Epidemiology and In Vitro Activity of Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam against KPC-Producing K. pneumoniae Collected from Bacteremic Patients, 2018 to 2020
Abstract
:1. Introduction
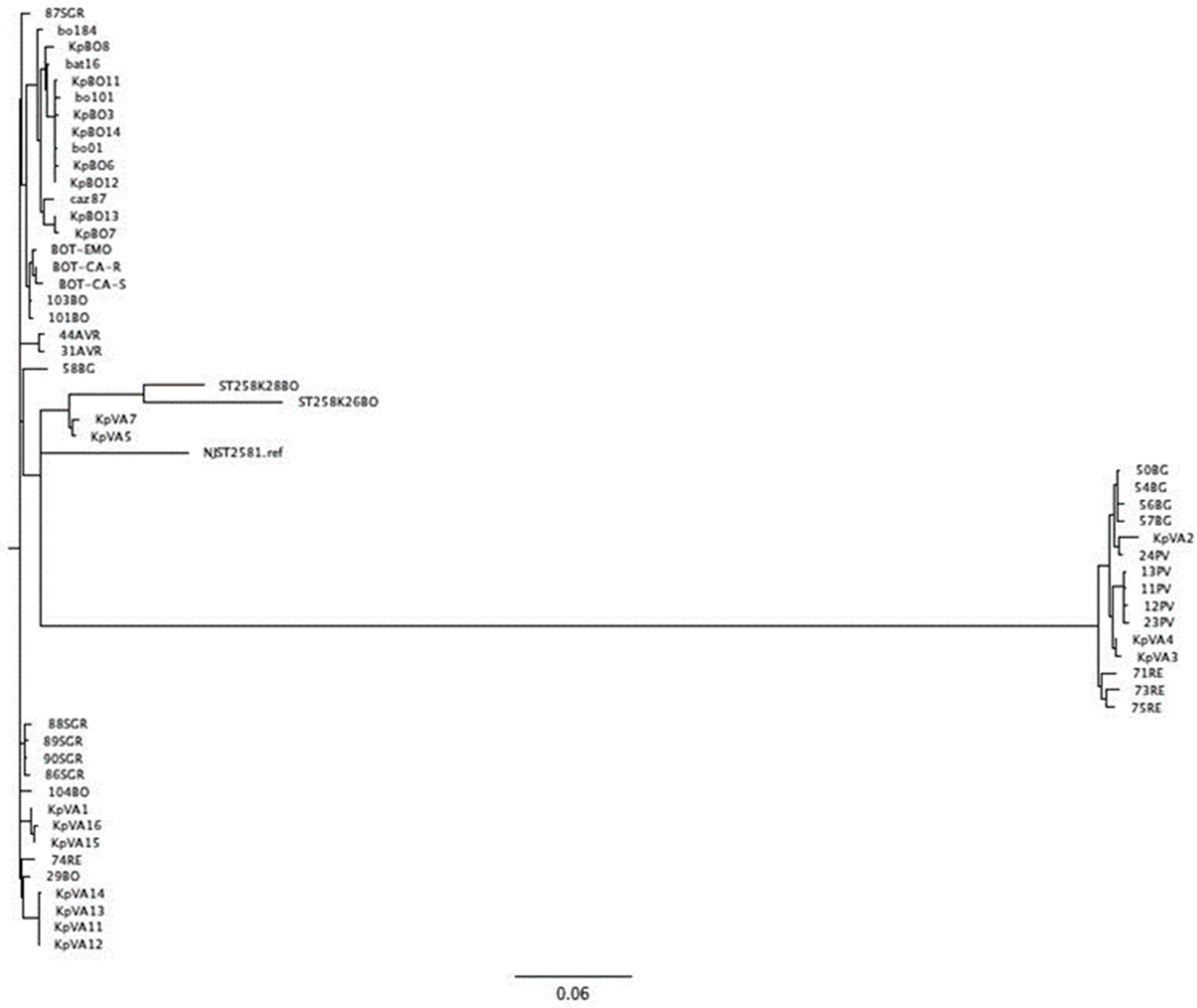
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Phenotypic Analysis
4.3. Whole Genome Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Lutgring, J.D.; Limbago, B.M. The problem of carbapenemase-producing-carbapenem-resistant-Enterobacteriaceae detection. J. Clin. Microbiol. 2016, 54, 529–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Ucha, J.C.; Arca-Suárez, J.; Bou, G.; Beceiro, A. New carbapenemase inhibitors: Clearing the way for the β-lactams. Int. J. Mol. Sci. 2020, 21, 9308. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Peghin, M. How to manage KPC infections. Ther. Adv. Infect. Dis. 2020, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef]
- Di Pilato, V.; Aiezza, N.; Viaggi, V.; Antonelli, A.; Principe, L.; Giani, T.; Luzzaro, F.; Rossolini, G.M. KPC-53, a KPC-3 variant of clinical origin associated with reduced susceptibility to ceftazidime-avibactam. Antimicrob. Agents Chemother. 2021, 65, e01429-20. [Google Scholar] [CrossRef]
- Munoz-Price, L.S.; Reeme, A.E.; Buchan, B.W.; Mettus, R.T.; Mustapha, M.M.; Van Tyne, D.; Shields, R.K.; Doi, Y. Patient-to-patient transmission of Klebsiella pneumoniae carbapenemase variants with reduced ceftazidime-avibactam susceptibility. Antimicrob. Agents Chemother. 2019, 63, e00955-19. [Google Scholar] [CrossRef]
- Novelli, A.; Del Giacomo, P.; Rossolini, G.M.; Tumbarello, M. Meropenem/vaborbactam: A next generation β-lactam β-lactamase inhibitor combination. Expert Rev. Anti-Infect. Ther. 2020, 18, 643–655. [Google Scholar] [CrossRef]
- Drawz, S.M.; Bonomo, R.A. Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 2010, 23, 160–201. [Google Scholar] [CrossRef] [Green Version]
- Gaibani, P.; Giani, T.; Bovo, F.; Lombardo, D.; Amadesi, S.; Lazzarotto, T.; Coppi, M.; Rossolini, G.M.; Ambretti, S. Resistance to Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam in Gram-Negative MDR Bacilli: Molecular Mechanisms and Susceptibility Testing. Antibiotics 2022, 11, 628. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Mack, A.R.; Taracila, M.A.; Bonomo, R.A. Resistance to Novel b-Lactam–b-Lactamase Inhibitor Combinations The “Price of Progress”. Infect. Dis. Clin. 2020, 34, 773–819. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, J.; Wang, R.; Cai, Y. Resistance to ceftazidime–avibactam and underlying mechanisms. J. Glob. Antimicrob. Resist. 2020, 22, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Momattin, H.; Al-Ali, A.Y.; Eljaaly, K.; Tirupathi, R.; Haradwala, M.B.; Areti, S.; Alhumaid, S.; Rabaan, A.A.; Mutair, A.A.; et al. Antibiotics in the pipeline: A literature review (2017–2020). Infection 2022, 50, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Meropenem/Vaborbactam: A Review in Complicated Urinary Tract Infections. Drugs 2018, 78, 1259–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theuretzbacher, U.; Carrara, E.; Conti, M.; Tacconelli, E. Role of new antibiotics for KPC-producing. Kleb. Pneumoniae. J. Antimicrob. Chemother. 2021, 76, i47–i54. [Google Scholar] [CrossRef] [PubMed]
- Campanella, T.A.; Gallagher, J.C. A Clinical Review and Critical Evaluation of Imipenem-Relebactam: Evidence to Date. Infect. Drug Resist. 2020, 13, 4297–4308. [Google Scholar] [CrossRef]
- Balabanian, G.; Rose, M.; Manning, N.; Landman, D.; Quale, J. Effect of Porins and blaKPC Expression on Activity of Imipenem with Relebactam in Klebsiella pneumoniae: Can Antibiotic Combinations Overcome Resistance? Microb. Drug Resist. 2018, 24, 877–881. [Google Scholar] [CrossRef]
- Gaibani, P.; Bovo, F.; Bussini, L.; Lazzarotto, T.; Amadesi, S.; Bartoletti, M.; Viale, P.; Ambretti, S. Dynamic evolution of imipenem/relebactam-resistance in a KPC-producing Klebsiella pneumoniae from single patient during ceftazidime/avibactam-based treatment. J. Antimicrob. Chemother. 2022, 77, 1570–1577. [Google Scholar] [CrossRef]
- Page, M.G. β-Lactamase inhibitors. Drug Resist. Updat. 2000, 3, 109–125. [Google Scholar] [CrossRef]
- Findlay, J.; Poirel, L.; Juhas, M.; Nordmann, P. KPC-mediated resistance to Ceftazidime-Avibactam and Collateral Effects in Klebsiella pneumoniae. Antimicrob. Agents Chemoter. 2021, 65, e00890-21. [Google Scholar] [CrossRef]
- Lomovskay, O.; Sun, D.; Rubio-Aparicio, D.; Nelson, K.; Tsivkovski, R.; Griffith, D.C.; Dudley, M.N. Vaborbactam: Spectrum of β-lactamase inhibition and impact of resistance mechanisms on activity in Enterobacteriales. Antimicrob. Agents Chemother. 2017, 61, e01443-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaibani, P.; Bianco, G.; Amadesi, S.; Boattini, M.; Ambretti, S.; Costa, C. Increased blaKPC Copy Number and OmpK35 and OmpK36 Porins Disruption Mediated Resistance to Imipenem/Relebactam and Meropenem/Vaborbactam in a KPC-Producing Klebsiella pneumoniae Clinical Isolate. Antimicob. Agent Chemother. 2022, 66, e0019122. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Lombardo, D.; Bussini, L.; Bovo, F.; Munari, B.; Giannella, M.; Bartoletti, M.; Viale, P.; Lazzarotto, T.; Ambretti, S. Epidemiology of Meropenem/Vaborbactam Resistance in KPC-Producing Klebsiella pneumoniae Causing Bloodstream Infections in Northern Italy, 2018. Antibiotics 2021, 10, 536. [Google Scholar] [CrossRef] [PubMed]
- Foschi, C.; Gaibani, P.; Lombardo, D.; Re, M.C.; Ambretti, S. Rectal screening of carbapenemase-producing Enterobacteriaceae: A proposed workflow. J. Glob. Antimicrob. Resist. 2020, 21, 86–90. [Google Scholar] [CrossRef]
- Opota, O.; Croxatto, A.; Prod’hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Gaibani, P.; Ambretti, S.; Tamburini, M.V.; Vecchio Nepita, E.; Re, M.C. Clinical application of Bruker Biotyper MALDI-TOF/MS system for real-time identification of KPC production in Klebsiella pneumoniae clinical isolates. J. Glob. Antimicrob. Resist. 2018, 12, 169–170. [Google Scholar] [CrossRef] [Green Version]
- Oviaño, M.; Bou, G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin. Microbiol. Rev. 2018, 32, e00037-18. [Google Scholar] [CrossRef] [Green Version]
- Gaibani, P.; Bussini, L.; Amadesi, S.; Bartoletti, M.; Bovo, F.; Lazzarotto, T.; Viale, P.; Ambretti, S. Successful Treatment of Bloodstream Infection due to a KPC-Producing Klebsiella Pneumoniae Resistant to Imipenem/Relebactam in a Hematological Patient. Microorganisms 2022, 10, 778. [Google Scholar] [CrossRef]
| Antimicrobial Agent | % Resistance a | |||
|---|---|---|---|---|
| Total of KPC-Kp Isolates (n = 116) | 2018 (n = 62) | 2019 (n =31) | 2020 (n = 23) | |
| Ceftazidime-avibactam | 8.6% 10/116 | 6.5% (4/62) | 12.9% (4/31) | 8.7% (2/23) |
| Meropenem-vaborbactam | 9.5% 11/116 | 14.5% (9/62) | 3.2% (1/31) | 4.3% (1/23) |
| Imipenem-relebactam | 2.6% 3/116 | 1.6% (1/62) | 3.2% (1/31) | 4.3% (1/23) |
| Isolate | Year | MIC (mg/L) | ||
|---|---|---|---|---|
| CAZ-AVI | MER-VAB | IMI-REL | ||
| BO1 | 2018 | 16 | 16 | 8 |
| BO3 | 2018 | 16 | >256 | 1.5 |
| BO6 | 2018 | 16 | 256 | 0.38 |
| BO7 | 2018 | 256 | 256 | 1 |
| BO8 | 2018 | 6 | 48 | 1 |
| BO11 | 2018 | 8 | 256 | 1 |
| BO12 | 2018 | 8 | 256 | 0.5 |
| BO13 | 2018 | 6 | 256 | 0.5 |
| BO14 | 2018 | 8 | 256 | 1.5 |
| BO77 | 2019 | >256 | <0.06 | 0.12 |
| BO101 | 2019 | >256 | >256 | >8 |
| BO184 | 2019 | 16 | 8 | 2 |
| BO204 | 2019 | >256 | 2 | 1 |
| BO302 | 2020 | 8 | 16 | 4 |
| BO628 | 2020 | >256 | 0.032 | 0.094 |
| BO630 | 2020 | 32 | 1 | 0.25 |
| Isolate | ST | Carbapenemase | β-Lactamases | Porins | Plasmid | |
|---|---|---|---|---|---|---|
| OmpK35 | OmpK36 | |||||
| BO1 | 512 | blaKPC-3 | blaSHV-11 | truncated at aa 42 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncX3 |
| BO3 | 512 | blaKPC-3 | blaSHV-182 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncX3 |
| BO6 | 512 | blaKPC-3 | blaSHV-182 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncX3 |
| BO7 | 1519 | blaKPC-3 | blaTEM-1A, blaSHV-182, blaOXA-9 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFIB(pQil), IncFII(K), IncX3 |
| BO8 | 512 | blaKPC-3 | blaTEM-1A, blaSHV-182, blaOXA-9 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFIB(pQil), IncFII(K), IncX3 |
| BO11 | 512 | blaKPC-3 | blaSHV-182 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFII(K), IncX3 |
| BO12 | 512 | blaKPC-3 | blaSHV-182 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFII(K), IncX3 |
| BO13 | 1519 | blaKPC-3 | blaSHV-182, blaOXA-9 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFIB(pQil), IncFII(K), IncX3 |
| BO14 | 512 | blaKPC-3 | blaSHV-182 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncX3 |
| BO77 | 307 | blaKPC-31 | blaTEM-1, blaSHV-28,blaOXA-1 | truncated at aa 229 | truncated at aa 182 | IncFIB(K), IncFIB(pQil), IncFII(K) |
| BO101 | 512 | blaKPC-3 | blaSHV-11 | truncated at aa 42 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncX3 |
| BO184 | 512 | blaKPC-3 | blaTEM-1, blaSHV-11 | truncated at aa 42 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1), IncFIB(pQil), IncFII(K) |
| BO204 | 512 | blaKPC-31 | blaTEM-1, blaSHV-11 | truncated at aa 42 | INS135GD | IncFIB(K), IncFIB(pKPHS1), IncFIB(pQil), IncFII(K), IncX3 |
| BO302 | 512 | blaKPC-3 | blaTEM-1, blaSHV-11 | truncated at aa 41 | INS135GD | ColRNAI, IncFIB(K), IncFIB(pKPHS1),IncFIB(pQil), IncFII(K), IncX3 |
| BO628 | 307 | blaKPC-31 | blaTEM-1, blaSHV-28, blaCTX-M-15 | truncated at aa 229 | truncated at aa 182 | IncFIB(K), IncFIB(pQil), IncFII(K) |
| BO630 | 1519 | blaKPC-86 | blaTEM-128, blaSHV-11 | truncated at aa 41 | INS135GD | Col(BS512), ColRNAI, IncFIB(pKPHS1), IncFIB(pQil), IncFII(K) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bovo, F.; Lombardo, D.; Lazzarotto, T.; Ambretti, S.; Gaibani, P. Epidemiology and In Vitro Activity of Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam against KPC-Producing K. pneumoniae Collected from Bacteremic Patients, 2018 to 2020. Antibiotics 2022, 11, 1621. https://doi.org/10.3390/antibiotics11111621
Bovo F, Lombardo D, Lazzarotto T, Ambretti S, Gaibani P. Epidemiology and In Vitro Activity of Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam against KPC-Producing K. pneumoniae Collected from Bacteremic Patients, 2018 to 2020. Antibiotics. 2022; 11(11):1621. https://doi.org/10.3390/antibiotics11111621
Chicago/Turabian StyleBovo, Federica, Donatella Lombardo, Tiziana Lazzarotto, Simone Ambretti, and Paolo Gaibani. 2022. "Epidemiology and In Vitro Activity of Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam against KPC-Producing K. pneumoniae Collected from Bacteremic Patients, 2018 to 2020" Antibiotics 11, no. 11: 1621. https://doi.org/10.3390/antibiotics11111621
APA StyleBovo, F., Lombardo, D., Lazzarotto, T., Ambretti, S., & Gaibani, P. (2022). Epidemiology and In Vitro Activity of Ceftazidime/Avibactam, Meropenem/Vaborbactam and Imipenem/Relebactam against KPC-Producing K. pneumoniae Collected from Bacteremic Patients, 2018 to 2020. Antibiotics, 11(11), 1621. https://doi.org/10.3390/antibiotics11111621







