Targeting SAM-I Riboswitch Using Antisense Oligonucleotide Technology for Inhibiting the Growth of Staphylococcus aureus and Listeria monocytogenes
Abstract
:1. Introduction
2. Results
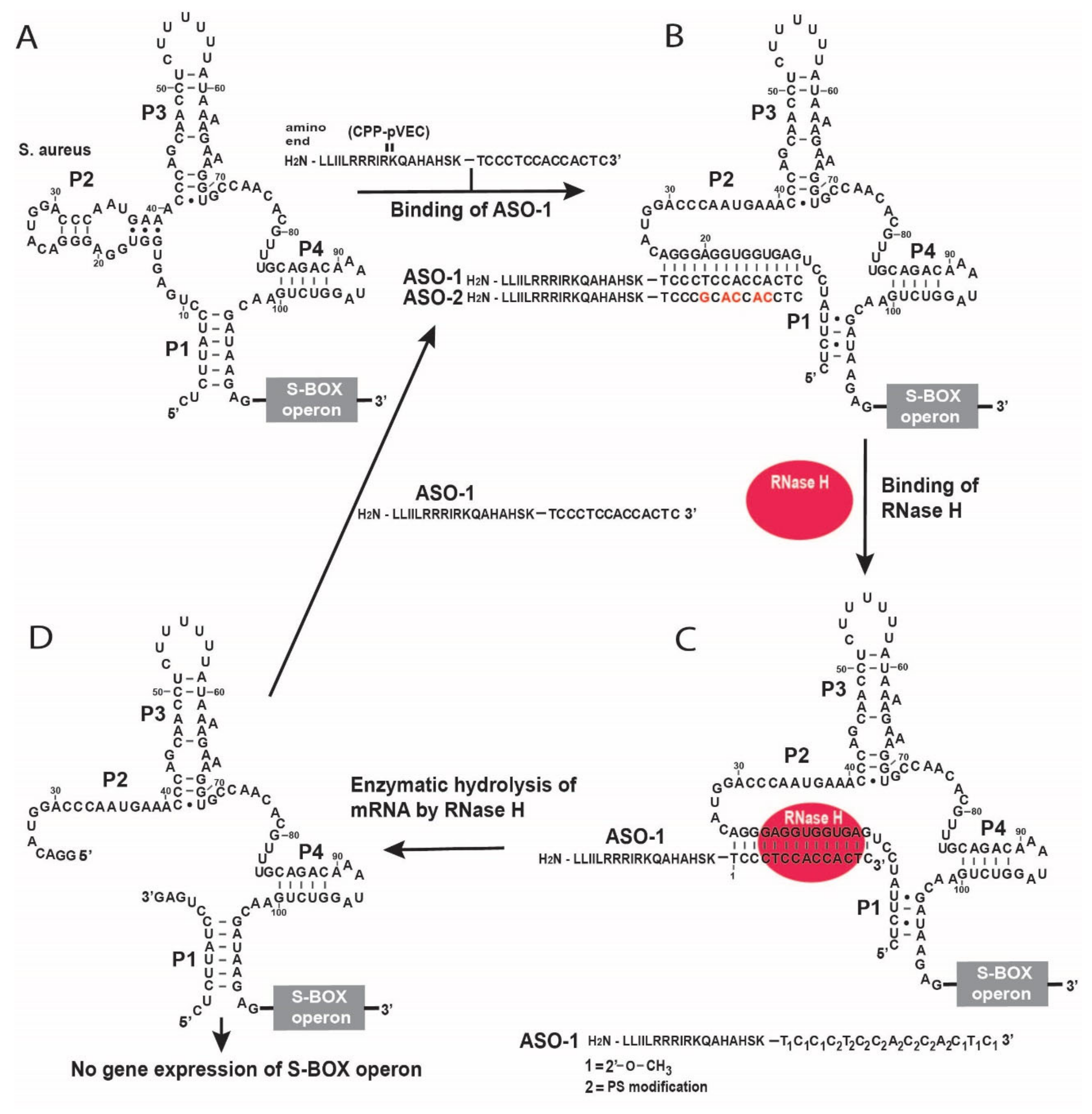
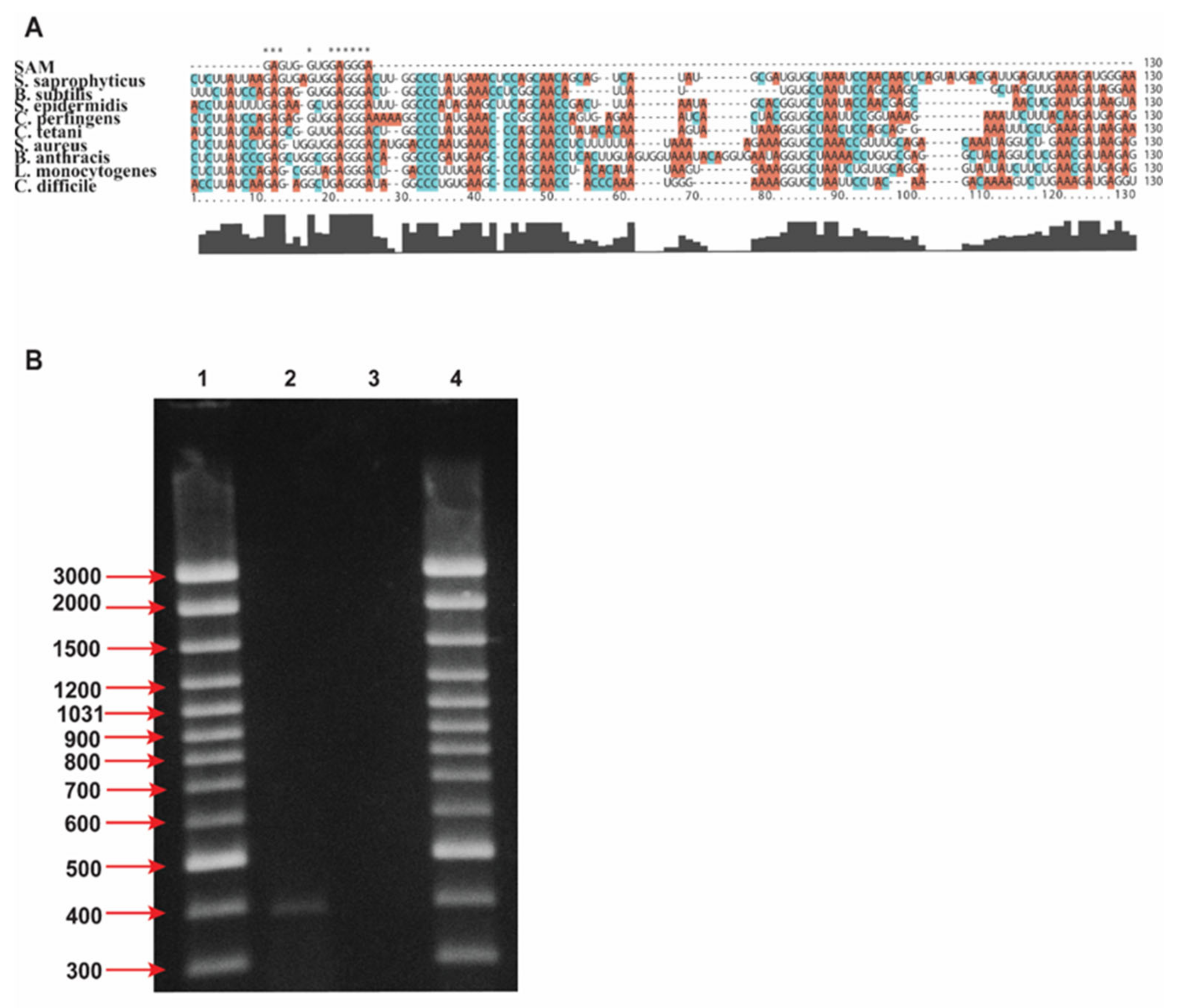
2.1. Design of Antisense Oligonucleotide Targeting SAM-I Riboswitches
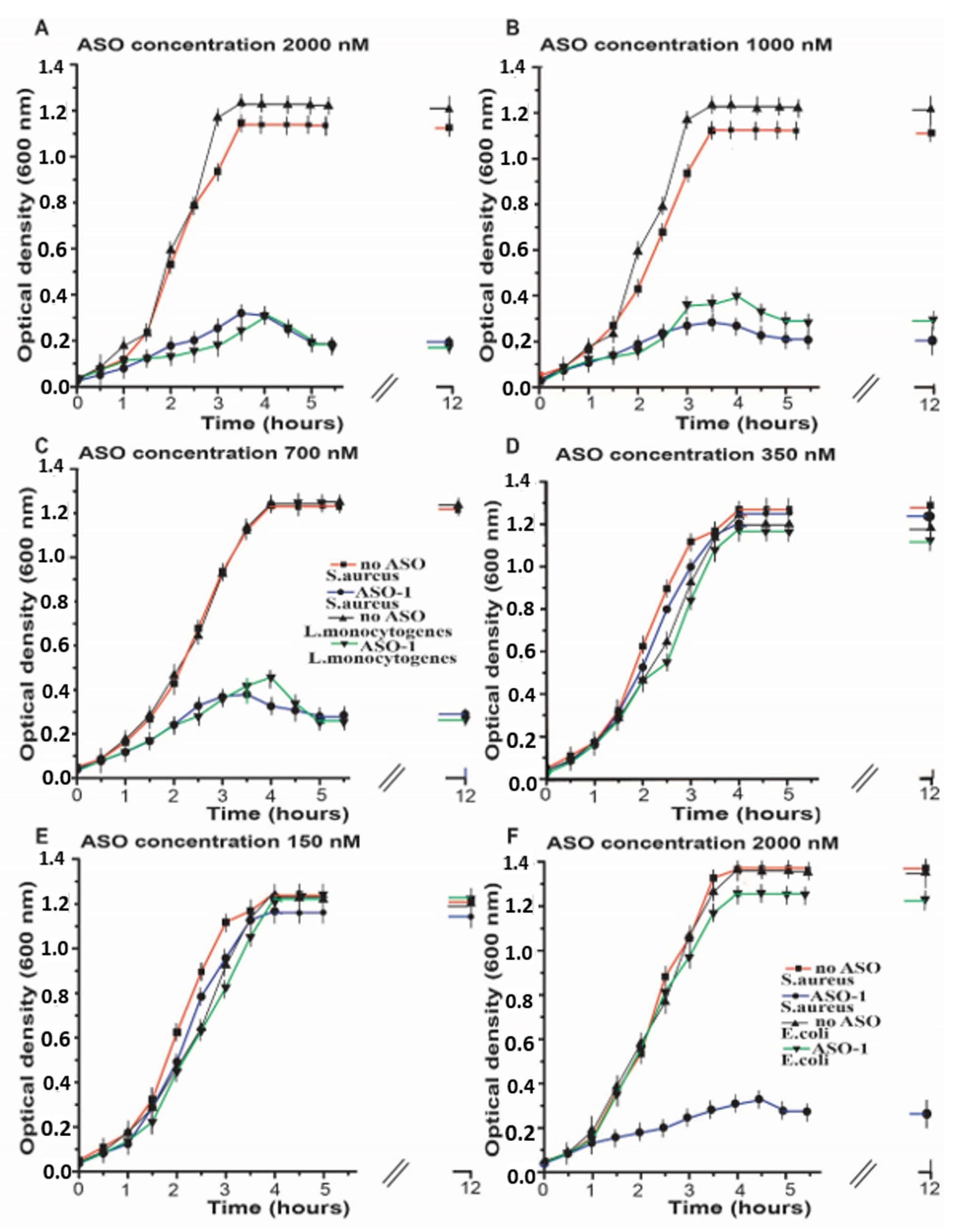
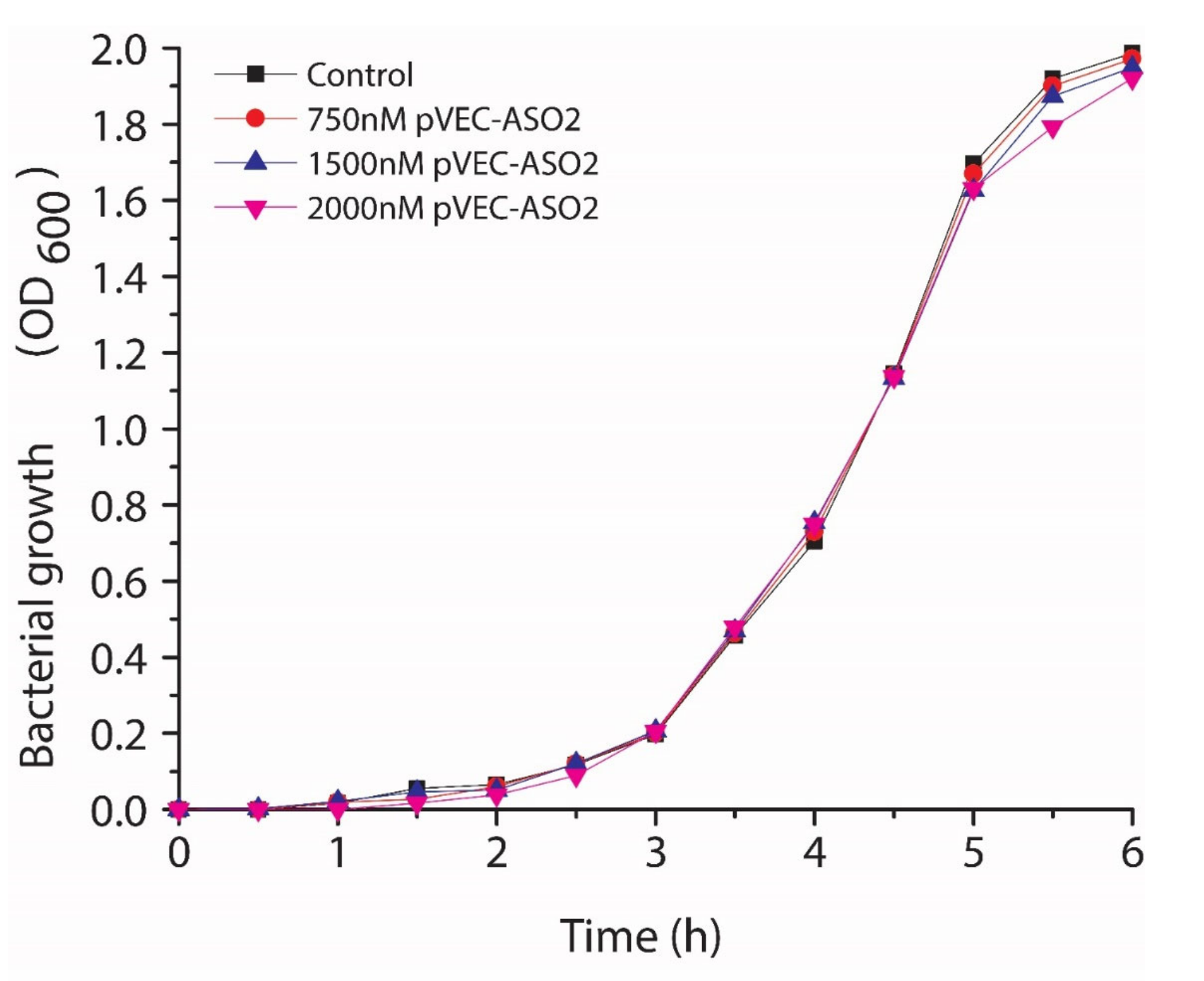
2.2. Inhibition of the Bacterial Growth Via the S-box mRNA with ASO That Targets the SAM-I Riboswitches
3. Discussion
4. Conclusions
5. Experimental Section
5.1. Databases for Bioinformatics Analysis
5.2. Antisense Oligonucleotide Sequence
5.3. PCR Amplification
5.4. Bacterial Strains and Media
5.5. Testing the ASO Toxicity in Human Cell Line
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Valsamatzi-Panagiotou, A.; Popova, K.B.; Penchovsky, R. Methods for prevention and constraint of antimicrobial resistance: A review. Environ. Chem. Lett. 2021, 19, 2005–2012. [Google Scholar] [CrossRef]
- Kaji, T.; Murai, M.; Itoh, H.; Yasukawa, J.; Hamamoto, H.; Sekimizu, K.; Inoue, M. Total Synthesis and Functional Evaluation of Fourteen Derivatives of Lysocin E: Importance of Cationic, Hydrophobic, and Aromatic Moieties for Antibacterial Activity. Chemistry 2016, 22, 16912–16919. [Google Scholar] [CrossRef] [PubMed]
- Penchovsky, R.; Traykovska, M. Designing drugs that overcome antibacterial resistance: Where do we stand and what should we do? Expert Opin. Drug Discov. 2015, 10, 631–650. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.; Penchovsky, R. Genome-wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria. Expert Opin. Ther. Targets 2019, 23, 631–643. [Google Scholar] [CrossRef]
- Pavlova, N.; Penchovsky, R. Bioinformatics and Genomic Analyses of the Suitability of Eight Riboswitches for Antibacterial Drug Targets. Antibiotics 2022, 11, 1177. [Google Scholar] [CrossRef]
- Traykovska, M.; Popova, K.B.; Penchovsky, R. Targeting glmS Ribozyme with Chimeric Antisense Oligonucleotides for Antibacterial Drug Development. ACS Synth. Biol. 2021, 10, 3167–3176. [Google Scholar] [CrossRef]
- Traykovska, M.; Penchovsky, R. Engineering antisense oligonucleotides as antibacterial agents that target FMN riboswitches and inhibit the growth of Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli. ACS Synth. Biol. 2022, 11, 1845–1855. [Google Scholar] [CrossRef]
- Traykovska, M.; Otcheva, L.A.; Penchovsky, R. Targeting TPP Riboswitches Using Chimeric Antisense Oligonucleotide Technology for Antibacterial Drug Development. ACS Appl. Bio. Mater. 2022, 5, 4896–4902. [Google Scholar] [CrossRef]
- Bai, H.; You, Y.; Yan, H.; Meng, J.; Xue, X.; Hou, Z.; Zhou, Y.; Ma, X.; Sang, G.; Luo, X. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials 2012, 33, 659–667. [Google Scholar] [CrossRef]
- Gheibi-Hayat, S.M.; Jamialahmadi, K. Antisense Oligonucleotide (AS-ODN) Technology: Principle, Mechanism and Challenges. Biotechnol. Appl. Biochem. 2020, 68, 1086–1094. [Google Scholar] [CrossRef]
- Kilanowska, A.; Nuckowski, L.; Studzinska, S. Studying in vitro metabolism of the first and second generation of antisense oligonucleotides with the use of ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Anal. Bioanal. Chem. 2020, 412, 7453–7467. [Google Scholar] [CrossRef] [PubMed]
- Papargyri, N.; Pontoppidan, M.; Andersen, M.R.; Koch, T.; Hagedorn, P.H. Chemical Diversity of Locked Nucleic Acid-Modified Antisense Oligonucleotides Allows Optimization of Pharmaceutical Properties. Mol. Ther. Nucleic Acids 2020, 19, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Lapuebla, A.; Landman, D.; Quale, J. In Vitro and In Vivo Activity of a Novel Antisense Peptide Nucleic Acid Compound Against Multidrug-Resistant Acinetobacter baumannii. Microb. Drug Resist. 2019, 25, 961–965. [Google Scholar] [CrossRef] [PubMed]
- Medina Escobar, A.; Bruno, V.; Sarna, J.R. Second-Generation Antisense Nucleotide Targeting Huntingtin Expression Found to Be Safe in Patients With Huntington’s Disease. Mov. Disord. Clin. Pract. 2019, 6, 434–435. [Google Scholar] [CrossRef]
- Lu, C.; Ding, F.; Chowdhury, A.; Pradhan, V.; Tomsic, J.; Holmes, W.M.; Henkin, T.M.; Ke, A. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-I riboswitch. J. Mol. Biol. 2010, 404, 803–818. [Google Scholar] [CrossRef] [Green Version]
- Penchovsky, R.; Stoilova, C.C. Riboswitch-based antibacterial drug discovery using high-throughput screening methods. Expert Opin. Drug Discov. 2013, 8, 65–82. [Google Scholar] [CrossRef]
- Panchal, V.; Brenk, R. Riboswitches as Drug Targets for Antibiotics. Antibiotics 2021, 10, 45. [Google Scholar] [CrossRef]
- Pavlova, N.; Kaloudas, D.; Penchovsky, R. Riboswitch distribution, structure, and function in bacteria. Gene 2019, 708, 38–48. [Google Scholar] [CrossRef]
- Stoddard, C.D.; Montange, R.K.; Hennelly, S.P.; Rambo, R.P.; Sanbonmatsu, K.Y.; Batey, R.T. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 2010, 18, 787–797. [Google Scholar] [CrossRef] [Green Version]
- Hennelly, S.P.; Sanbonmatsu, K.Y. Tertiary contacts control switching of the SAM-I riboswitch. Nucleic Acids Res. 2011, 39, 2416–2431. [Google Scholar] [CrossRef]
- Boyapati, V.K.; Huang, W.; Spedale, J.; Aboul-ela, F. Basis for ligand discrimination between ON and OFF state riboswitch conformations: The case of the SAM-I riboswitch. RNA 2012, 18, 1230–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Aboul-ela, F.; Kim, J.; Jha, S.; Boyapati, V. Dynamics differences of SAM-I riboswitch aptamer between SAM bound and without SAM: Insight into conformational rearrangement. FASEB J. 2009, 23, 842.1. [Google Scholar] [CrossRef]
- Berthold, P.R.; Shiraishi, T.; Nielsen, P.E. Cellular delivery and antisense effects of peptide nucleic acid conjugated to polyethyleneimine via disulfide linkers. Bioconjug. Chem. 2010, 21, 1933–1938. [Google Scholar] [CrossRef] [PubMed]
- Kaloudas, D.; Pavlova, N.; Penchovsky, R. EBWS: Essential Bioinformatics Web Services for Sequence Analyses. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 942–953. [Google Scholar] [CrossRef]
- Fluiter, K.; Mook, O.R.; Vreijling, J.; Langkjaer, N.; Højland, T.; Wengel, J.; Baas, F. Filling the gap in LNA antisense oligo gapmers: The effects of unlocked nucleic acid (UNA) and 4′-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. Biosyst. 2009, 5, 838–843. [Google Scholar] [CrossRef]
- Frontiñan-Rubio, J.; González, V.J.; Vázquez, E.; Durán-Prado, M. Rapid and efficient testing of the toxicity of graphene-related materials in primary human lung cells. Sci. Rep. 2022, 12, 7664. [Google Scholar] [CrossRef]
- Penchovsky, R.; Pavlova, N.; Kaloudas, D. RSwitch: A Novel Bioinformatics Database on Riboswitches as Antibacterial Drug Targets. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 804–808. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 8. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traykovska, M.; Penchovsky, R. Targeting SAM-I Riboswitch Using Antisense Oligonucleotide Technology for Inhibiting the Growth of Staphylococcus aureus and Listeria monocytogenes. Antibiotics 2022, 11, 1662. https://doi.org/10.3390/antibiotics11111662
Traykovska M, Penchovsky R. Targeting SAM-I Riboswitch Using Antisense Oligonucleotide Technology for Inhibiting the Growth of Staphylococcus aureus and Listeria monocytogenes. Antibiotics. 2022; 11(11):1662. https://doi.org/10.3390/antibiotics11111662
Chicago/Turabian StyleTraykovska, Martina, and Robert Penchovsky. 2022. "Targeting SAM-I Riboswitch Using Antisense Oligonucleotide Technology for Inhibiting the Growth of Staphylococcus aureus and Listeria monocytogenes" Antibiotics 11, no. 11: 1662. https://doi.org/10.3390/antibiotics11111662




