Abstract
Klebsiella pneumoniae (K. pneumoniae) is involved in several hospital and community-acquired infections. The prevalence of K. pneumoniae-producing-carbapenemase (KPC) resistance genes rapidly increases and threatens public health worldwide. This study aimed to assess the antibiotic resistance level of K. pneumoniae isolates from Makkah Province, Saudi Arabia, during the Islamic ‘Umrah’ ritual and to identify the plasmid types, presence of genes associated with carbapenem hydrolyzing enzymes, and virulence factors. The phenotypic and genotypic analyses based on the minimum inhibitory concentration (MIC), biofilm formation, PCR, and characterization of KPC-encoding plasmids based on the replicon typing technique (PBRT) were explored. The results showed that most isolates were resistant to carbapenem antibiotics and other antibiotics classes. This study identified sixteen different replicons of plasmids in the isolates and multiple genes encoding carbapenem factors, with blaVIM and blaOXA-48 being the most prevalent genes identified in the isolates. However, none of the isolates exhibited positivity for the KPC production activity. In addition, this study also identified six virulence-related genes, including kfu, wabG, uge, rmpA, fimH, and a capsular polysaccharide (CPS). Together, the data reported in this study indicate that the isolated K. pneumoniae during the pilgrimage in Makkah were all resistant to carbapenem antibiotics. Although the isolates lacked KPC production activity, they carried multiple carbapenem-resistant genes and virulence factors, which could drive their resistant phenotype. The need for specialized methods for KPC detection, monitoring the possibility of nosocomial transmission, and diverse therapeutic alternatives are necessary for controlling the spreading of KPC. This study can serve as a reference for clinicians and researchers on types of K. pneumoniae commonly found during religious gathering seasons in Saudi Arabia.
1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is an encapsulated, non-motile gram-negative nosocomial pathogenic [1,2]. This bacterium involves several disseminated hospital and community-acquired infections, such as pneumonia, septicemia, surgical site infections, and urinary tract infections (UTIs) [3]. K. pneumoniae belongs to the ESKAPE pathogenic group, which includes Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species, which escapes the antimicrobial drugs through multi-action mechanisms [4,5]. The evolution of multidrug-resistant infections (MDR) has been facilitated by the improper use of antimicrobial medications over time, which led to limited treatment options [6]. Given this information, previous research indicated that the mortality rate from severe infections exceeded 40%. This high prevalence is associated with carbapenem-resistant K. pneumoniae (CRKP) strains, which are primarily hospital-acquired infections (HAIs) [7,8,9]. In 2017, the World Health Organization (WHO) classified and listed CRKP among the global pathogens that pose an unprecedented challenge in developing new antibiotics [10].
K. pneumoniae-producing-carbapenemase (KPC) was first identified in the first decade of the 21st century [11]. Since then, the KPC-encoding gene has been noticed frequently in several regions, including Europe, Asia, and the Middle East [12]. The emergence of K. pneumoniae harboring carbapenemase resistance genes posed a significant threat to global public health [13]. In recent years, extensive dissemination, rapid spreading, and failure to treat KPC infections have been associated with multiple resistance mechanisms. For instance, alteration of the bacterial target site, inactivation of the antimicrobial metabolic pathways, reduction of antibiotic accumulation through efflux pump systems, and biofilm formation [14]. It should be noted that plasmid-mediated carbapenemase genes are one of the most concerning mechanisms of antibiotic resistance by K. pneumoniae [15]. Its ability to anchor and rapidly spread MDR elements allows it to attain numerous β-lactamase enzymes that can break down the β-lactam ring in strong antibiotics, such as carbapenems used as the final line of infection therapy [16].
Based on the Ambler classification system, carbapenemases are divided into A, B, and D β-lactamase classes [17]. Class C possesses chromosome-encoded cephalosporinase, producing insignificant effects against carbapenems [17]. Class A carbapenemases are encoded by chromosomes, such as Sme, SFC-1, IMI-1, and NmcA. The plasmid encodes other classes, namely KPCs, GES, and IMI-2 [18]. These carbapenemases are effective against carbapenems, but the most clinically prevalent is the KPC [19]. From the molecular level of class B β-lactamases, these enzymes have active-site zinc, making them metalloenzymes [20]. The most common families of metallo-β-lactamases are GIM, IMP, SIM, and VIM. Another metallo-β-lactamase, NDM-1, was discovered in 2009 in clinical isolates of K. pneumoniae and Escherichia coli in India [21]. This new family of carbapenems enzymes poses a high threat to community health due to the rapid dissemination of the blaNDM gene in isolates [22]. Class D β-lactamases contain more than 350 OXA-enzymes, also known as oxacillinases (OXA-type enzymes) [23]. This class of β-lactamases includes carbapenem-hydrolyzing class D β-lactamases (CHDLs) or Class D β-lactamases that are resistant carbapenem antibiotics. Several subgroups of CHDLs are identified according to their amino acid sequence identity. The most disseminated enzyme subgroups of CHDLs in bacterial pathogens are OXA-23, OXA-24/40, OXA-48, OXA-51, OXA-58, and OXA-143 [14]. OXA-48 carbapenemase is the enzyme with the highest virulence factors among other subgroups and is presented in K. pneumoniae and other Enterobacteriaceae family members. The rest of the enzyme subgroups can mainly be found in Acinetobacter isolates [24].
Virulence genes play a vital role in K. pneumoniae’s pathogenicity. These genes encode virulence factors such as hypermucoviscosity, lipopolysaccharide formation, capsule synthesis, adhesion, and iron uptake systems [25,26]. A positive string test identifies hypervirulent K. pneumoniae (hvKp) isolates, which harbor hypermucoviscosity colonies [27]. This phenotype was confirmed by two genes, rmpA and magA, that regulate capsule polysaccharide synthesis and mucoviscosity [28]. Among the 79 defined capsular serotypes in previous studies, capsule serotypes K1 and K2 are the most common and frequently associated with the rmpA gene, allowing bacteria to evade the immune system [29]. Another gene that regulates capsule formation, hypermucoviscosity, and iron transport system is the Klebsiella ferric iron uptake (Kfu) [30]. wabG and uge are two other hypervirulence genes that promote lipopolysaccharides synthesis and robust K. pneumoniae resistance [31]. Moreover, the fimH gene regulates fimbriae protein and the bacteria cell’s adhesion [32]. Consequently, the most concerning aspect of K. pneumoniae is the dual-risk isolates which produce carbapenemases and carry hypervirulence genes [33].
This study describes the characteristics of the antibiotic-resistant K. pneumoniae isolates from Makkah Province, Saudi Arabia, during the Islamic pilgrimage ‘Umrah’ ritual. It aims to identify the plasmid types, the presence of genes associated with carbapenem hydrolyzing enzymes and virulence factors, and to determine antibiotic susceptibility by performing the minimum inhibitory concentration (MIC) assay. The study may serve as a potential reference for clinicians and research scientists to understand the nature of K. pneumoniae KPC, that are common in religious gathering seasons in Saudi Arabia. This study also highlights the need for rapid methods for KPC detection, monitoring the possibility of nosocomial transmission, and exploring novel therapeutic alternatives for controlling the spreading of KPC.
2. Results and Discussion
2.1. Bacterial Identification and Antibiotic Susceptibility Test
A total of 23 carbapenems-resistant K. pneumoniae (CRKP) strains were collected from different patients in King Faisal Hospital in Makkah province, Saudi Arabia, during the religious gathering season of Umrah. Samples were isolated from sputum, wounds, urine, blood, and central venous puncture, with ratios of 43%, 22%, 17%, 13%, and 4%, respectively. The isolates’ identification confirmation was processed using a MicroScan and VITEK2. The MIC of the nine antibiotics used against the isolates and the ATCC reference strain (BAA 1705) showed a remarkable resistance to carbapenems drugs. Most isolates exhibited resistance against FOX, CPM, AZT, CAZ, CTC, CIP, AMP, and most importantly against MEM and IMI, all demonstrating MIC of >1024 µg/mL, as shown in Table 1. However, there was only one isolate (K5) that showed an intermediate susceptibility for FOX (MIC = 16 µg/mL) and CIP (MIC = 2 µg/mL), while the MIC was <0.5 µg/mL against AZT, CAZ, and CPM. The same isolate had above-the-threshold resistance levels for CTC, IMI, and AMP (MIC ≥ 1024 µg/mL). Two strains, K5 and K21, are susceptible to MEM (MIC < 0.5 µg/mL) among the 23 isolates. Additionally, isolate K9 exhibited a sensitivity for IMI at MIC = 2 µg/mL, as shown in Table 1. The study showed that MEM was considered more effective than IMI, which 33% of samples resistant to IMI (MIC of >1024 µg/mL). These results are consistent with a study by Subash et al., who reported that the sensitivity of Gram-negative bacilli to MEM is more than IMI due to the overuse of the latter antibiotic (for many years) as an empirical treatment option [34].

Table 1.
MIC values of different antibiotics against carbapenemase-producing K. pneumoniae isolates. Antibiotic resistance (R) values were indicated according to the Clinical and Laboratory Standards Institute (CLSI) (M100-S24) [35].
2.2. Biofilm Formation of K. pneumoniae
All 23 K. pneumoniae isolates and the ATCC reference strain (700603) were tested for their ability to form biofilms, as shown in Table 2. The biofilm formation of the 23 isolates was interpreted by four standard criteria for producing biofilm, as shown in Table 2. The findings showed that nine isolates (≈39.13%) were non-biofilm-forming strains, while eleven were weak biofilm-forming strains (≈8.83%). In addition, only three isolates out of the 23 (≈13.04%) formed biofilm moderately. There was no evidence of strong biofilm formation for all 23 isolates (0%) (Table 2). The ATCC strain (700603) showed moderate biofilm formation ability. More details on the biofilm formation level of all K. pneumoniae strains are shown in the Supplementary Materials Section (Table S1).

Table 2.
The frequency of carbapenemase-resistant K. pneumoniae isolates forming biofilm (n = 23).
This table shows that 13% of all collected isolates could moderately form a biofilm, which they initially collected from the sputum and wound. By comparison, other isolates (≈87%) were collected from the sputum, blood, wound, urine, and central venous puncture. There was no relationship between the source of isolates and the biofilms-forming. Ashwath et al. reported that the ability of bacteria to form biofilms is not necessarily involved in the source of isolation [36]. It is important to note that 39% of the non-biofilm-forming isolates have high resistance to carbapenem antibiotics, which could indicate a lack of association between biofilm formation and resistance to carbapenem antibiotics. This latter might occur due to other antibiotic-resistance factors. Further studies are needed to confirm the irrelevance between biofilm formation and carbapenem antibiotics resistance.
2.3. Detection of Plasmids by PCR-Based Replicon Typing (PBRT)
Thirty replicons were amplified using eight multiplex PCR assays, as shown in Table 3. for plasmid identification in K. pneumonia isolates owing to its efficiency and low cost compared to traditional methods, such as conjugation or molecular cloning [37]. PBRT uses a set of primers and a probe specific to the target plasmid to generate amplicons of different molecular weights. The different amplicon sizes correspond to different plasmids and can be used to identify a particular plasmid. Here, PBRT was used to determine the plasmids in KPC K. pneumoniae isolates. Sixteen replicons (with their percentage of appearance) out of thirty were presented in this study as follows: I1Y (57%), FIIK (43%), A/C (39%), HIB-M (39%), FIIS (35%), IncW L (30%), FIB KN (30%), FIB-M (30%), FIB KQ (26%), FII (26%), X1 (22%), R (13%), N2 (9%), HI3 (4%), IncA/C L (4%), and U (4%) (Figure 1). More detailed results are shown in the Supplementary Materials Section—Table S2. A previous study showed seven types of replicons in the K. pneumoniae isolates [38].

Table 3.
This table shows the type of samples and the patient demographics for the carbapenems-resistant K. pneumoniae.
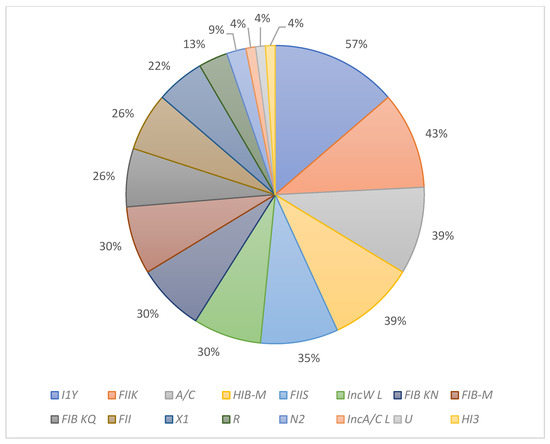
Figure 1.
The percentage of replicons in K. pneumoniae KPC isolates.
Plasmids are considered one of the significant factors of antibiotic resistance, including the carbapenem group [39,40]. The study samples showed that they contain a high number of plasmids. A recent study on K. pneumoniae pathogenic strains’ genetic characteristics demonstrated that such bacterium usually has many plasmids that can cause diseases [39]. A previous study conducted in Riyadh, Saudi Arabia, between 2011 and 2012 observed that K. pneumoniae isolates contained four plasmids, the most popular was FIIK, reaching 69% [38]. However, in our study, this plasmid appeared in 43% of the tested isolates. Additionally, the study identified a group of isolates containing more than seven plasmids, which reached ten plasmids in one isolate. It is worth noting that four isolates do not contain plasmids (K5, K7, K11, and K13) and have shown high resistances to both MEM and IMI, which may be due to the limited plasmids that can be monitored through the PBRT mechanism used in this study, which needs further investigation.
2.4. Genotyping Characterization for Harboring Carbapenems Genes
The presence of gene-encoding carbapenems factors (blaKPC, blaOXA-48, blaVIM, blaNDM-1, blaIMP-variants, and blaOXA-23-like) was investigated by PCR. Gene blaVIM was the most prevalent among the studied isolates, and it was presented in all tested isolates (100%), followed by blaOXA-48 gene in 87% of the isolates, blaNDM-1 gene in 30% of the isolates, and blaOXA-23-like gene in 4% of the isolates (Table 4). Both blaKPC and blaIMP genes are missing in all tested isolates (0%).

Table 4.
Distribution of carbapenem genes in 23 isolates.
A reference study conducted in Saudi Arabia on a group of samples collected during the years 2010 to 2018 indicated a low prevalence of the VIM gene among the study K. pneumoniae isolates; during this study, a percentage of gene presence of 100% for all study samples. On the other hand, no isolates exhibited positivity for the KPC production activity, as shown in Table 5. The KPC enzyme is considered one of the non-dominant genes in the K. pneumoniae bacterium isolated in Saudi Arabia, which was previously monitored in studies conducted on wastewater [41,42].

Table 5.
Detection of carbapenemase genes in 23 isolates.
Several studies have shown the possibility of K. pneumoniae carrying more than one carbapenem resistance gene in isolates from China, Singapore, India, Europe, and the Middle East [43,44,45]. In this study, all isolates showed the coexistence of at least two carbapenems genes, except for isolates K5 and K21, which produced the blaVIM gene only. Another study in Egypt showed the appearance of OXA-48 and NDM genes in 13 of the study K. pneumoniae isolates [46]. In contrast, this study recorded that five isolates (K2, K6, K8, K10 and K12) carried three carbapenem-resistant genes, blaOXA-48, blaVIM and blaNDM-1 (Table 5). A study conducted in the Makkah region in Saudi Arabia reported that K. pneumoniae contains only three carbapenem-resistant genes [44]. However, our study indicated that this bacterium could hold more than three resistant genes (isolate K22), which held four carbapenem resistance genes blaOXA-48, blaVIM, blaNDM-1, and blaOXA-23. More investigations are required to connect the number and type of genes against antibiotic resistance.
2.5. Detection of Virulence Factors
Virulence factors indicate chromosomally encoded genes involved in the bacterium’s attachment to host tissues and the ability to produce toxins that cause diseases. This study identified six virulence-related genes (kfu, wabG, uge, rmpA, fimH, and CPS), with a percentage of gene presence of 65%, 43%, 30%, 26%, 22%, and 13%, respectively (Figure 2).
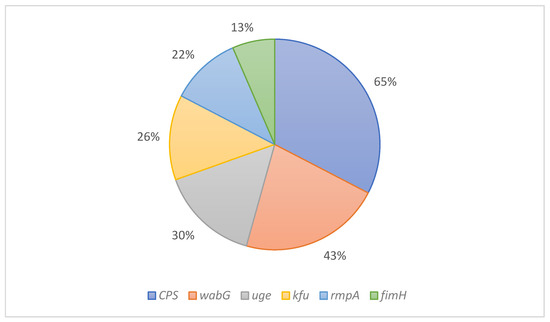
Figure 2.
The percentage of virulence factors in K. pneumoniae KPC isolates.
The capsular polysaccharide gene (i.e., CPS) is a shared gene within K. pneumoniae isolates [29]. One study showed that this gene was present in all isolates (100%) of the study (n = 65 isolates) [47], while it appeared in 65% of our study isolates. wabG gene in K. pneumoniae is a virulence factor involved in the attachment of bacterium to host tissues [48], and it is present in 43% of the isolates in this study. Another study conducted on 23 samples in Saudi Arabia isolated from 2011 to 2015 showed that the prevalence of the Kfu gene was 35% [49]. However, our study exhibited a decrease in the number of isolates carrying the kfu gene to 26%. In recent years, few studies in Saudi Arabia shed light on the rmpA gene, which demonstrated a higher presence rate (35%) than what was shown in this study (22%). Finally, the fimH gene was presented in 13% of the isolates in our study, which appeared in 22% in a previous study conducted in the Western region of Saudi Arabia [50]. All six genes appeared in all tested isolates but varied ratios. Further research is required to identify more virulence factor genes using whole genome sequencing on a more significant number of isolates than in this study (n = 23 isolates).
3. Materials and Methods
3.1. Materials
Mueller-Hinton broth (MHB) was purchased from Scharlau (Barcelona, Spain), while LB agar was bought from Invitrogen (USA). Nine antibiotics, including cefoxitin (FOX), cefepime (CPM), aztreonam (AZT), ceftazidime (CAZ), cefotaxime (CTC), ciprofloxacin (CIP), meropenem (MEM), imipenem (IMI), and ampicillin (AMP) were obtained from Sigma-Aldrich (Gillingham, UK), except for AMP which was bought from USB (Cleveland, OH, USA). Acetic acid and ethanol were purchased from BDH (Prolabo, UK) and Scharlau (Barcelona, Spain), respectively. TAE 10X buffer was obtained from Thermo Fisher Scientific (Waltham, MA, USA). Distilled water was generated using Milli-Q® IQ 7005 Purification System (Millipore SAS, Molsheim, France) and was used throughout this study.
3.2. Bacterial Inoculum Preparation
Twenty-three carbapenems-resistant K. pneumoniae (CRKP) strains were isolated from blood, sputum, wounds, central venous puncture, and urine culture from King Faisal Hospital in Makkah, Saudi Arabia (Table 3). An ATCC BAA 1705 strain (with KPC gene) and ATCC 700603 were used as positive controls. The isolates’ identification and confirmation were assessed using MicroScan (Beckman Coulter, CA, USA) and VITEK 2 (bioMérieux, Marcy-l′Étoile, France) according to the manufacturer’s instructions.
3.3. Minimum Inhibitory Concentration (MIC) Assay
The MICs of nine antibiotics that include stock solutions of FOX (5000 μg/mL), CPM (5000 μg/mL), AZT (5000 μg/mL), CAZ (5000 μg/mL), CTC (5000 μg/mL), CIP (5000 μg/mL), IMI (5000 μg/mL), MEM (10,000 μg/mL), and AMP (10,000 μg/mL) against K. pneumoniae were determined using the microdilution method. A serial dilution of the drugs, as a half-fold dilution from 1024 to 0.5 μg/mL, in MHB was added into 96-well microtiter plates at a final volume of 100 μL in each well. Then, single pure colonies from all isolates, to create bacterial inoculums using 0.5 McFarland standard, giving a cell density of 1.5 × 108 colonies forming unit (CFU)/mL, were measured by DensiChek Plus Instrument (bioMérieux, Marcy L’Etoile, France) at 600 nm. The bacterial inoculums were added to each well (100 μL) to attain a final inoculum of 1 × 106 CFU/mL. All 96-well microtiter plates were incubated overnight at 37 °C with a continuous shaking speed of 140 RPM. The endpoints of the MIC were measured at a UV absorbance of 600 nm using a PowerWave XS2 plate reader (bioMérieux, Marcy L’Etoile, France) [51]. All the results were evaluated according to the Clinical and Laboratory Standards Institute (CLSI) criteria for Antimicrobial Susceptibility Testing [35]. Wells contained bacterium, and medium only were used as positive and negative controls, respectively.
3.4. Biofilm Formation of K. pneumoniae
Biofilm formation assay was performed by allowing the cells to adhere to the walls and the bottom of the 96-well microtiter plates following a modified method [52,53]. K. pneumoniae bacterial strains (23 isolates) were cultured on an LB agar plate and incubated overnight at 37 °C. Then, a 106 CFU/mL dilution in LB broth was inoculated into a 96-well microtiter plate and incubated at 37 °C overnight. After the incubation, the plate was washed three times with distilled water using BioTek ELx50 Microplate Strip Washer (BioTek, Winooski, VT, USA) to remove the bacterial suspension and unattached cells. A 125 μL of 0.1% crystal violet (CV) solution was added to each well. The plate was incubated at room temperature for 10–15 min, followed by three thorough washings with distilled water. Finally, the plate was turned upside-down to dry completely at room temperature for 30 min. To quantify the biofilm formation, a 125 μL of 30% acetic acid was added to each well to dissolve the CV-stained biofilm and incubated at room temperature for 10–15 min. The 125 μL solubilized CV-stained biofilm was transferred to a new 96-well microtiter plate, and the absorbance was measured using a PowerWave XS2 plate reader (BioTek, Winooski, VT, USA) at 550 nm. The biofilm formation was quantified by comparing the optical densities cut-off (ODc) with the negative control following the previous studies [54,55]:
ODc = average OD of negative control + (3 × standard deviations (SD) of negative control OD)
The classification of biofilm formation, along with the optical density (OD), for all bacterial isolates was interpreted according to the criteria of [52,53,54], as shown in Table 6.

Table 6.
Criteria of classification for biofilm formation.
3.5. Bacterial DNA Extraction
The bacterial genomic DNA extraction was obtained using a modified boiling method [56]. Pure bacterial colonies were collected from all isolates and added to 100 µL of sterilized distilled water. The bacterial inoculums were boiled using Eppendorf Thermomixer (Thermo Fisher Scientific, Waltham, MA, USA) at 99 °C for 5 min. The tubes were placed on ice for 3, then centrifuged at 1300 rpm for 20 min. An 80 µL of the supernatant containing DNA was transferred into new Eppendorf tubes, and 160 µL cold absolute ethanol was added; the mixture was centrifuged at 1300 rpm for 5 min. The supernatant was removed, 100 µL of 70% ethanol was added to wash the pellet, followed by further centrifugation at 1300 rpm for 5 min, and then the supernatant was discarded. The pellet-containing genomic DNA was allowed to dry out completely, then re-suspended in 50 µL of nuclease-free water. The concentrations and DNA purities were evaluated spectrophotometrically using QuickDrop (Molecular Devices, San Jose, CA, USA).
3.6. Detection of Plasmids by PCR-Based Replicon Typing (PBRT)
PCR-based replicon typing technique (PBRT) package was used to detect the resistant plasmids of the K. pneumoniae isolates using PBRT 2.0 kit (Diatheva, Italy). Whole bacterial DNA was obtained via the boiling method and was used as a PCR template. Thirty Replicons were amplified using eight multiplex PCR assays, including HI-1, HI-2, I-1 alpha, I-2, X-1, X-2, X-3, X-4, L, I1y, N, FI-A, FI-B, FI-C, FII, FII-S, FII-K, FIB KN, FIB KQ, W, Y, P1, A/C, T, K, U, R, B/O, HIBM, and FIBM (Table 7). These replicons demonstrate the main plasmid incompatibility groups and replicase genes identified in the resistance plasmids among Enterobacteriaceae. The kit also contained positive controls for all particular replicons. PCR conditions were conducted in a thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) with some modifications as follows: initial denaturation at 95 °C for 10 min, followed by 30 cycles of denaturation at 95 °C for 60 s, annealing at 60 °C for 30 s, extension step at 72 °C for 60 s, and a final extension step at 72 °C for 5 min. The final PCR products were analyzed using electrophoresis at 80 V for 45 min in a 2.5% agarose gel with SYPR safe (Thermo Fisher Scientific, Waltham, MA, USA). The bands were visualized using a UV transilluminator (Bio-Rad Laboratories, Hercules, CA, USA).

Table 7.
PCRs Mix and amplicon sizes (bp) for detecting plasmids in K. pneumoniae isolates.
3.7. Detection of Carbapenem Genes by Polymerase Chain Reaction (PCR)
Conventional PCR was used to detect the presence of carbapenem resistance genes of CRKP isolates, including KPC, IMP, OXA 48, OXA 23, VIM, and NDM-1 (Macrogen, Seoul, Republic of Korea). The primers sequence and amplicon sizes represented in this study are shown in Table 8. Multiplex PCR mixture 1 (KPC, OXA-48, VIM) and Multiplex PCR mixture 2 (IMP, OXA-23, NDM) carried out in 25 µL contained 1X PuRE Taq™ Ready-To-Go™ PCR beads Master Mix (GE Healthcare, Amersham, UK),1 µL of each forward primer (10 pmol), 1 µL of each reverse primer (10 pmol), 1 µL of the DNA template, and 18 µL nuclease-free water. The PCR conditions were conducted in a thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) with some modifications as follows: multiplex PCR 1, initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 56 °C, extension step at 72 °C for 1 min, and a final extension step at 72 °C for 5 min. For multiplex PCR 2, initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 1 min, annealing at 52 °C, extension step at 72 °C for 1 min, and a final extension step at 72 °C for 5 min [57]. The final PCR products were analyzed using electrophoresis at 80 V for 45 min in a 2% agarose gel with SYPR safe (Thermo Fisher Scientific, Waltham, MA, USA). The bands were visualized using a UV transilluminator (Bio-Rad Laboratories, Hercules, CA, USA).

Table 8.
Primer sequence and amplicon sizes (bp) for detecting carbapenem genes in K. pneumoniae isolates.
3.8. Detection of Virulence Genes by Polymerase Chain Reaction (PCR)
Conventional PCR was used to detect the presence of virulence genes of CRKP isolates, including kfu, wab G, uge, rmpA, fimH, magA, and CPS (Macrogen, Seoul, Republic of Korea). The primers sequence and amplicon sizes represented in this study are shown in Table 9. PCR mixture carried out in 25 µL contained 1X PuRE Taq™ Ready-To-Go™ PCR beads Master Mix (GE Healthcare, Amersham, UK), 1 µL of the forward primer (10 pmol), 1 µL of the reverse primer (10 pmol), 1 µL of the DNA template, and 22 µL nuclease-free water. The PCR conditions were conducted in a thermal cycler (Thermo Fisher Scientific, Waltham, MA, USA) with some modifications: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C, extension step at 72 °C for 5 min, and a final extension step at 72 °C for 7 min [59]. The final PCR products were analyzed using electrophoresis at 80 V for 45 min in a 2% agarose gel with SYPR safe (Thermo Fisher Scientific, Waltham, MA, USA). The bands were visualized using a UV transilluminator (Bio-Rad Laboratories, Hercules, CA, USA).

Table 9.
Primer sequence and amplicon sizes (bp) for detecting virulence genes in K. pneumoniae isolates.
4. Conclusions
The prevalence of antibiotic-resistant K. pneumoniae with KPC genes is rapidly growing worldwide and may represent a global threat to healthcare systems in the near future. This study investigated the resistant potential of K. pneumoniae isolated from Makkah Province, Saudi Arabia, during the Islamic pilgrimage ‘Umrah’ and identified carbapenem-resistant genes and virulence factors. The significant findings of this study demonstrated that the isolated K. pneumoniae were resistant to most carbapenem antibiotics (MEM and IMI), in addition to FOX, CPM, AZT, CAZ, CTC, CIP, and AMP. Although the isolates lacked the presence of the blaKPC gene, the data indicated the presence of multiple genes encoding carbapenem factors, with the blaVIM and blaOXA-48 being the most prevalent genes. The study also found many plasmids and identified numerous virulence factors in the isolated K. pneumoniae, which could be attributed to their resistant phenotype. The reported data in this study shed light on the nature of K. pneumoniae, commonly found in the religious gathering seasons in Saudi Arabia. They may serve as a reference to clinicians and scientists in the region.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11111627/s1. Table S1: The raw data and biofilm formation level of all 23 K. pneumoniae isolates, along with the ATCC reference strain (700603); Table S2: Detailed results of all replicons appeared in all 23 K. pneumoniae isolates.
Author Contributions
Conceptualization, E.J.A.; methodology, R.Y.B. and B.K.S.; formal analysis, R.Y.B., M.H.A., M.A.A., A.A.H. and A.A.A.; investigation, R.Y.B., M.H.A., M.A.A., A.A.H., A.N.A., M.I.A., M.K.A., A.A.B., A.A.A. and F.S.B.; resources, E.A.T., F.S.B., M.S.A. and E.J.A.; writing—original draft preparation, R.Y.B., A.N.A., M.I.A., M.K.A., A.A.B., E.A.T. and N.B.A.; writing—review and editing, E.A.T., N.B.A., M.S.A., E.J.A. and B.K.S.; visualization, R.Y.B.; supervision, E.A.T., N.B.A., M.S.A., E.J.A. and B.K.S.; project administration, B.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Industrial Development and Logistics Program (NIDLP) through the Health Initiative and the Technology Leader Program Initiative, projects 20-0103 and 20-0051.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lu, B.; Lin, C.; Liu, H.; Zhang, X.; Tian, Y.; Huang, Y.; Yan, H.; Qu, M.; Jia, L.; Wang, Q. Molecular Characteristics of Klebsiella pneumoniae Isolates from Outpatients in Sentinel Hospitals, Beijing, China, 2010–2019. Front. Cell. Infect. Microbiol. 2020, 10, 85. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Fatahian Kelishadrokhi, A.; Chehelgerdi, M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect. Drug Resist. 2019, 12, 603–611. [Google Scholar] [CrossRef]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Ragheb, S.M.; Tawfick, M.M.; El-Kholy, A.A.; Abdulall, A.K. Phenotypic and Genotypic Features of Klebsiella pneumoniae Harboring Carbapenemases in Egypt: OXA-48-Like Carbapenemases as an Investigated Model. Antibiotics 2020, 9, 852. [Google Scholar] [CrossRef]
- Kopotsa, K.; Mbelle, N.M.; Osei Sekyere, J. Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb. Genom. 2020, 6, mgen000474. [Google Scholar] [CrossRef]
- Imtiaz, W.; Syed, Z.; Rafaque, Z.; Andrews, S.C.; Dasti, J.I. Analysis of Antibiotic Resistance and Virulence Traits (Genetic and Phenotypic) in Klebsiella pneumoniae Clinical Isolates from Pakistan: Identification of Significant Levels of Carbapenem and Colistin Resistance. Infect. Drug Resist. 2021, 14, 227–236. [Google Scholar] [CrossRef]
- Wu, C.; Zheng, L.; Yao, J. Analysis of Risk Factors and Mortality of Patients with Carbapenem-Resistant Klebsiella pneumoniae Infection. Infect. Drug Resist. 2022, 15, 2383–2391. [Google Scholar] [CrossRef]
- Xu, L.; Sun, X.; Ma, X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 18. [Google Scholar] [CrossRef]
- World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; 9240026436; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Thaden, J.T. Epidemiology and Mechanisms of Resistance of Extensively Drug Resistant Gram-Negative Bacteria. Antibiotics 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Bi, Y.; Liu, S.; Li, X.; Dong, S.; Ju, M. Predictors of mortality in patients with carbapenem-resistant Klebsiella pneumoniae infection: A meta-analysis and a systematic review. Ann. Palliat. Med. 2021, 10, 7340–7350. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Kharazmkia, A.; Amirizadeh, M.; Goudarzi, Z.; Birjandi, M.; Barfipoursalar, A.; Mir, S. Prevalence of KPC-producing bacteria in negative gram of clinical samples obtained from patients. Ann. Med. Surg. 2022, 77, 103690. [Google Scholar] [CrossRef] [PubMed]
- Brink, A.J. Epidemiology of carbapenem-resistant Gram-negative infections globally. Curr. Opin. Infect. Dis. 2019, 32, 609–616. [Google Scholar] [CrossRef]
- Ambler, R.P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Bedenić, B.; Sardelić, S. Carbapenemases. In Growing and Handling of Bacterial Cultures; IntechOpen: London, UK, 2018. [Google Scholar]
- Walsh, T.R.; Toleman, M.A.; Poirel, L.; Nordmann, P. Metallo-beta-lactamases: The quiet before the storm? Clin. Microbiol. Rev. 2005, 18, 306–325. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Gong, Y.; Lu, Y.; Xue, D.; Wei, Y.; Li, Q.; Li, G.; Lu, S.; Wang, J.; Wang, Y.; Peng, Y.; et al. Emergence of a Carbapenem-Resistant Klebsiella pneumoniae Isolate Co-harbouring Dual bla NDM- 6 -Carrying Plasmids in China. Front. Microbiol. 2022, 13, 900831. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G. OXA beta-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Antunes, N.T.; Lamoureaux, T.L.; Toth, M.; Stewart, N.K.; Frase, H.; Vakulenko, S.B. Class D beta-lactamases: Are they all carbapenemases? Antimicrob. Agents Chemother. 2014, 58, 2119–2125. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.J.A.; Alaa, H.A.A. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. Afr. J. Microbiol. Res. 2016, 10, 829–843. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Perini, M.; Mauri, C.; Comandatore, F.; Meroni, E.; Luzzaro, F.; Principe, L. Antimicrobial Susceptibility, Virulence, and Genomic Features of a Hypervirulent Serotype K2, ST65 Klebsiella pneumoniae Causing Meningitis in Italy. Antibiotics 2022, 11, 261. [Google Scholar] [CrossRef]
- Muraya, A.; Kyany’a, C.; Kiyaga, S.; Smith, H.J.; Kibet, C.; Martin, M.J.; Kimani, J.; Musila, L. Antimicrobial Resistance and Virulence Characteristics of Klebsiella pneumoniae Isolates in Kenya by Whole-Genome Sequencing. Pathogens 2022, 11, 545. [Google Scholar] [CrossRef]
- Pan, Y.J.; Lin, T.L.; Chen, C.T.; Chen, Y.Y.; Hsieh, P.F.; Hsu, C.R.; Wu, M.C.; Wang, J.T. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015, 5, 15573. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 642484. [Google Scholar] [CrossRef] [PubMed]
- Ballen, V.; Gabasa, Y.; Ratia, C.; Ortega, R.; Tejero, M.; Soto, S. Antibiotic Resistance and Virulence Profiles of Klebsiella pneumoniae Strains Isolated From Different Clinical Sources. Front. Cell. Infect. Microbiol. 2021, 11, 738223. [Google Scholar] [CrossRef]
- Remya, P.A.; Shanthi, M.; Sekar, U. Characterisation of virulence genes associated with pathogenicity in Klebsiella pneumoniae. Indian J. Med. Microbiol. 2019, 37, 210–218. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Dela Cruz, C.S.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Front. Microbiol. 2021, 12, 750662. [Google Scholar] [CrossRef] [PubMed]
- Paudel, S.; Adhikari, P.; KC, S.S.; Shrestha, U.T.; Shah, P.K. Antibiogram and Biofilm Development among Klebsiella pneumoniae from Clinical Isolates. Tribhuvan Univ. J. Microbiol. 2021, 8, 83–92. [Google Scholar] [CrossRef]
- Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing. In Twenty-Fourth Informational Supplement, M100–S24; Clinical Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2011; Volume 31, pp. 100–121. [Google Scholar]
- Ashwath, P.; Deekshit, V.K.; Rohit, A.; Dhinakaran, I.; Karunasagar, I.; Karunasagar, I.; Akhila, D.S. Biofilm Formation and Associated Gene Expression in Multidrug-Resistant Klebsiella pneumoniae Isolated from Clinical Specimens. Curr. Microbiol. 2022, 79, 73. [Google Scholar] [CrossRef] [PubMed]
- Hawkey, J.; Wyres, K.L.; Judd, L.M.; Harshegyi, T.; Blakeway, L.; Wick, R.R.; Jenney, A.W.; Holt, K.E. ESBL plasmids in Klebsiella pneumoniae: Diversity, transmission and contribution to infection burden in the hospital setting. Genome Med. 2022, 14, 97. [Google Scholar] [CrossRef]
- Zaman, T.U.; Alrodayyan, M.; Albladi, M.; Aldrees, M.; Siddique, M.I.; Aljohani, S.; Balkhy, H.H. Clonal diversity and genetic profiling of antibiotic resistance among multidrug/carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect. Dis. 2018, 18, 205. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Iriarte, A.; Reyes-Lamothe, R.; Sherratt, D.J.; Tolmasky, M.E. Small Klebsiella pneumoniae Plasmids: Neglected Contributors to Antibiotic Resistance. Front. Microbiol. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, K.; Chen, W.; Chen, J.; Zheng, J.; Liu, C.; Cheng, L.; Zhou, W.; Shen, H.; Cao, X. Epidemiological characteristics of carbapenem-resistant Enterobacteriaceae collected from 17 hospitals in Nanjing district of China. Antimicrob. Resist. Infect. Control 2020, 9, 15. [Google Scholar] [CrossRef]
- Alghoribi, M.F.; Binkhamis, K.; Alswaji, A.A.; Alhijji, A.; Alsharidi, A.; Balkhy, H.H.; Doumith, M.; Somily, A. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: A new public health concern in Saudi Arabia. J. Infect. Public Health 2020, 13, 647–650. [Google Scholar] [CrossRef]
- Alhazmi, W.; Al-Jabri, A.; Al-Zahrani, I. The Molecular Characterization of Nosocomial Carbapenem-Resistant Klebsiella pneumoniae Co-Harboring blaNDM and blaOXA-48 in Jeddah. Microbiol. Res. 2022, 13, 753–764. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F.; et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Khan, M.A.; Mohamed, A.M.; Faiz, A.; Ahmad, J. Enterobacterial infection in Saudi Arabia: First record of Klebsiella pneumoniae with triple carbapenemase genes resistance. J. Infect. Dev. Ctries 2019, 13, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Kumari, N.; Kumar, M.; Katiyar, A.; Kumar, A.; Priya, P.; Kumar, B.; Biswas, N.R.; Kaur, P. Genome-wide identification of carbapenem-resistant Gram-negative bacterial (CR-GNB) isolates retrieved from hospitalized patients in Bihar, India. Sci. Rep. 2022, 12, 8477. [Google Scholar] [CrossRef] [PubMed]
- ElMahallawy, H.; Zafer, M.M.; Al-Agamy, M.; Amin, M.A.; Mersal, M.M.; Booq, R.Y.; Alyamani, E.; Radwan, S. Dissemination of ST101 blaOXA-48 producing Klebsiella pneumoniae at tertiary care setting. J. Infect. Dev. Ctries 2018, 12, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Nahavandinejad, M.; Asadpour, L. Mucoviscosity determination and detection of magA and rmpA genes in clinical isolates of Klebsiella pneumoniae in Northern Iran. Crescent J. Med. Biol. Sci. 2017, 4, 104–107. [Google Scholar]
- Izquierdo, L.; Coderch, N.; Pique, N.; Bedini, E.; Corsaro, M.M.; Merino, S.; Fresno, S.; Tomas, J.M.; Regue, M. The Klebsiella pneumoniae wabG gene: Role in biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 2003, 185, 7213–7221. [Google Scholar] [CrossRef]
- Uz Zaman, T.; Albladi, M.; Siddique, M.I.; Aljohani, S.M.; Balkhy, H.H. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect. Drug Resist. 2018, 11, 1183–1187. [Google Scholar] [CrossRef]
- Alsanie, W.F. Molecular diversity and profile analysis of virulence-associated genes in some Klebsiella pneumoniae isolates. Pract. Lab. Med. 2020, 19, e00152. [Google Scholar] [CrossRef]
- Arabaci, C.; Dal, T.; Basyigit, T.; Genisel, N.; Durmaz, R. Investigation of carbapenemase and mcr-1 genes in carbapenem-resistant Klebsiella pneumoniae isolates. J. Infect. Dev. Ctries 2019, 13, 504–509. [Google Scholar] [CrossRef]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Hassan, A.; Usman, J.; Kaleem, F.; Omair, M.; Khalid, A.; Iqbal, M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz. J. Infect. Dis. 2011, 15, 305–311. [Google Scholar] [CrossRef]
- Singh, A.K.; Prakash, P.; Achra, A.; Singh, G.P.; Das, A.; Singh, R.K. Standardization and Classification of In vitro Biofilm Formation by Clinical Isolates of Staphylococcus aureus. J. Glob. Infect. Dis. 2017, 9, 93–101. [Google Scholar] [CrossRef]
- Ahmed, O.B.; Dablool, A. Quality improvement of the DNA extracted by boiling method in gram negative bacteria. Int. J. Bioassays 2017, 6, 5347–5349. [Google Scholar] [CrossRef]
- Candan, E.D.; Aksoz, N. Klebsiella pneumoniae: Characteristics of carbapenem resistance and virulence factors. Acta Biochim. Polonica 2015, 62, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Amudhan, S.M.; Sekar, U.; Arunagiri, K.; Sekar, B. OXA beta-lactamase-mediated carbapenem resistance in Acinetobacter baumannii. Indian J. Med. Microbiol. 2011, 29, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Fevre, C.; Passet, V.; Deletoile, A.; Barbe, V.; Frangeul, L.; Almeida, A.S.; Sansonetti, P.; Tournebize, R.; Brisse, S. PCR-based identification of Klebsiella pneumoniae subsp. rhinoscleromatis, the agent of rhinoscleroma. PLoS Negl. Trop. Dis. 2011, 5, e1052. [Google Scholar] [CrossRef]
- Ma, L.C.; Fang, C.T.; Lee, C.Z.; Shun, C.T.; Wang, J.T. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J. Infect. Dis. 2005, 192, 117–128. [Google Scholar] [CrossRef]
- Yu, W.L.; Ko, W.C.; Cheng, K.C.; Lee, H.C.; Ke, D.S.; Lee, C.C.; Fung, C.P.; Chuang, Y.C. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 2006, 42, 1351–1358. [Google Scholar] [CrossRef]
- Turton, J.F.; Perry, C.; Elgohari, S.; Hampton, C.V. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 2010, 59, 541–547. [Google Scholar] [CrossRef]
- Yu, W.L.; Ko, W.C.; Cheng, K.C.; Lee, C.C.; Lai, C.C.; Chuang, Y.C. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 2008, 62, 1–6. [Google Scholar] [CrossRef]
- Abdul-Razzaq, M.S.; Al-Khafaji, J.K.T.; Al-Maamory, E.H.K.-A. Molecular characterization of capsular polysaccharide genes of Klebsiella pneumoniae in Iraq. Int. J. Curr. Microbiol. App Sci. 2014, 3, 224–234. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).