Deep Learning and Antibiotic Resistance
Abstract
:1. Introduction
2. History
3. Present
3.1. Antimicrobial Peptides Testing
3.2. Detection of AR Genes
3.3. Other Measures to Decrease AR
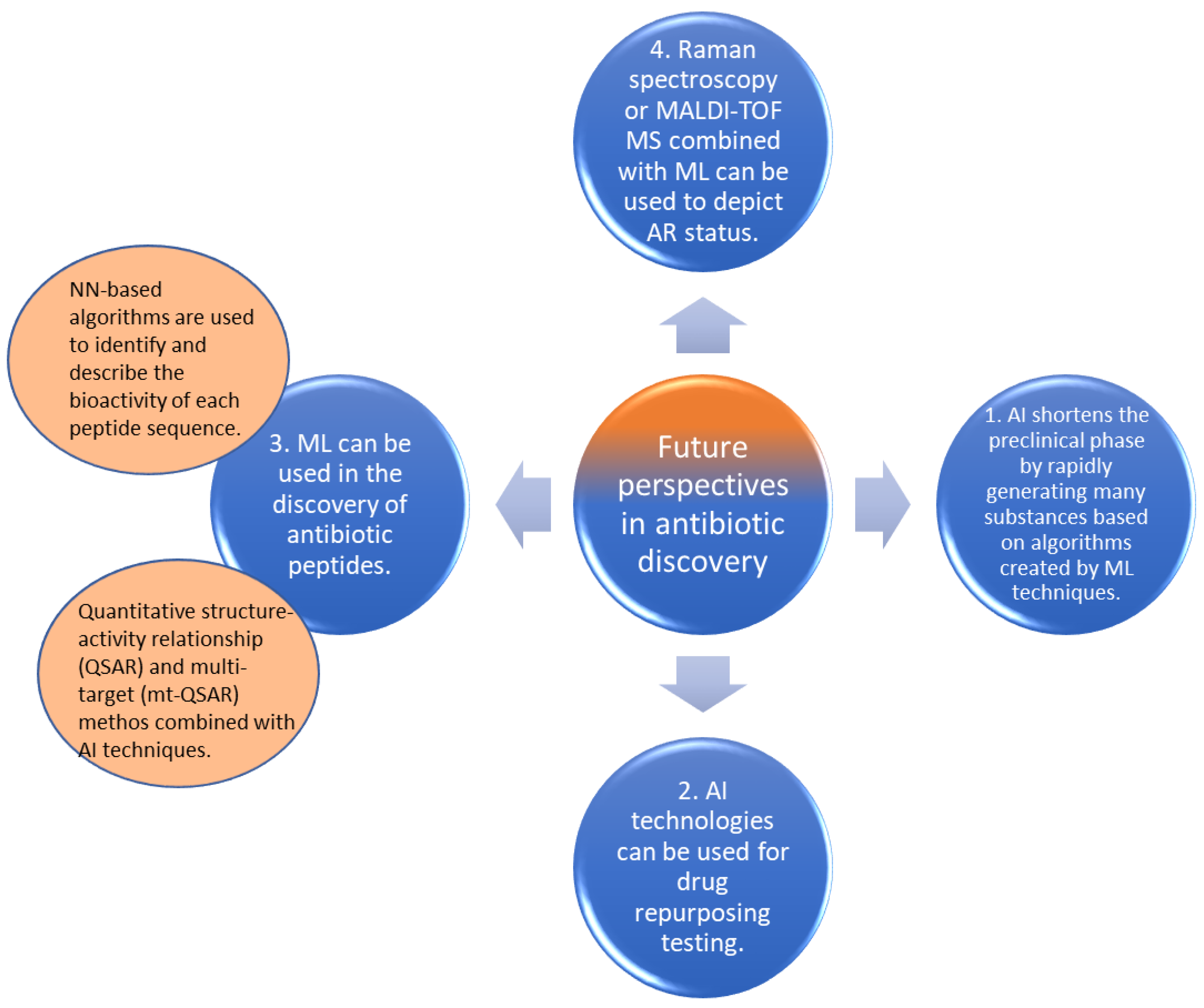
4. Future Perspectives
4.1. Critical Findings Concerning AI in Antibiotics Development
4.2. Drug Repurposing Testing
4.3. Discovery of Antibiotic Peptides
4.4. Other Applications Combined with AI for Antibiotic Discovery
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Perry, J.; Waglechner, N.; Wright, G. The Prehistory of Antibiotic Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- A Scientific Roadmap for Antibiotic Discovery. Available online: http://pew.org/26ZYUqA (accessed on 15 September 2022).
- Wilson, L.A.; Rogers Van Katwyk, S.; Fafard, P.; Viens, A.M.; Hoffman, S.J. Lessons learned from COVID-19 for the post-antibiotic future. Glob. Health 2020, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Gould, I.M.; Bal, A.M. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence 2013, 4, 185–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watkins, R.R.; Bonomo, R.A. Overview: Global and Local Impact of Antibiotic Resistance. Infect. Dis. Clin. N. Am. 2016, 30, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar]
- Jukič, M.; Bren, U. Machine Learning in Antibacterial Drug Design. Front. Pharmacol. 2022, 13, 864412. [Google Scholar] [CrossRef]
- Martorell-Marugán, J.; Tabik, S.; Benhammou, Y.; del Val, C.; Zwir, I.; Herrera, F.; Carmona-Sáez, P. Deep Learning in Omics Data Analysis and Precision Medicine. In Computational Biology; Chapter 3; Husi, H., Ed.; Codon Publications: Brisbane, Australia, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK550335/ (accessed on 21 September 2022). [CrossRef] [Green Version]
- David, L.; Brata, A.M.; Mogosan, C.; Pop, C.; Czako, Z.; Muresan, L.; Ismaiel, A.; Dumitrascu, D.I.; Leucuta, D.C.; Stanculete, M.F.; et al. Artificial Intelligence and antibiotic discovery. Antibiotics 2021, 10, 1376. [Google Scholar] [CrossRef]
- Gupta, R.; Srivastava, D.; Sahu, M.; Tiwari, S.; Ambasta, R.K.; Kumar, P. Artificial Intelligence to deep learning: Machine intelligence approach for drug discovery. Mol. Divers. 2021, 25, 1315–1360. [Google Scholar] [CrossRef]
- Iskandar, K.; Murugaiyan, J.; Halat, D.H.; El Hage, S.; Chibabhai, V.; Adukkadukkam, S.; Roques, C.; Molinier, L.; Salameh, P.; Van Dongen, M. Antibiotic Discovery and Resistance: The Chase and the Race. Antibiotics 2022, 11, 182. [Google Scholar] [CrossRef]
- Jacoby, G.A. History of Drug-Resistant Microbes. In Antimicrobial Drug Resistance: Mechanisms of Drug Resistance; Infectious Disease; Mayers, D.L., Ed.; Humana Press: Totowa, NJ, USA, 2009; pp. 3–7. [Google Scholar] [CrossRef]
- Petrova, M.; Gorlenko, Z.; Mindlin, S. Molecular structure and translocation of a multiple antibiotic resistance region of a Psychrobacter psychrophilus permafrost strain. FEMS Microbiol. Lett. 2009, 296, 190–197. [Google Scholar] [CrossRef] [Green Version]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic resistance is ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef]
- CDC Newsroom. CDC. 2016. Available online: https://www.cdc.gov/media/releases/2016/p0503-unnecessary-prescriptions.html (accessed on 20 September 2022).
- Sengupta, S.; Barman, P.; Lo, J. Opportunities to Overcome Implementation Challenges of Infection Prevention and Control in Low-Middle Income Countries. Curr. Treat. Options Infect. Dis. 2019, 11, 267–280. [Google Scholar] [CrossRef] [Green Version]
- Ruiz Puentes, P.; Henao, M.C.; Cifuentes, J.; Muñoz-Camargo, C.; Reyes, L.H.; Cruz, J.C.; Arbeláez, P. Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence. Membranes 2022, 12, 708. [Google Scholar] [CrossRef]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive deep learning model for discovery of novel antimicrobial peptides effective against WHO priority pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Lin, T.-T.; Yang, L.-Y.; Lu, I.-H.; Cheng, W.-C.; Hsu, Z.-R.; Chen, S.-H.; Lin, C.-Y. AI4AMP: An Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning. mSystems 2021, 6, e00299-21. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Z.; Han, W.; Cao, H.; Umarov, R.; Yan, A.; Fan, M.; Chen, H.; Duarte, C.M.; Li, L.; et al. HMD-ARG: Hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome 2021, 9, 40. [Google Scholar]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Steiner, M.C.; Gibson, K.M.; Crandall, K.A. Drug Resistance Prediction Using Deep Learning Techniques on HIV-1 Sequence Data. Viruses 2020, 12, 560. [Google Scholar]
- Mathur, P. Hand hygiene: Back to the basics of infection control. Indian J. Med. Res. 2011, 134, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Curtis, L.T. Prevention of hospital-acquired infections: Review of non-pharmacological interventions. J. Hosp. Infect. 2008, 69, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Neiderud, C.J. How urbanization affects the epidemiology of emerging infectious diseases. Infect. Ecol. Epidemiol. 2015, 5, 27060. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Yang, Y.; Cao, S.; Liu, H.; Li, X.; Sun, J.; Li, F.; Ishfaq, M.; Zhang, X. Prevalence and Characteristic of Swine-Origin mcr-1-Positive Escherichia coli in Northeastern China. Front. Microbiol. 2021, 12, 712707. [Google Scholar] [CrossRef] [PubMed]
- Chroboczek, T.; Boisset, S.; Rasigade, J.P.; Meugnier, H.; Akpaka, P.E.; Nicholson, A.; Nicolas, M.; Olive, C.; Bes, M.; Vandenesch, F.; et al. Major West Indies MRSA clones in human beings: Do they travel with their hosts? J. Travel Med. 2013, 20, 283–288. [Google Scholar] [CrossRef] [Green Version]
- Boyd, N.K.; Teng, C.; Frei, C.R. Brief Overview of Approaches and Challenges in New Antibiotic Development: A Focus on Drug Repurposing. Front. Cell. Infect. Microbiol. 2021, 11, 684515. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; Macnair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A deep learning approach to antibiotic discovery. Cell 2020, 180, 688–702.e13. [Google Scholar] [CrossRef] [Green Version]
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar] [CrossRef]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [Green Version]
- Pandey, V.; Tiwari, P.; Gangopadhyay, A.N.; Gupta, D.K.; Sharma, S.P.; Kumar, V. Propranolol for Infantile Haemangiomas: Experience from a Tertiary Center. J. Cutan. Aesthetic Surg. 2014, 7, 37–41. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alhumaid, S.; Al Mutair, A.; Garout, M.; Abulhamayel, Y.; Halwani, M.A.; Alestad, J.H.; Al Bshabshe, A.; Sulaiman, T.; AlFonaisan, M.K.; et al. Application of Artificial Intelligence in Combating High Antimicrobial Resistance Rates. Antibiotics 2022, 11, 784. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Cao, J.; de la Fuente-Nunez, C.; Ou, W.R.; Torres, T.M.D.; Pande, G.S.; Sinskey, J.A.; Lu, T.K. Yeast-Based Synthetic Biology Platform for Antimicrobial Peptide Production. ACS Synth. Biol. 2018, 7, 896–902. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Kwon, S.; Bae, H.; Jo, J.; Yoon, S. Comprehensive ensemble in QSAR prediction for drug discovery. BMC Bioinform. 2019, 20, 521. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Chen, J.; Liu, C.; Zeng, Y.; Sun, Q.; Li, J.; Shen, Z.; Chen, S.; Zhang, R. Identification of antibiotic resistance and virulence-encoding factors in Klebsiella pneumoniae by Raman spectroscopy and deep learning. Microb. Biotechnol. 2022, 15, 1270–1280. [Google Scholar] [CrossRef]
- Brincat, A.; Hofmann, M. Automated extraction of genes associated with antibiotic resistance from the biomedical literature. Database 2022, 2022, baab077. [Google Scholar] [CrossRef]
- Singh, T.; Choudhary, P.; Singh, S. Antimicrobial Peptides: Mechanism of Action. In Insights on Antimicrobial Peptides; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- Fjell, C.D.; Jenssen, H.; Hilpert, K.; Cheung, W.A.; Panté, N.; Hancock, R.E.W.; Cherkasov, A. Identification of Novel Antibacterial Peptides by Chemoinformatics and Machine Learning. J. Med. Chem. 2009, 52, 2006–2015. [Google Scholar] [CrossRef]
- Kleandrova, V.V.; Ruso, J.M.; Speck-Planche, A.; Dias Soeiro Cordeiro, M.N. Enabling the Discovery and Virtual Screening of Potent and Safe Antimicrobial Peptides. Simultaneous Prediction of Antibacterial Activity and Cytotoxicity. ACS Comb. Sci. 2016, 18, 490–498. [Google Scholar] [CrossRef]
- Kleandrova, V.V.; Scotti, M.T.; Speck-Planche, A. Computational Drug Repurposing for Antituberculosis Therapy: Discovery of Multi-Strain Inhibitors. Antibiotics 2021, 10, 1005. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; You, R.; Zhu, S.; Zhou, X.J.; Sun, F. ARG-SHINE: Improve antibiotic resistance class prediction by integrating sequence homology, functional information and deep convolutional neural network. NAR Genom. Bioinform. 2021, 3, lqab066. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Abbas, A.; Kim, M.; Shin, J.; Kim, Y.M.; Cho, K.H. Prediction of antibiotic-resistance genes occurrence at a recreational beach with deep learning models. Water Res. 2021, 196, 117001. [Google Scholar] [CrossRef] [PubMed]
- Legenza, L.; Barnett, S.; Lacy, J.P.; See, C.; Desotell, N.; Eibergen, A.; Piccirillo, J.F.; Rose, W.E. Geographic mapping of Escherichia coli susceptibility to develop a novel Clinical Decision Support Tool. Antimicrob. Agents Chemother. 2019, 63, e00048-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, T.-Y.; Chuan, C.-N.; Teng, S.-H. Current status of MALDI-TOF mass spectrometry in clinical microbiology. J. Food Drug Anal. 2019, 27, 404–414. [Google Scholar] [CrossRef]
- Idelevich, E.A.; Becker, K. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 2021, 59, e0181419. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popa, S.L.; Pop, C.; Dita, M.O.; Brata, V.D.; Bolchis, R.; Czako, Z.; Saadani, M.M.; Ismaiel, A.; Dumitrascu, D.I.; Grad, S.; et al. Deep Learning and Antibiotic Resistance. Antibiotics 2022, 11, 1674. https://doi.org/10.3390/antibiotics11111674
Popa SL, Pop C, Dita MO, Brata VD, Bolchis R, Czako Z, Saadani MM, Ismaiel A, Dumitrascu DI, Grad S, et al. Deep Learning and Antibiotic Resistance. Antibiotics. 2022; 11(11):1674. https://doi.org/10.3390/antibiotics11111674
Chicago/Turabian StylePopa, Stefan Lucian, Cristina Pop, Miruna Oana Dita, Vlad Dumitru Brata, Roxana Bolchis, Zoltan Czako, Mohamed Mehdi Saadani, Abdulrahman Ismaiel, Dinu Iuliu Dumitrascu, Simona Grad, and et al. 2022. "Deep Learning and Antibiotic Resistance" Antibiotics 11, no. 11: 1674. https://doi.org/10.3390/antibiotics11111674
APA StylePopa, S. L., Pop, C., Dita, M. O., Brata, V. D., Bolchis, R., Czako, Z., Saadani, M. M., Ismaiel, A., Dumitrascu, D. I., Grad, S., David, L., Cismaru, G., & Padureanu, A. M. (2022). Deep Learning and Antibiotic Resistance. Antibiotics, 11(11), 1674. https://doi.org/10.3390/antibiotics11111674








