef] [PubMed]
- Tetard, A.; Zedet, A.; Girard, C.; Plésiat, P.; Llanes, C. Cinnamaldehyde induces expression of efflux pumps and multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2019, 63, e01081-19. [Google Scholar] [CrossRef] [PubMed]
- Blanco, P.; Hernando-Amado, S.; Reales-Calderon, J.A.; Corona, F.; Lira, F.; Alcalde-Rico, M.; Bernardini, A.; Sanchez, M.B.; Martinez, J.L. Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms 2016, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Becerril, R.; Nerin, C.; Gomez-Lus, R. Evaluation of bacterial resistance to essential oils and antibiotics after exposure to oregano and cinnamon essential oils. Foodborne Pathog. Dis. 2012, 9, 699–705. [Google Scholar] [CrossRef]
- Lu, M.; Dai, T.; Murray, C.K.; Wu, M.X. Bactericidal property of oregano oil against multidrug-resistant clinical isolates. Front. Microbiol. 2018, 9, 2329. [Google Scholar] [CrossRef]
- Utchariyakiat, I.; Surassmo, S.; Jaturanpinyo, M.; Khuntayaporn, P.; Chomnawang, M.T. Efficacy of cinnamon bark oil and cinnamaldehyde on anti-multidrug resistant Pseudomonas aeruginosa and the synergistic effects in combination with other antimicrobial agents. BMC Complement. Altern. Med. 2016, 16, 158. [Google Scholar] [CrossRef]
- Oliver, A.; Baquero, F.; Blazquez, J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: Molecular characterization of naturally occurring mutants. Mol. Microbiol. 2002, 43, 1641–1650. [Google Scholar] [CrossRef]
- Oliver, A.; Canton, R.; Campo, P.; Baquero, F.; Blazquez, J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 2000, 288, 1251–1254. [Google Scholar] [CrossRef]
- Daigle, D.M.; Cao, L.; Fraud, S.; Wilke, M.S.; Pacey, A.; Klinoski, R.; Strynadka, N.C.; Dean, C.R.; Poole, K. Protein modulator of multidrug efflux gene expression in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 5441–5451. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martinez-Bueno, M.; Molina-Henares, A.J.; Teran, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef]
- Li, X.Z.; Plésiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [PubMed]
- Schniederberend, M.; Williams, J.F.; Shine, E.; Shen, C.; Jain, R.; Emonet, T.; Kazmierczak, B.I. Modulation of flagellar rotation in surface-attached bacteria: A pathway for rapid surface-sensing after flagellar attachment. PLoS Pathog. 2019, 15, e1008149. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A.; Kolter, R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 1998, 30, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Marmont, L.S.; Whitfield, G.B.; Rich, J.D.; Yip, P.; Giesbrecht, L.B.; Stremick, C.A.; Whitney, J.C.; Parsek, M.R.; Harrison, J.J.; Howell, P.L. PelA and PelB proteins form a modification and secretion complex essential for Pel polysaccharide-dependent biofilm formation in Pseudomonas aeruginosa. J. Biol. Chem. 2017, 292, 19411–19422. [Google Scholar] [CrossRef]
- Fadli, M.; Chevalier, J.; Hassani, L.; Mezrioui, N.E.; Pages, J.M. Natural extracts stimulate membrane-associated mechanisms of resistance in Gram-negative bacteria. Lett. Appl. Microbiol. 2014, 58, 472–477. [Google Scholar] [CrossRef]
- Pesingi, P.V.; Singh, B.R.; Pesingi, P.K.; Bhardwaj, M.; Singh, S.V.; Kumawat, M.; Sinha, D.K.; Gandham, R.K. MexAB-OprM efflux pump of Pseudomonas aeruginosa offers resistance to carvacrol: A herbal antimicrobial agent. Front. Microbiol. 2019, 10, 2664. [Google Scholar] [CrossRef]
- Ghosh, S.; Cremers, C.M.; Jakob, U.; Love, N.G. Chlorinated phenols control the expression of the multidrug resistance efflux pump MexAB-OprM in Pseudomonas aeruginosa by interacting with NalC. Mol. Microbiol. 2011, 79, 1547–1556. [Google Scholar] [CrossRef]
- Starr, L.M.; Fruci, M.; Poole, K. Pentachlorophenol induction of the Pseudomonas aeruginosa mexAB-oprM efflux operon: Involvement of repressors NalC and MexR and the antirepressor ArmR. PLoS ONE 2012, 7, e32684. [Google Scholar] [CrossRef]
- Braz, V.S.; Furlan, J.P.; Fernandes, A.F.; Stehling, E.G. Mutations in NalC induce MexAB-OprM overexpression resulting in high level of aztreonam resistance in environmental isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2016, 363, fnw166. [Google Scholar] [CrossRef]
- Llanes, C.; Hocquet, D.; Vogne, C.; Benali-Baitich, D.; Neuwirth, C.; Plésiat, P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 2004, 48, 1797–1802. [Google Scholar] [CrossRef] [PubMed]
- Wada, A.; Prates, E.T.; Hirano, R.; Werner, A.Z.; Kamimura, N.; Jacobson, D.A.; Beckham, G.T.; Masai, E. Characterization of aromatic acid/proton symporters in Pseudomonas putida KT2440 toward efficient microbial conversion of lignin-related aromatics. Metab. Eng. 2021, 64, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Rundell, E.A.; Commodore, N.; Goodman, A.L.; Kazmierczak, B.I. A Screen for antibiotic resistance determinants reveals a fitness cost of the flagellum in Pseudomonas aeruginosa. J. Bacteriol. 2020, 202, e00682-19. [Google Scholar] [CrossRef] [PubMed]
- Valentin, J.D.P.; Straub, H.; Pietsch, F.; Lemare, M.; Ahrens, C.H.; Schreiber, F.; Webb, J.S.; van der Mei, H.C.; Ren, Q. Role of the flagellar hook in the structural development and antibiotic tolerance of Pseudomonas aeruginosa biofilms. ISME J. 2022, 16, 1176–1186. [Google Scholar] [CrossRef]
- Rosche, W.A.; Foster, P.L. Determining mutation rates in bacterial populations. Methods 2000, 20, 4–17. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020; Volume M100. [Google Scholar]
- Richardot, C.; Juarez, P.; Jeannot, K.; Patry, I.; Plésiat, P.; Llanes, C. Amino acid substitutions account for most MexS alterations in clinical nfxC mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 2302–2310. [Google Scholar] [CrossRef]
- Llanes, C.; Pourcel, C.; Richardot, C.; Plésiat, P.; Fichant, G.; Cavallo, J.D.; Merens, A.; Group, G.S. Diversity of beta-lactam resistance mechanisms in cystic fibrosis isolates of Pseudomonas aeruginosa: A French multicentre study. J. Antimicrob. Chemother. 2013, 68, 1763–1771. [Google Scholar] [CrossRef]
- Joshi, N.; Sickle, F. A Sliding-Window, Adaptaive, Quality-Based Trimming Tool for FastQ Files, Version 1.33. 2011. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5 6 1–5 6 37. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, P.; Soscia, C.; Voulhoux, R.; Filloux, A. PelC is a Pseudomonas aeruginosa outer membrane lipoprotein of the OMA family of proteins involved in exopolysaccharide transport. Biochimie 2007, 89, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Liberati, N.T.; Urbach, J.M.; Miyata, S.; Lee, D.G.; Drenkard, E.; Wu, G.; Villanueva, J.; Wei, T.; Ausubel, F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 2833–2838. [Google Scholar] [CrossRef]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in Gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef]
- Lacks, S.; Greenberg, B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol. 1977, 114, 153–168. [Google Scholar] [CrossRef]
- Ditta, G.; Stanfield, S.; Corbin, D.; Helinski, D.R. Broad host range DNA cloning system for gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 1980, 77, 7347–7351. [Google Scholar] [CrossRef]
- Kaniga, K.; Delor, I.; Cornelis, G.R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: Inactivation of the blaA gene of Yersinia enterocolitica. Gene 1991, 109, 137–141. [Google Scholar] [CrossRef]
- Hocquet, D.; Nordmann, P.; El Garch, F.; Cabanne, L.; Plésiat, P. Involvement of the MexXY-OprM efflux system in emergence of cefepime resistance in clinical strains of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006, 50, 1347–1351. [Google Scholar] [CrossRef]
- Jeannot, K.; Sobel, M.L.; El Garch, F.; Poole, K.; Plésiat, P. Induction of the MexXY efflux pump in Pseudomonas aeruginosa is dependent on drug-ribosome interaction. J. Bacteriol. 2005, 187, 5341–5346. [Google Scholar] [CrossRef] [PubMed]
- Dumas, J.L.; van Delden, C.; Perron, K.; Köhler, T. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol. Lett. 2006, 254, 217–225. [Google Scholar] [CrossRef] [PubMed]

| Strains/Mutants a | Sequence | Transcript Levels d | MICs (µg/mL) of Antibiotics and CNA e | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nalC | mexB | armR | mexY | mexC | mexE | TIC | AZT | CIP | IPM | CST | GEN | CNA | |
| PA14 | WT | 1 | 1 | 1 | 1 | 1 | 32 | 4 | 0.06 | 0.5 | 1 | 0.5 | 700 |
| PA14 + CNA b | WT | 3.2 | 43 | 5.5 | 118 | 100 | 64 | 16 | 1 | 0.5 | 2 | 2 | - |
| PA14mexAB+ c | WT | 6.5 | 1 | ND | ND | ND | 128 | 16 | 0.25 | 0.5 | 1 | 0.5 | 900 |
| PA14∆AB | WT | - | ND | ND | ND | ND | 16 | ≤0.5 | 0.06 | 0.5 | 1 | 0.5 | 500 |
| CNA-A1 | −A486 | 3.4 | 61 | 0.2 | 0.5 | 1.3 | 128 | 32 | 0.25 | 0.5 | 0.5 | 0.125 | 900 |
| CNA-A2 | T24P | 2.5 | 24 | 0.1 | 0.1 | 0.2 | 256 | 16 | 0.25 | 0.5 | 0.5 | 0.25 | 900 |
| CNA-A3 | WT | 1.3 | 1 | 0.5 | 0.7 | 0.4 | 64 | 8 | 0.06 | 0.5 | 0.25 | 0.125 | 800 |
| CNA-B1 | +C450 | 4.9 | 164 | 0.3 | 0.4 | 0.8 | >256 | 64 | 0.5 | 0.5 | 0.5 | 0.25 | 1000 |
| CNA-B2 | WT | 1.2 | 1 | 0.5 | 0.6 | 0.5 | 64 | 16 | 0.06 | 0.5 | 0.5 | 0.25 | 800 |
| CNA-B3 | +C450 | 5.7 | 146 | 1.1 | 0.4 | 0.9 | >256 | 64 | 1 | 1 | 4 | 1 | 1000 |
| CNA-B4 | +C450 | 3.3 | 130 | 0.4 | 0.5 | 0.9 | >256 | 64 | 0.5 | 2 | 1 | 0.125 | 1100 |
| CNA-B5 | +C450 | 6.0 | 208 | 1.1 | 0.7 | 0.9 | >256 | 64 | 0.5 | 2 | 4 | 1 | 1000 |
| CNA-B6 | +C450 | 5.0 | 136 | 0.7 | 0.6 | 0.8 | >256 | 64 | 0.5 | 1 | 1 | 0.5 | 1100 |
| CNA-B7 | +C450 | 7.6 | 423 | 2.5 | 0.6 | 1.6 | >256 | 64 | 1 | 2 | 4 | 1 | 1100 |
| Proteins | A-Type Mutants (CNA MIC in µg/mL) | B-Type Mutants (CNA MIC in µg/mL) | Protein Functions | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CNA-A1 (900) | CNA-A2 (900) | CNA-A3 (800) | CNA-B1 (1000) | CNA-B2 (800) | CNA-B3 (1000) | CNA-B4 (1100) | CNA-B5 (1000) | CNA-B6 (1100) | CNA-B7 (1100) | ||
| DNA mismatch repair | |||||||||||
| MutS | −9 pb | Q589R | −9 pb | −9 pb | −9 pb | −9 pb | −9 pb | DNA mismatch repair protein MutS | |||
| Transmembrane transport | |||||||||||
| NalC | -A486 | T24P | +C450 | +GG442 | +C450 | +C450 | +C450 | +C450 | Transcriptional repressor NalC of MexAB-OprM pump | ||
| DppB | G114D | G114D | G114D | G114D | G114D | G114D | Dipeptide ABC transporter permease DppB | ||||
| Energy production | |||||||||||
| AtpD | P305S | F0F1 ATP synthase beta subunit | |||||||||
| AtpA | D290G | D290G | F0F1 ATP synthase alpha subunit | ||||||||
| AtpE | T23A | F0F1 ATP synthase C subunit | |||||||||
| Response to oxidative stress | |||||||||||
| MxtR | W1071* | W1071* | L1068P | Orphan sensor kinase MxtR | |||||||
| YbbN | F86S | F86S | F86S | F86S | F86S | Thioredoxin (oxidoreductase) | |||||
| Response to chemicals | |||||||||||
| PcaB | E239G | E239G | E239G | 3-carboxy-cis, cis-muconate cycloisomerase | |||||||
| TlpQ | L561P | L561P | L561P | L561P | L561P | L561P | Chemotaxis transducer TlpQ | ||||
| Motility | |||||||||||
| FliE | Q54* | Q54* | Q54* | Flagellar hook-basal body protein FliE | |||||||
| FlgJ | Q341R | Q341R | Flagellar rod assembly protein FlgJ | ||||||||
| FlgG | P83L | Flagellar basal body rod protein FlgG | |||||||||
| PilJ | T403A | T403A | Twitching motility protein | ||||||||
| Nucleotide–sugar metabolic process | |||||||||||
| RbsR | V320M | V320M | V320M | V320M | V320M | Ribose operon repressor RbsR | |||||
| Unknown functions | |||||||||||
| PA0841 | A304V | A304V | Hypothetical protein | ||||||||
| PA3283 | N142S | N142S | N142S | Hypothetical protein | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tetard, A.; Gaillot, S.; Dubois, E.; Aarras, S.; Valot, B.; Phan, G.; Plésiat, P.; Llanes, C. Exposure of Pseudomonas aeruginosa to Cinnamaldehyde Selects Multidrug Resistant Mutants. Antibiotics 2022, 11, 1790. https://doi.org/10.3390/antibiotics11121790
Tetard A, Gaillot S, Dubois E, Aarras S, Valot B, Phan G, Plésiat P, Llanes C. Exposure of Pseudomonas aeruginosa to Cinnamaldehyde Selects Multidrug Resistant Mutants. Antibiotics. 2022; 11(12):1790. https://doi.org/10.3390/antibiotics11121790
Chicago/Turabian StyleTetard, Alexandre, Susie Gaillot, Eline Dubois, Soumaya Aarras, Benoît Valot, Gilles Phan, Patrick Plésiat, and Catherine Llanes. 2022. "Exposure of Pseudomonas aeruginosa to Cinnamaldehyde Selects Multidrug Resistant Mutants" Antibiotics 11, no. 12: 1790. https://doi.org/10.3390/antibiotics11121790
APA StyleTetard, A., Gaillot, S., Dubois, E., Aarras, S., Valot, B., Phan, G., Plésiat, P., & Llanes, C. (2022). Exposure of Pseudomonas aeruginosa to Cinnamaldehyde Selects Multidrug Resistant Mutants. Antibiotics, 11(12), 1790. https://doi.org/10.3390/antibiotics11121790







