Molecular Epidemiology of Carbapenem-Resistant K. pneumoniae Clinical Isolates from the Adult Patients with Comorbidities in a Tertiary Hospital, Southern Saudi Arabia
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Antimicrobial Susceptibility Testing
2.3. Genetic Analysis of MDR Klebsiella pneumoniae Isolates
2.3.1. Detection of Antibiotic Resistance Genes
2.3.2. DNA Extraction
2.4. Statistical Analysis
3. Results
3.1. Sociodemographic and Clinical Characteristics of Study Participants
3.2. Antimicrobial Susceptibility Testing
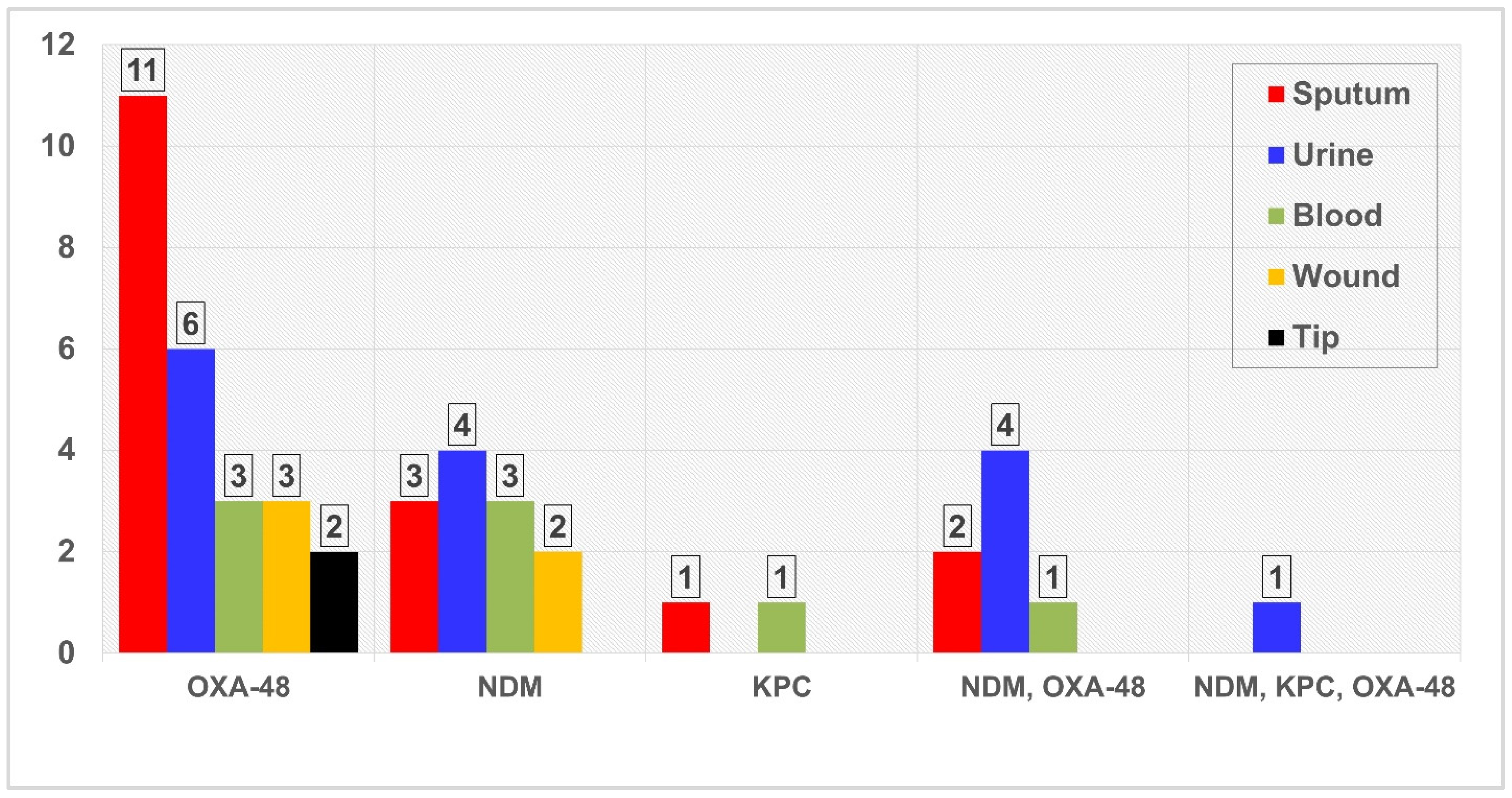
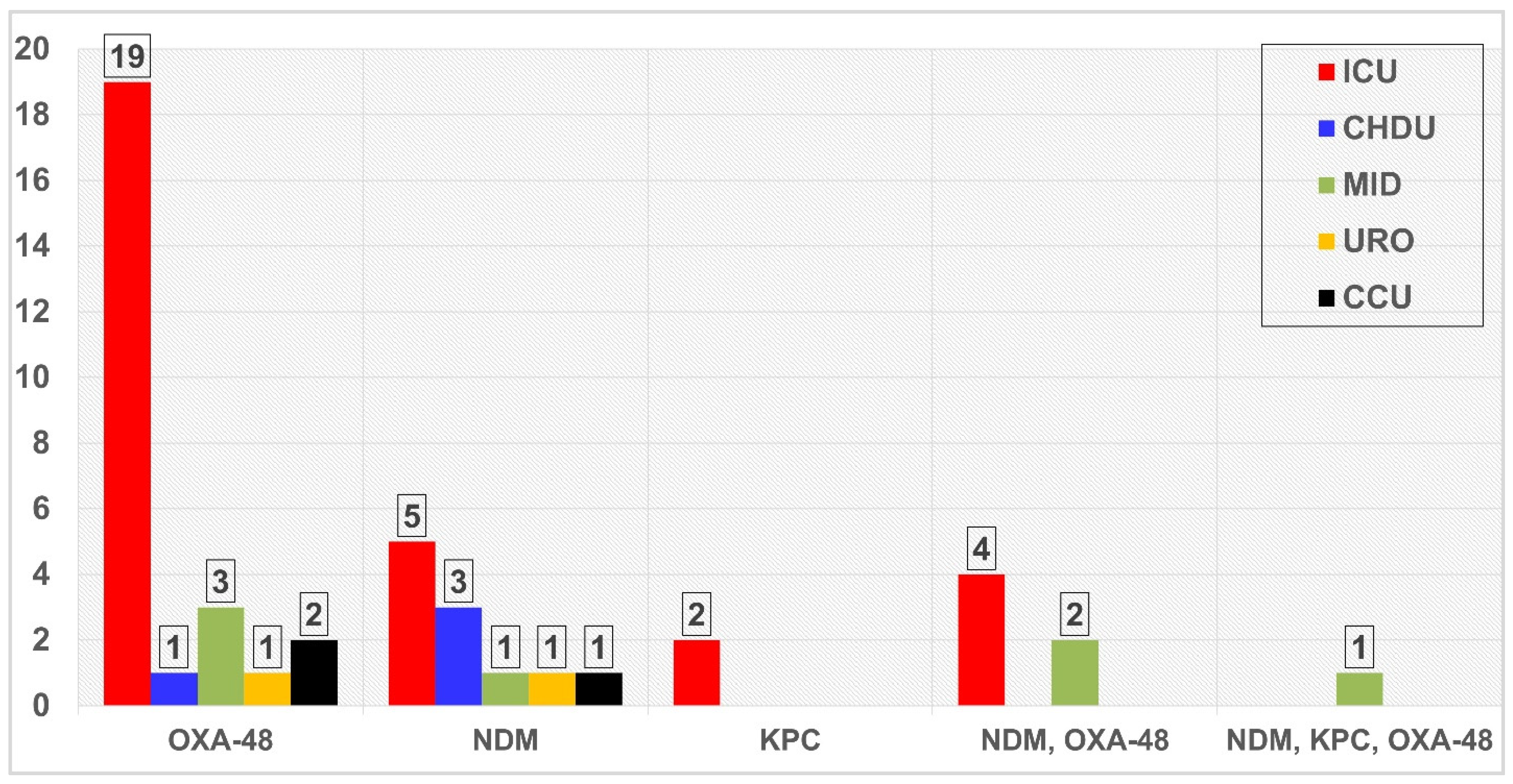
3.3. Screening of PCR for Resistance Genes
3.4. Factors Associated with Presence of Resistance Genes
4. Discussion
Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moradigaravand, D.; Martin, V.; Peacock, S.J.; Parkhill, J. Evolution and Epidemiology of Multidrug-Resistant Klebsiella Pneumoniae in the United Kingdom and Ireland. MBio 2017, 8, e01976-16. [Google Scholar] [CrossRef]
- Abdalhamid, B.; Elhadi, N.; Albunayan, S.; Alsamman, K.; Aljindan, R. First Description of Methyltransferases in Extensively Drug-Resistant Klebsiella Pneumoniae Isolates from Saudi Arabia. J. Med. Microbiol. 2017, 66, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Palmeiro, J.K.; De Souza, R.F.; Schörner, M.A.; Passarelli-Araujo, H.; Grazziotin, A.L.; Vidal, N.M.; Venancio, T.M.; Dalla-costa, L.M. Molecular Epidemiology of Multidrug-Resistant Klebsiella Pneumoniae Isolates in a Brazilian Tertiary Hospital. Front. Microbiol. 2019, 10, 1669. [Google Scholar] [CrossRef] [PubMed]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella Pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef] [PubMed]
- Shibl, A.; Al-agamy, M.; Memish, Z.; Senok, A. The Emergence of OXA-48- and NDM-1-Positive Klebsiella Pneumoniae in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2013, 17, e1130–e1133. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Assiri, A.; Almasri, M.; Roshdy, H.; Hathout, H.; Kaase, M.; Gatermann, S.G.; Yezli, S. Molecular Characterization of Carbapenemase Production among Gram-Negative Bacteria in Saudi Arabia. Microb. Drug Resist. 2015, 21, 307–314. [Google Scholar] [CrossRef]
- uz Zaman, T.; Aldrees, M.; Al Johani, S.M.; Alrodayyan, M.; Aldughashem, F.A.; Balkhy, H.H. Multi-Drug Carbapenem-Resistant Klebsiella Pneumoniae Infection Carrying the OXA-48 Gene and Showing Variations in Outer Membrane Protein 36 Causing an Outbreak in a Tertiary Care Hospital in Riyadh, Saudi Arabia. Int. J. Infect. Dis. 2014, 28, e186–e192. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Mohamed, A.M.; Faiz, A.; Ahmad, J. Enterobacterial Infection in Saudi Arabia: First Record of Klebsiella Pneumoniae with Triple Carbapenemase Genes Resistance. J. Infect. Dev. Ctries. 2019, 13, 334–341. [Google Scholar] [CrossRef] [PubMed]
- The, H.C.; Karkey, A.; Thanh, D.P.; Boinett, C.J.; Cain, A.K.; Ellington, M.; Baker, K.S.; Dongol, S.; Thompson, C.; Harris, S.R.; et al. A High-Resolution Genomic Analysis of Multidrug- Resistant Hospital Outbreaks of Klebsiella Pneumoniae. EMBO Mol. Med. 2015, 7, 227–239. [Google Scholar]
- Alraddadi, B.M.; Heaphy, E.L.G.; Aljishi, Y.; Ahmed, W.; Eljaaly, K.; Al-Turkistani, H.H.; Alshukairi, A.N.; Qutub, M.O.; Alodini, K.; Alosaimi, R.; et al. Molecular Epidemiology and Outcome of Carbapenem-Resistant Enterobacterales in Saudi Arabia. BMC Infect. Dis. 2022, 22, 254. [Google Scholar] [CrossRef]
- Candido, L.; Barbosa, G.; Silva, A.; Bordoni, G.P.; Barbosa, G.D.O.; Carneiro, L.C. Elevated Mortality Risk from CRKp Associated with Comorbidities: Systematic Review and Meta-Analysis. Antibiotics 2022, 11, 874. [Google Scholar]
- Zhang, H.; Yang, M. Risk Factors and Prognosis of Carbapenem-Resistant Klebsiella Pneumoniae Infections in Respiratory Intensive Care Unit: A Retrospective Study. Infect. Drug Resist. 2021, 14, 3297–3305. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Suppl. M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Magiorakos, A.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F. Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2011, 18, 268–281. [Google Scholar] [CrossRef]
- Favier, C.; Arlet, G.; Dallenne, C.; Da Costa, A.; Decre, D.; Curie, M. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important b-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Magzoub, M.A.; Bilal, N.E.; Hamid, M.E. Distribution of Class I Integrons and Their Effect on the Prevalence of Multi-Drug Resistant Escherichia Coli Clinical Isolates from Sudan. Saudi Med. J. 2013, 34, 240–247. [Google Scholar] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Heinz, E.; Brindle, R.; Morgan-Mccalla, A.; Peters, K.; Thomson, N.R. Caribbean Multi-Centre Study of Klebsiella Pneumoniae: Whole-Genome Sequencing, Antimicrobial Resistance and Virulence Factors. Microb. Genom. 2019, 5, e000266. [Google Scholar] [CrossRef] [PubMed]
- Radwan, E.; Al-dughmani, H.; Koura, B.; Bader, M.; Al Deen, M.; Bueid, A.; Elgaher, M.; Alghoribi, M.F.; Albarrag, M.; Somily, M. Molecular Characterization of Carbapenem-Resistant Enterobacterales in Thirteen Tertiary Care Hospitals in Saudi Arabia. Ann. Saudi Med. 2021, 48, 63–70. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Sartor, A.L.; Balkhy, H.H.; Walsh, T.R.; Al Johani, S.M.; AlJindan, R.Y.; Alfaresi, M.; Ibrahim, E.; Al-Jardani, A.; Al-Abri, S.; et al. Molecular Characterization of Carbapenemase-Producing Escherichia Coli and Klebsiella Pneumoniae in the Countries of the Gulf Cooperation Council: Dominance of OXA-48 and NDM Producers. Antimicrob. Agents Chemother. 2014, 58, 3085–3090. [Google Scholar] [CrossRef]
- Al-agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of Carbapenemases, ESBLs, and Plasmid-Mediated Quinolone Determinants in Carbapenem-Insensitive Escherichia Coli and Klebsiella Pneumoniae in Riyadh Hospitals. J. Infect. Public Health 2018, 11, 64–68. [Google Scholar] [CrossRef]
- Alsharapy, S.A.; Gharout-Sait, A.; Muggeo, A.; Guillard, T.; Cholley, P.; Brasme, L.; Bertrand, X.; Moghram, G.S.; Touati, A.; De Champs, C. Characterization of Carbapenem-Resistant Enterobacteriaceae Clinical Isolates in Al Thawra University Hospital, Sana’a, Yemen. Microb. Drug Resist. 2019, 26, 211–217. [Google Scholar] [CrossRef]
- Jamal, W.; Rotimi, V.; Albert, M.; Khodakhast, F.; Udo, E.; Poirel, L. Emergence of Nosocomial New Delhi Metallo-Beta-Lactamase-1 (NDM-1)-Producing Klebsiella Pneumoniae in Patients Admitted to a Tertiary Care Hospital in Kuwait. Int. J. Antimicrob. Agents 2012, 39, 183–184. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Al Maskari, Z.; Al Rashdi, F.; Bernabeu, S.; Nordmann, P. NDM-1-Producing Klebsiella Pneumoniae Isolated in the Sultanate of Oman. J. Antimicrob. Chemother. 2011, 66, 304–306. [Google Scholar] [CrossRef] [PubMed]
- Shoja, S.; Ansari, M.; Faridi, F.; Azad, M.; Davoodian, P.; Javadpour, S.; Farahani, A.; Mobarrez, B.D.; Karmostaji, A. Identification of Carbapenem-Resistant Klebsiella Pneumoniae with Emphasis. Microb. Drug Resist. 2017, 24, 447–454. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Shibl, A.M.; Elkhizzi, N.A.; Meunier, D.; Turton, J.F.; Livermore, D.M. Persistence of Klebsiella Pneumoniae Clones with OXA-48 or NDM Carbapenemases Causing Bacteraemias in a Riyadh Hospital. Diagn. Microbiol. Infect. Dis. 2013, 76, 214–216. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Algak, T.B.; Abbas, M.; Elamin, B.K. Emergence of BlaTEM, BlaCTX-M, BlaSHV and BlaOXA Genes in Multidrug-resistant Enterobacteriaceae and Acinetobacter Baumannii in Saudi Arabia. Exp. Ther. Med. 2021, 22, 1450. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Gentile, B.; Pagani, C.; Di Gregorio, A.; Anselmo, A.; Palozzi, A.M.; Fortunato, A.; Pittiglio, V.; Ridolfo, A.L.; Gismondo, M.R. Whole Genome Sequencing for the Molecular Characterization of Carbapenem- Resistant Klebsiella Pneumoniae Strains Isolated at the Italian ASST Fatebenefratelli Sacco Hospital, 2012–2014. BMC Infect. Dis. 2017, 17, 666. [Google Scholar] [CrossRef]
- Al-agamy, M.H.; El-mahdy, T.S.; Radwan, H.H.; Poirel, L. Cooccurrence of NDM-1, ESBL, RmtC, AAC(6′)-Ib, and QnrB in Clonally Related Klebsiella Pneumoniae Isolates Together with Coexistence of CMY-4 and AAC(6′)-Ib in Enterobacter Cloacae Isolates from Saudi Arabia. BioMed Res. Int. 2019, 2019, 6736897. [Google Scholar] [CrossRef]
- Zheng, S.; Cao, S.; Xu, H.; Feng, D.; Wan, L.; Wang, G.; Xiao, X. Risk Factors, Outcomes and Genotypes of Carbapenem-Nonsusceptible Klebsiella Pneumoniae Bloodstream Infection: A Three-Year Retrospective Study in a Large Tertiary Hospital in Northern China Risk Factors, Outcomes and Genotypes of Carbapenem-Nonsuscept. Infect. Dis. (Auckl.) 2018, 50, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Ramsamy, Y.; Mlisana, K.P.; Allam, M.; Amoako, D.G.; Abia, A.L.K.; Ismail, A.; Singh, R.; Kisten, T.; Essack, S.Y. Genomic Analysis of Carbapenemase-Producing Extensively Drug-Resistant Klebsiella Pneumoniae Isolates Reveals the Horizontal Spread of P18-43_01 Plasmid Encoding Bla NDM-1 in South Africa. Microorganisms 2020, 8, 137. [Google Scholar] [CrossRef]
- Rodr, E.; Garza-Gonza, E.; Bocanegra-Ibarias, P.; Paz-Velarde, B.A.; Morf, R.; Leo, G. A Case–Control Study of Infections Caused by Klebsiella Pneumoniae Producing New Delhi Metallo-Beta-Lactamase-1: Predictors and Outcomes. Front. Cell. Infect. Microbiol. 2022, 12, 867347. [Google Scholar] [CrossRef]
- Dirar, M.H.; Bilal, N.E.; Ibrahim, M.E.; Hamid, M.E. Prevalence of Extended-Spectrum β-Lactamase (ESBL) and Molecular Detection of BlaTEM, BlaSHV and BlaCTX-M Genotypes among Enterobacteriaceae Isolates from Patients in Khartoum, Sudan Maha. Pan Afr. Med. J. 2020, 37, 213. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Hu, T.; Hu, J.; Song, N.; Zhang, Y.; Chen, Y. Five-Year Change of Prevalence and Risk Factors for Infection and Mortality of Carbapenem-Resistant Klebsiella Pneumoniae Bloodstream Infection in a Tertiary Hospital in North China. Antimicrob. Resist. Infect. Control 2020, 9, 79. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.H.; Mchugh, T.D.; Roulston, K.; Arruda, L.B.; Sadouki, Z.; Riaz, S. Detection of Carbapenemases and Extended-Spectrum-β-Lactamase Bla OXA1-Bla SHV-Bla TEM Genes in Gram-Negative Bacterial Isolates from ICU Burns Patients. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 43. [Google Scholar] [CrossRef]
- Zhu, W.; Yuan, Z.; Zhou, H. Risk Factors for Carbapenem-Resistant Klebsiella Pneumoniae Infection Relative to Two Types of Control Patients: A Systematic Review and Meta-Analysis. Antimicrob. Agents Chemother. 2020, 9, 23. [Google Scholar] [CrossRef]

| Gene | Primers | Sequence | Band/bp | Reference |
|---|---|---|---|---|
| blaNDM-1 | NDM-F NDM-R | GGTTTGGCGATCTGGTTTTC CGGAATGGCTCATCACGATC | 621 | [17] |
| blaIMP | IMP-F IMP-R | TTGACACTCCATTTACDG GATYGAGAATTAAGCCACYCT | 139 | [15] |
| blaVIM | VIM-F VIM-R | GATGGTGTTTGGTCGCATA CGAATGCGCAGCACCAG | 390 | [15] |
| blaKPC | KPC-F KPC-R | CATTCAAGGGCTTTCTTGCTGC ACGACGGCATAGTCATTTGC | 538 | [15] |
| blaOXA-48-like | OXA-F OXA-R | GCTTGATCGCCCTCGATT GATTTGCTCCGTGGCCGAAA | 281 | [15] |
| Agent | Overall, n (%) | ICU, n (%) | Medical, n (%) | CCU, n (%) | CHDU, n (%) | Urology, n (%) |
|---|---|---|---|---|---|---|
| Amikacin | 43 (60.6) | 28 (75.7) | 8 (57.1) | 2 (28.6) | 4 (44.4) | 1 (25) |
| Ampicillin | 71 (100) | (100) | (100) | (100) | (100) | (100) |
| Amoxicillin/clavulanate | 54(76.1) | 33 (89.2) | 10 (71.4) | 4 (57.1) | 5 (55.6) | 2 (50) |
| Aztreonam | 58 (81.7) | 34 (91.9) | 12 (85.7) | 4 (57.1) | 6 (66.7) | 2 (50) |
| Ceftriaxone | 61 (85.9) | 36 (97.3) | 11 (78.6) | 4 (57.1) | 7 (77.8) | 3 (75) |
| Cefepime | 57 (80.3) | 35 (94.6) | 9 (64.3) | 4 (57.1) | 6 (66.7) | 3 (75) |
| Cefuroxime | 64 (90.1) | 36 (97.3) | 13 (92.9) | 5 (71.4) | 7 (77.8) | 3 (75) |
| Ciprofloxacin | 49 (69) | 30 (81.1) | 8 (57.1) | 4 (57.1) | 6 (66.7) | 1 (25) |
| trimethoprim/sulfamethoxazole | 58 (81.7) | 32 (86.5) | 11 (78.6) | 6 (85.7) | 6 (66.7) | 3 (75) |
| Colistin | 9 (12.7) | 9 (24.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Gentamicin | 45(63.4) | 29 (78.4) | 7 (50) | 3 (42.9) | 4 (44.4) | 2 (50) |
| Imipenem | 40 (56.3) | 29 (78.4) | 4 (28.6) | 3 (42.9) | 3 (33.3) | 1 (25) |
| Meropenem | 49 (69) | 33 (89.2) | 6 (42.9) | 2 (28.6) | 6 (66.7) | 2 (50) |
| Piperacillin/tazobactam | 57 (80.3) | 35 (94.6) | 9 (64.3) | 4 (57.1) | 6(66.7) | 3 (75) |
| Tigecycline | 4 (5.6) | 3 (8.1) | 1 (7.1) | 0 0.0 (0.0) | 0 0.0 (0.0) | 0 0.0 (0.0) |
| Comorbidity | Number of Patients | Frequency of Resistance-Gene-Carrying K. pneumoniae, n (%) |
|---|---|---|
| Single | 23 | 17(73.9) |
| COPD | 8 | 7 (87.5) |
| DM | 7 | 4 (57.1) |
| HTN | 5 | 3 (60) |
| CVA | 2 | 2 (100) |
| HD | 1 | 1 (100) |
| Two | 25 | 13 (52) |
| DM, HTN | 8 | 5 (62.5) |
| DM, CVA | 4 | 0 (0.0) |
| DM, COPD | 3 | 3 (100) |
| HTN, COPD | 3 | 3 (100) |
| HD, CVA | 3 | 1 (33.3) |
| HTN, CVA | 2 | 00.0 |
| DM, CKD | 1 | 00.0 |
| HTN, CKD | 1 | 1 (100) |
| Three | 10 | 7 (70) |
| DM, HTN, CKD | 4 | 3 (75) |
| DM, HTN, CVA | 4 | 2 (50) |
| DM, HTN, CVD | 1 | 1 (100) |
| DM, HTN, HD | 1 | 1 (100) |
| Four | 11 | 9 (81.8) |
| DM, HTN, CKD, HD | 5 | 4(80) |
| DM, HTN, HD, CVA | 2 | 2 (100) |
| DM, HTN, CKD, COPD | 1 | 1 (100) |
| DM, HTN, HD, COPD | 1 | 0 (0.0) |
| DM, HTN, CKD, CVD | 1 | 1 (100) |
| DM, HTN, CVD, CVA | 1 | 1 (100) |
| Five | 2 | 1 (50) |
| DM, HTN, CKD, HD, COPD | 1 | 1 (100) |
| DM, HTN, CKD, CVD, CVA | 1 | 00.0 |
| Variable | Total | n (%) of Resistant Gene | χ2 | p Value |
|---|---|---|---|---|
| Gender | 5.94 | 0.015 | ||
| Male | 38 | 30 (78.9) | ||
| Female | 33 | 17 (51.5) | ||
| Age group, year | 1.706 | 0.426 | ||
| ≤45 | 11 | 9 (81.8) | ||
| 46 to 65 | 16 | 11 (68.8) | ||
| >65 | 44 | 27 (61.4) | ||
| ICU admission | 7.649 | 0.002 | ||
| Yes | 37 | 30 (81.1) | ||
| No | 34 | 17 (50) | ||
| Diabetes Mellitus | 0.446 | 0.581 | ||
| Yes | 46 | 29(63) | ||
| No | 25 | 18 (72) | ||
| Hypertension | 0.541 | 0.373 | ||
| Yes | 42 | 29(69) | ||
| No | 29 | 18 (62.1) | ||
| Chronic kidney diseases | 0.433 | 0.511 | ||
| Yes | 15 | 11 (73.3) | ||
| No | 56 | 36 (64.3) | ||
| Heart diseases | 0.213 | 0.644 | ||
| Yes | 14 | 10 (71.4) | ||
| No | 57 | 37 (64.9) | ||
| Cardiovascular diseases | 0.147 | 0.702 | ||
| Yes | 4 | 3 (75) | ||
| No | 67 | 44 (65.7) | ||
| Cerebrovascular accident | 6.729 | 0.009 | ||
| Yes | 19 | 8 (42.1) | ||
| No | 52 | 39 (75) | ||
| Chronic obstructive pulmonary diseases | 4.851 | 0.028 | ||
| Yes | 17 | 15 (88.2) | ||
| No | 54 | 32(59.3) | ||
| Number of comorbidities | 3.476 | 0.176 | ||
| One | 23 | 17(73.9) | ||
| Two | 25 | 13 (52) | ||
| Three or more | 23 | 17(73.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshahrani, A.M.; Ibrahim, M.E.; Aldossary, A.K.; Alghamdi, M.A.; Ahmed, O.B.; Bin Abdulhak, A.A. Molecular Epidemiology of Carbapenem-Resistant K. pneumoniae Clinical Isolates from the Adult Patients with Comorbidities in a Tertiary Hospital, Southern Saudi Arabia. Antibiotics 2022, 11, 1697. https://doi.org/10.3390/antibiotics11121697
Alshahrani AM, Ibrahim ME, Aldossary AK, Alghamdi MA, Ahmed OB, Bin Abdulhak AA. Molecular Epidemiology of Carbapenem-Resistant K. pneumoniae Clinical Isolates from the Adult Patients with Comorbidities in a Tertiary Hospital, Southern Saudi Arabia. Antibiotics. 2022; 11(12):1697. https://doi.org/10.3390/antibiotics11121697
Chicago/Turabian StyleAlshahrani, Abdullah M., Mutasim E. Ibrahim, Ahmed K. Aldossary, Mushabab A. Alghamdi, Omar B. Ahmed, and Aref A. Bin Abdulhak. 2022. "Molecular Epidemiology of Carbapenem-Resistant K. pneumoniae Clinical Isolates from the Adult Patients with Comorbidities in a Tertiary Hospital, Southern Saudi Arabia" Antibiotics 11, no. 12: 1697. https://doi.org/10.3390/antibiotics11121697
APA StyleAlshahrani, A. M., Ibrahim, M. E., Aldossary, A. K., Alghamdi, M. A., Ahmed, O. B., & Bin Abdulhak, A. A. (2022). Molecular Epidemiology of Carbapenem-Resistant K. pneumoniae Clinical Isolates from the Adult Patients with Comorbidities in a Tertiary Hospital, Southern Saudi Arabia. Antibiotics, 11(12), 1697. https://doi.org/10.3390/antibiotics11121697






