Multidrug-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections among Patients Attending Health Facilities in Mwanza and Dar es Salaam, Tanzania
Abstract
1. Introduction
2. Results
2.1. Patients’ Socio-Demographic and Clinical Characteristics
2.2. Prevalence of Causing Community Acquired Urinary Tract Infections and Distribution of Causative Pathogens
2.3. Percentages of Antibiotic-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections
2.4. Prevalence and Patterns of MDR Bacteria Causing Community Acquired Urinary Tract Infections
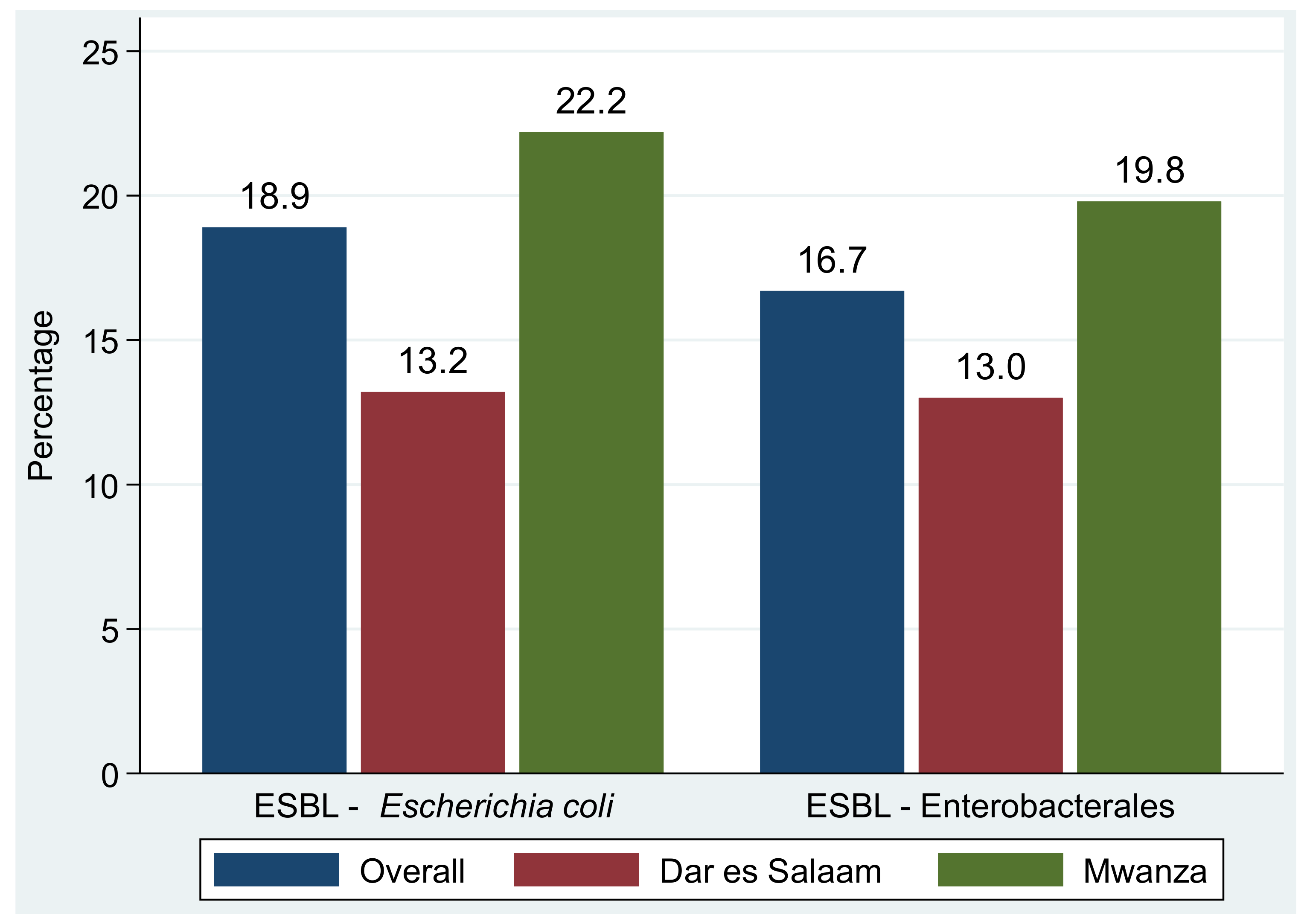
2.5. Prevalence and Types of MDR Phenotypes Causing Community Acquired Urinary Tract Infections
3. Discussion
4. Materials and Methods
4.1. Study Design and Setting
4.2. Study Population
4.3. Data and Sample Collection
4.4. Laboratory Procedures
4.4.1. Quantitative Urine Culture
4.4.2. Bacteria Identification
4.4.3. Antibiotic Susceptibility Testing
4.4.4. Quality Control measures
4.4.5. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donkor, E.S.; Horlortu, P.Z.; Dayie, N.T.; Obeng-Nkrumah, N.; Labi, A.K. Community acquired urinary tract infections among adults in Accra, Ghana. Infect. Drug. Resist. 2019, 12, 2059. [Google Scholar] [CrossRef] [PubMed]
- Erdem, I.; Ali, R.K.; Ardic, E.; Omar, S.E.; Mutlu, R.; Topkaya, A.E. Community-acquired Lower Urinary Tract Infections: Etiology, Antimicrobial Resistance, and Treatment Results in Female Patients. J. Glob. Infect. Dis. 2018, 10, 129–132. [Google Scholar] [CrossRef] [PubMed]
- Kabugo, D.; Kizito, S.; Ashok, D.D.; Kiwanuka, A.G.; Nabimba, R.; Namunana, S.; Najjuka, F.C. Factors associated with community-acquired urinary tract infections among adults attending assessment centre, Mulago Hospital Uganda. Afr. Health Sci. 2016, 16, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Shalini, M.A.; Prabhakar, K.; Lakshmi, S.Y. A Study on prevalence and evaluation of clinical isolates from community acquired infections using different media in semiurban areas. World J. Med. Sci. 2010, 5, 49–53. [Google Scholar]
- Wagnleher, F.; Naber, K. Treatment of bacterial urinary tract infection: Presence and Future. Eur. Urol. 2005, 49, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Kranz, J.; Schmidt, S.; Lebert, C.; Schneidewind, L.; Schmiemann, G.; Wagenlehner, F. Uncomplicated Bacterial Community-acquired Urinary Tract Infection in Adults: Epidemiology, Diagnosis, Treatment, and Prevention. Dtsch. Arztebl. Int. 2017, 114, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Durkin, M.J.; Keller, M.; Butler, A.M.; Kwon, J.H.; Dubberke, E.R.; Miller, A.C.; Olsen, M.A. An Assessment of Inappropriate Antibiotic Use and Guideline Adherence for Uncomplicated Urinary Tract Infections. Open Forum Infect. Dis. 2018, 5, ofy198. [Google Scholar] [CrossRef]
- Larson, E. Community factors in the development of antibiotic resistance. Annu. Rev. Public Health 2007, 28, 435–447. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Masenga, G.G.; Shayo, B.C.; Msuya, S.; Rasch, V. Urinary incontinence and its relation to delivery circumstances: A population-based study from rural Kilimanjaro, Tanzania. PLoS ONE 2019, 14, e0208733. [Google Scholar] [CrossRef]
- The United Republic of Tanzania Ministry of Health and Social Welfare. Standard Treatment Guidelines and Essential Medicines List, 4th ed. 2013. Available online: https://www.pascar.org/uploads/files/Tanzania_-_Standard_Treatment_Guidelines_and_Essential_Medicines_List_-_Fourth_Edition.pdf (accessed on 5 May 2022).
- Horumpende, P.G.; Sonda, T.B.; van Zwetselaar, M.; Antony, M.L.; Tenu, F.F.; Mwanziva, C.E.; Chilongola, J.O. Prescription and non-prescription antibiotic dispensing practices in part I and part II pharmacies in Moshi Municipality, Kilimanjaro Region in Tanzania: A simulated clients approach. PLoS ONE 2018, 13, e0207465. [Google Scholar] [CrossRef]
- Ndaki, P.M.; Mushi, M.F.; Mwanga, J.R.; Konje, E.T.; Ntinginya, N.E.; Mmbaga, B.T.; Benitez-Paez, F. Dispensing antibiotics without prescription at community pharmacies and accredited drug dispensing outlets in Tanzania: A cross-sectional study. Antibiotics 2021, 10, 1025. [Google Scholar] [CrossRef]
- Schmider, J.; Bühler, N.; Mkwatta, H.; Lechleiter, A.; Mlaganile, T.; Utzinger, J.; Mzee, T.; Kazimoto, T.; Becker, S.L. Microbiological Characterisation of Community-Acquired Urinary Tract Infections in Bagamoyo, Tanzania: A Prospective Study. Trop. Med. Infect. Dis. 2022, 7, 100. [Google Scholar] [CrossRef]
- Barry, M.; Diallo, B.; Kanté, D.; Diallo, I. Antimicrobial susceptibility profile of community-acquired urinary tract infection in adults: A seven months prospective cross-sectional study in Dakar Town, Senegal. Afr. J. Urol. 2017, 23, 166–171. [Google Scholar] [CrossRef]
- Magliano, E.; Grazioli, V.; Deflorio, L.; Leuci, A.I.; Mattina, R.; Romano, P.; Cocuzza, C.E. Gender and age-dependent etiology of community-acquired urinary tract infections. Sci. World J. 2012, 2012, 349597. [Google Scholar] [CrossRef]
- Sood, S.; Gupta, R. Antibiotic resistance pattern of community acquired uropathogens at a tertiary care hospital in Jaipur, Rajasthan. Indian J. Community Med. 2012, 37, 39–44. [Google Scholar] [CrossRef]
- Akoachere, J.F.T.K.; Yvonne, S.; Akum, N.H.; Seraphine, E.N. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res. Notes 2012, 5, 219. [Google Scholar] [CrossRef]
- Pickett, D.A.; Welch, D.F. Recognition of Staphylococcus saprophyticus in urine cultures by screening colonies for production of phosphatase. J. Clin. Microbiol. 1985, 21, 310–313. [Google Scholar] [CrossRef]
- Rupp, M.E.; Fey, P.D. Staphylococcus epidermidis and other coagulase-negative staphylococci. Mandell Douglas Bennett’s Princ. Pract. Infect. Dis. 2014, 2, 2272–2282.e5. [Google Scholar] [CrossRef]
- Falagas, M.; Kastoris, A.; Vouloumanou, E.; Dimopoulos, G. Community-acquired Stenotrophomonas maltophilia infections: A systematic review. Eur. J. Clin. Microbiol. Infect. Dis. 2009, 28, 719–730. [Google Scholar] [CrossRef]
- Lo, S.; Thiam, I.; Fal, B.; Ba-Diallo, A.; Diallo, O.F.; Diagne, R.; Sow, A.I. Urinary tract infection with Corynebacterium aurimucosum after urethroplasty stricture of the urethra: A case report. J. Med. Case Rep. 2015, 9, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.; Burd, E. Other Gram-Negative and Gram-Variable Bacilli. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2015; pp. 2667–2683. [Google Scholar] [CrossRef]
- Stepanovic, S.; Jezek, P.; Vukovic, D.; Dakic, I.; Petráš, P. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J. Clin. Microbiol. 2003, 41, 5262–5264. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Falsen, E.; Pot, B.; Jantzen, E.; Hoste, B.; Vandamme, P.; De Ley, J. Acidovorax, a new genus for Pseudomonas facilis, Pseudomonas delafieldii, E. Falsen (EF) group 13, EF group 16, and several clinical isolates, with the species Acidovorax facilis comb. nov., Acidovorax delafieldii comb. nov., and Acidovorax temperans sp. nov. Int. J. Syst. Bacteriol. 1990, 40, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Rai, A.; Jaiswal, C.; Xess, A.; Shahi, S. Coagulase negative Staphylococci as causative agents of urinary tract infections-prevalence and resistance status in IGIMS, Patna. Indian. J. Pathol. Microbiol. 2001, 44, 415–419. [Google Scholar]
- Vikesland, P.; Garner, E.; Gupta, S.; Kang, S.; Maile-Moskowit, A.; Zhu, N. Differential drivers of antimicrobial resistance across the world. Acc. Chem. Res. 2019, 52, 916–924. [Google Scholar] [CrossRef]
- Malmros, K.; Huttner, B.D.; McNulty, C.A.M.; Rodríguez-Baño, J.; Pulcini, C.; Tängdén, T. Comparison of antibiotic treatment guidelines for urinary tract infections in 15 European countries: Results of an online survey. Int. J. Antimicrob. Agents 2019, 54, 478–486. [Google Scholar] [CrossRef]
- Mboya, E.A.; Sanga, L.A.; Ngocho, J.S. Irrational use of antibiotics in the Moshi Municipality Northern Tanzania: A cross sectional study. Pan Afr. Med. J. 2018, 31, 165. [Google Scholar] [CrossRef]
- CDC Urinary Tract Infection, 9 August 2022. Available online: https://www.cdc.gov/antibiotic-use/uti.html (accessed on 6 October 2021).
- Barrow, G.I.; Feltham, R.K.A. Cowan and Steel’s Manual for the Identification of Medical Bacteria, 3rd ed.; Cambridge University Press: Cambridge, MA, USA, 1993; p. 216. [Google Scholar] [CrossRef]
- Alghamdi, A.; Almajid, M.; Alalawi, R.; Alganame, A.; Alanazi, S.; Alghamdi, G.; Alharthi, S.; Alghamdi, I. Evaluation of asymptomatic bacteruria management before and after antimicrobial stewardship program implementation: Retrospective study. BMC. Infect. Dis. 2021, 21, 869. [Google Scholar] [CrossRef]
- Procop, D.A.; Gary, W.; Deirdre, L.C.; Geraldine, S.H.; William, M.J.; Koneman, E.W.; Schreckenberger, P.C. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology, 7th ed.; Wolters Kluwer Health: Philadelphia, PA, USA, 2017. [Google Scholar]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 55–63. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]

| Variables | DAR, n = 649 | MWANZA, n = 678 | Overall, n = 1327 | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Type of patient | Adult males | 103 (15.9%) | 70 (10.3%) | 173 (13.0%) |
| Adult females non-pregnant | 273 (42.1%) | 318 (46.9%) | 591 (44.5%) | |
| Adult females pregnant | 195 (30.0%) | 240 (35.4%) | 435 (32.8%) | |
| Children | 78 (12.0%) | 50 (7.4%) | 128 (9.7%) | |
| Health centre | Buguruni HC | 226 (34.8%) | NA | 226 (34.8%) |
| Magomeni HC | 423 (65.2%) | NA | 423 (65.2%) | |
| Buzuruga HC | NA | 338 (49.8%) | 338 (49.8%) | |
| Karume HC | NA | 340 (50.2%) | 340 (50.2%) | |
| Sex | Male | 139 (21.4%) | 89 (13.1%) | 228 (17.2%) |
| Female | 510 (78.6%) | 589 (86.9%) | 1099 (82.8%) | |
| Residency | Urban | 649 (100.0%) | 338 (49.8%) | 987 (74.4%) |
| Rural | 0 (0.0%) | 340 (50.2%) | 340 (25.6%) | |
| Median [IQR] age in years | 27 [22–38] | 28 [23–39] | 28 [22–39] | |
| Type of toilet used at home | Pit latrine | 144 (22.2%) | 106 (15.6%) | 250 (18.8%) |
| Flush latrine | 503 (77.5%) | 567 (83.6%) | 1070 (80.6%) | |
| Others | 2 (0.3%) | 5 (0.7%) | 7 (0.5%) | |
| Occupations | Farmer | 6 (0.9%) | 184 (27.1%) | 190 (14.3%) |
| Business | 259 (39.9%) | 174 (25.7%) | 433 (32.6%) | |
| Civil servant | 33 (5.1%) | 34 (5.0%) | 67 (5.0%) | |
| Housewife | 126 (19.4%) | 181 (26.7%) | 307 (23.1%) | |
| Not working | 137 (21.1%) | 49 (7.2%) | 186 (14.0%) | |
| Still on studies | 88 (13.6%) | 56 (8.3%) | 144 (10.8%) | |
| Source of water for domestic use | Tap water | 477 (73.5%) | 427 (63%) | 904 (68.1%) |
| Well water | 171 (26.3%) | 107 (15.8%) | 278 (21.0%) | |
| Lake | 1 (0.1%) | 144 (21.2%) | 145 (10.9%) | |
| Marital status | Single | 149 (24.7%) | 125 (19.6%) | 274 (22.1%) |
| Married | 454 (75.3%) | 511 (80.3%) | 965 (77.9%) | |
| Previous history of UTIs | No | 263 (40.5%) | 461 (68%) | 724 (54.6%) |
| Yes | 379 (58.4%) | 214 (31.6%) | 593 (44.7%) | |
| Unknown | 7 (1.1%) | 3 (0.4%) | 10 (0.7%) | |
| Previous antibiotic use past 3 months | Yes | 252 (38.8%) | 151 (22.2%) | 403 (30.4%) |
| No | 396 (61.2%) | 528 (77.8%) | 924 (69.6%) | |
| Currently prescribed antibiotic | Yes | 322 (49.6%) | 149 (22.0%) | 471 (35.5%) |
| No | 327 (50.4%) | 529 (78.0%) | 856 (64.5%) | |
| Variables | Frequencies | |||
|---|---|---|---|---|
| Mwanza, n (%) | Dar es Salaam, n (%) | Total, n (%) | ||
| Culture results | SB | 180 (26.5%) | 184 (28.4%) | 364 (27.4%) |
| NMG and NSB | 498 (73.5%) | 465 (71.6%) | 964 (72.6%) | |
| Isolated uropathogens | E. coli | 90 (43.7%) | 68 (33.1%) | 158 (38.3%) |
| Enterococcus spp. | 17 (8.3%) | 10 (4.9%) | 27 (6.6%) | |
| S. aureus | 14 (6.8%) | 18 (8.7%) | 32 (7.8%) | |
| K. pneumoniae | 14 (6.8%) | 10 (4.9%) | 24 (5.8%) | |
| S. haemolyticus | 11 (5.3%) | 21 (10.2%) | 32 (7.8%) | |
| S. pyogenes | 11 (5.3%) | 2 (0.9%) | 13 (3.2%) | |
| S. epidermidis | 8 (3.9%) | 12 (5.8%) | 20 (4.9%) | |
| Candida spp. (yeast) | 8 (3.9%) | 10 (4.9%) | 18 (4.4%) | |
| K. aerogenes | 6 (2.9%) | 0 (0.0%) | 6 (1.5%) | |
| C. aurimucosum | 5 (2.4%) | 4 (1.9%) | 9 (2.2%) | |
| A. junii | 0 (0.0%) | 5 (2.4%) | 5 (1.2%) | |
| S. saprophyticus | 0 (0.0%) | 5(2.4%) | 5 (1.2%) | |
| Other GNB | 14 (6.8%) | 23 (11.2%) | 37 (8.9%) | |
| Other GPB | 8 (3.9%) | 12 (5.8%) | 20 (4.9%) | |
| Total uropathogens | 206 | 206 | 412 | |
| Antibiotic | E. coli | Other Enterobacterales | Non-Enterobacterales | ||||||
|---|---|---|---|---|---|---|---|---|---|
| DAR (n = 53) | MWZ (n = 90) | Overall (n = 143) | DAR (n = 47) | MWZ (n = 31) | Overall (n = 78) | DAR (n = 14) | MWZ (n = 3) | Overall (n = 17) | |
| AMP | 83.0% | 87.8% | 86.0% | 61.7% | 96.8% | 75.6% | NA | NA | NA |
| SXT | 81.1% | 84.4% | 83.2% | 61.7% | 54.8% | 58.9% | 25.0% * | 50.0% * | 30.0% * |
| TCY | 79.3% | 74.4% | 76.2% | 40.4% | 54.8% | 46.2% | 55.6% * | 0.0% * | 45.5% * |
| AMC | 47.2% | 50.2% | 48.9% | 48.9% | 58.1% | 52.6% | NA | NA | NA |
| CIP | 50.9% | 50.0% | 50.4% | 44.7% | 22.6% | 35.9% | 14.3% | 33.3% | 17.7% |
| FEP | 28.3% | 28.9% | 28.7% | 36.2% | 22.6% | 30.8% | 0.0% | 33.3% | 5.9% |
| CAZ | 28.3% | 27.8% | 27.9% | 31.9% | 25.8% | 29.5% | 35.7% | 0.0% | 29.4% |
| CRO | 26.4% | 27.8% | 27.3% | 34.0% | 25.8% | 30.8% | 50.0% * | 0.0% * | 40.0% * |
| GEN | 26.4% | 20.0% | 22.4% | 27.7% | 9.7% | 20.5% | 0.0% | 33.3% | 5.9% |
| NIT | 15.1% | 38.9% | 30.1% | 40.4% | 58.1% | 47.4% | NA | NA | NA |
| IMP | 0.0% | 1.1% | 0.7% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| MEM | 0.0% | 1.1% | 0.7% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% |
| Antibiotic Agents | S. aureus | Enterococcus spp. | Coagulase-Negative Staphylococci (*CoNS) | Streptococcus spp. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DAR (n = 16) | MWZ (n = 14) | Overall (n = 30) | DAR (n = 10) | MWZ (n = 18) | Overall (n = 28) | DAR (n = 47) | MWZ (n = 24) | Overall (n = 71) | DAR (n = 9) | MWZ (n = 13) | Overall (n = 22) | |
| AMP | NA | NA | NA | 10.0% | 16.7% | 14.3% | NA | NA | NA | NA | NA | NA |
| FOX | 50.0% | 57.1% | 53.3% | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| GEN | 37.5% | 50.5% | 43.3% | NA | NA | NA | 44.7% | 50.0% | 46.5% | NA | NA | NA |
| CIP | 43.8% | 78.6% | 60.0% | 70.0% | 50.0% | 57.1% | 57.5% | 62.5% | 59.2% | 77.8% | 69.2% | 72.7% |
| CLI | 31.3% | 71.5% | 50.0% | NA | NA | NA | 42.6% | 70.8% | 52.1% | 22.2% | 0.0% | 9.1% |
| ERY | 68.8% | 78.5% | 73.4% | 90.0% | 88.9% | 89.3% | 80.9% | 83.3% | 81.7% | 55.6% | 53.9% | 54.6% |
| NIT | 43.8% | 7.1% | 26.6% | 40.0% | 27.8% | 32.2% | 14.9% | 20.8% | 16.9% | NA | NA | NA |
| LNZ | 18.8% | 21.4% | 20.0% | 40.0% | 38.9% | 39.3% | 4.3% | 0.0% | 2.8% | 0.0% | 7.7% | 4.6% |
| TCY | 50.1% | 64.3% | 56.7% | NA | NA | NA | 61.7% | 66.7% | 63.4% | 77.8% | 92.3% | 86.4% |
| SXT | 31.3% | 64.3% | 46.6% | 40.0% | 72.3% | 60.7% | 78.7% | 95.8% | 84.5% | 33.3% | 38.5% | 36.4% |
| Isolate | MDR Patterns | Classes Resisted | Frequency | ||
|---|---|---|---|---|---|
| Mwanza (n = 61) | DAR (n = 47) | Overall (n = 108) | |||
| E. coli | CIP-NIT-TCY | 3 | 2 | 1 | 3 |
| CIP-GEN-TCY | 3 | 0 | 1 | 1 | |
| AMP-GEN-TCY | 3 | 3 | 2 | 5 | |
| AMP-NIT-TCY | 3 | 5 | 1 | 6 | |
| AMP-GEN-NIT | 3 | 0 | 1 | 1 | |
| AMP-CIP-TCY | 3 | 12 | 11 | 23 | |
| AMP-NIT-TCY | 3 | 0 | 1 | 1 | |
| AMP-CIP-GEN | 3 | 1 | 0 | 1 | |
| AMP-CIP-NIT-TCY | 4 | 13 | 1 | 14 | |
| AMP-CIP-GEN-TCY | 4 | 8 | 8 | 16 | |
| AMP-CIP-GEN-NIT | 4 | 0 | 1 | 1 | |
| AMP-CIP-GEN-NIT-TCY | 5 | 5 | 1 | 6 | |
| AMP-CIP-GEN-MEM-NIT-TCY | 6 | 1 | 0 | 1 | |
| K. pneumoniae | AMP-CIP-NIT | 3 | 0 | 1 | 1 |
| AMP-CIP-TCY | 3 | 2 | 0 | 2 | |
| AMP-NIT-TCY | 3 | 1 | 0 | 1 | |
| AMP-CIP-NIT-TCY | 4 | 3 | 3 | 6 | |
| AMP-GEN-NIT-TCY | 4 | 1 | 0 | 1 | |
| Other GNB | AMP-NIT-TCY | 3 | 1 | 0 | 1 |
| CIP-GEN-TCY | 3 | 0 | 1 | 1 | |
| AMP-CIP-GEN | 3 | 0 | 4 | 4 | |
| CIP-NIT-TCY | 3 | 0 | 1 | 1 | |
| AMP-CIP-GEN-TCY | 4 | 0 | 2 | 2 | |
| AMP-CIP-GEN-NIT-TCY | 5 | 0 | 2 | 2 | |
| K. oxytoca | AMP-CIP-TCY | 3 | 1 | 0 | 1 |
| AMP-CIP-NIT | 3 | 0 | 1 | 1 | |
| AMP-CIP-GEN | 3 | 0 | 1 | 1 | |
| AMP-CIP-GEN-TCY | 4 | 0 | 1 | 1 | |
| M. morganii | AMP-NIT-TCY | 3 | 1 | 0 | 1 |
| GEN-NIT-TCY | 3 | 0 | 1 | 1 | |
| E. hormaechei | AMP-NIT-TCY | 3 | 1 | 0 | 1 |
| Total | 33 | 61 | 47 | 108 | |
| Isolate | MDR Patterns | Classes Resisted | Frequency | ||
|---|---|---|---|---|---|
| Mwanza (n = 18) | DAR (n = 17) | Overall (n = 35) | |||
| S. aureus | CIP-NIT-TCY | 3 | 1 | 1 | 2 |
| CIP-GEN-TCY | 3 | 6 | 2 | 8 | |
| CIP-GEN-NIT | 3 | 0 | 1 | 1 | |
| CIP-GEN-NIT-TCY | 4 | 0 | 1 | 1 | |
| Other GPC | CIP-NIT-TCY | 3 | 1 | 0 | 1 |
| CIP-GEN-TCY | 3 | 1 | 0 | 1 | |
| CoNS | CIP-NIT-TCY | 3 | 3 | 1 | 4 |
| CIP-GEN-TCY | 3 | 5 | 8 | 13 | |
| CIP-GEN-NIT | 3 | 0 | 1 | 1 | |
| CIP-GEN-NIT-TCY | 4 | 1 | 2 | 3 | |
| Total | 10 | 18 | 17 | 35 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silago, V.; Moremi, N.; Mtebe, M.; Komba, E.; Masoud, S.; Mgaya, F.X.; Mirambo, M.M.; Nyawale, H.A.; Mshana, S.E.; Matee, M.I. Multidrug-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections among Patients Attending Health Facilities in Mwanza and Dar es Salaam, Tanzania. Antibiotics 2022, 11, 1718. https://doi.org/10.3390/antibiotics11121718
Silago V, Moremi N, Mtebe M, Komba E, Masoud S, Mgaya FX, Mirambo MM, Nyawale HA, Mshana SE, Matee MI. Multidrug-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections among Patients Attending Health Facilities in Mwanza and Dar es Salaam, Tanzania. Antibiotics. 2022; 11(12):1718. https://doi.org/10.3390/antibiotics11121718
Chicago/Turabian StyleSilago, Vitus, Nyambura Moremi, Majigo Mtebe, Erick Komba, Salim Masoud, Fauster X. Mgaya, Mariam M. Mirambo, Helmut A. Nyawale, Stephen E. Mshana, and Mecky Isaac Matee. 2022. "Multidrug-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections among Patients Attending Health Facilities in Mwanza and Dar es Salaam, Tanzania" Antibiotics 11, no. 12: 1718. https://doi.org/10.3390/antibiotics11121718
APA StyleSilago, V., Moremi, N., Mtebe, M., Komba, E., Masoud, S., Mgaya, F. X., Mirambo, M. M., Nyawale, H. A., Mshana, S. E., & Matee, M. I. (2022). Multidrug-Resistant Uropathogens Causing Community Acquired Urinary Tract Infections among Patients Attending Health Facilities in Mwanza and Dar es Salaam, Tanzania. Antibiotics, 11(12), 1718. https://doi.org/10.3390/antibiotics11121718







