Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria
Abstract
1. Introduction
2. Results and Discussion
2.1. Designing and Modeling of α-Helical Peptides
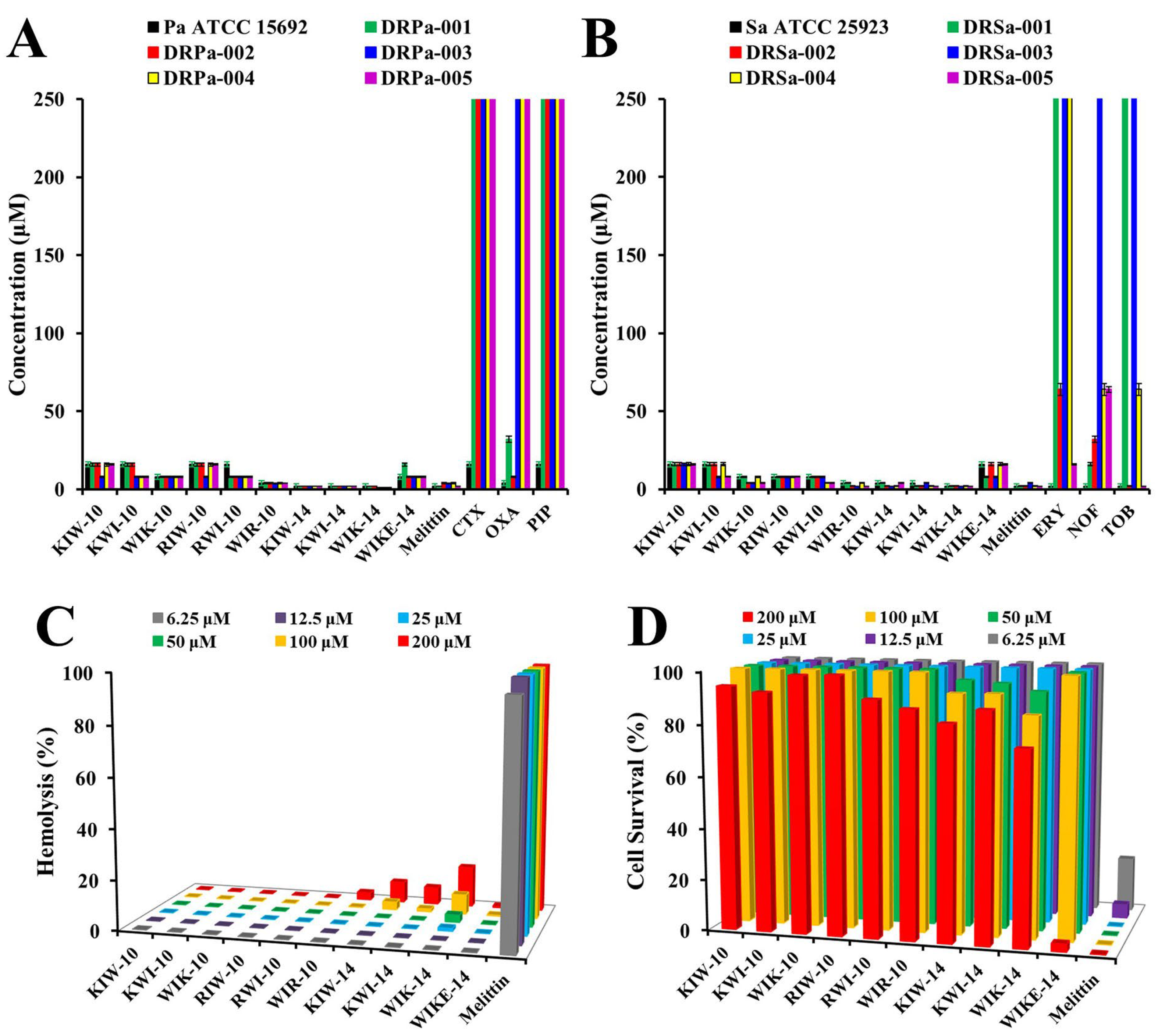
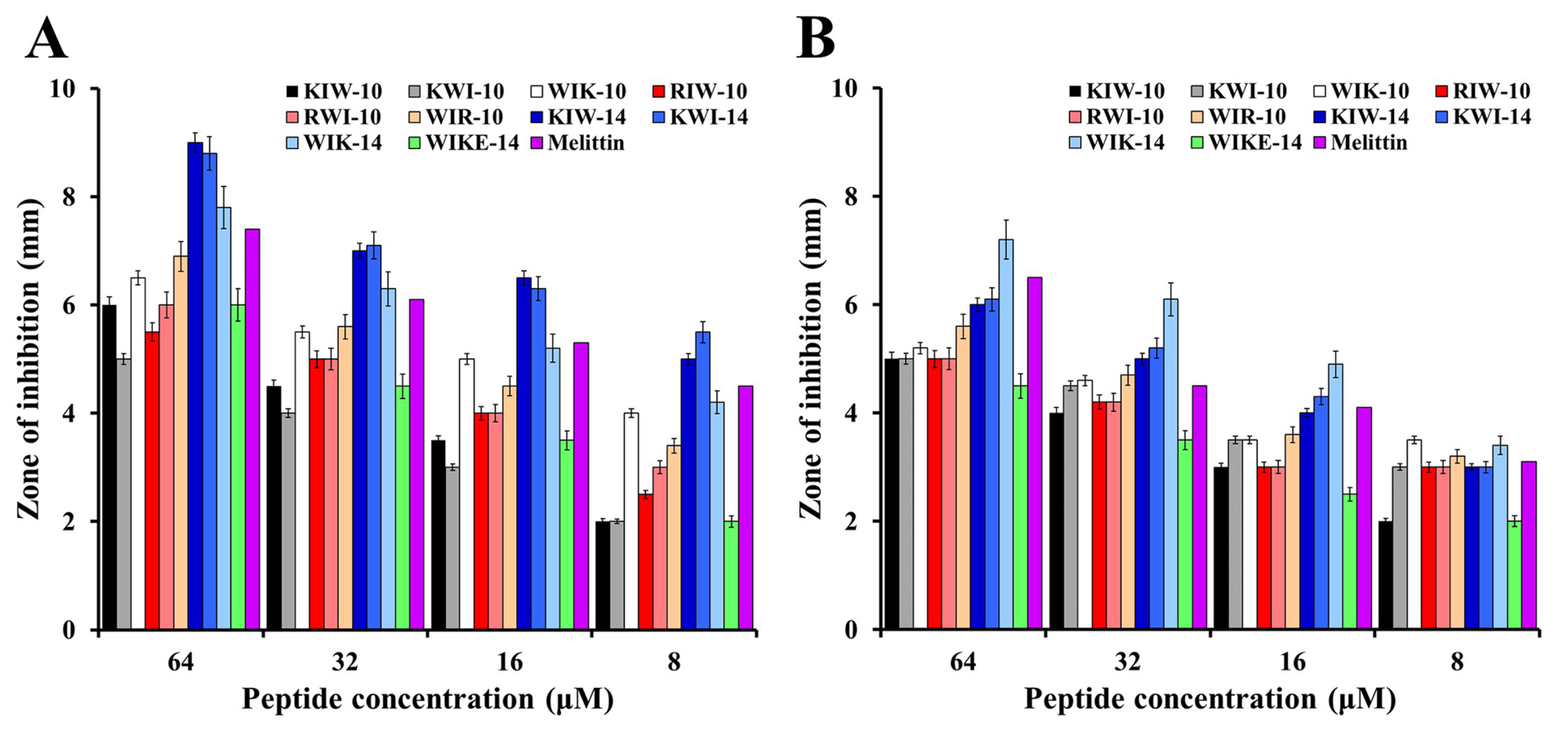
2.2. Cell-Selective Effects of Designed Peptides against Bacterial Strains and Mammalian Cells
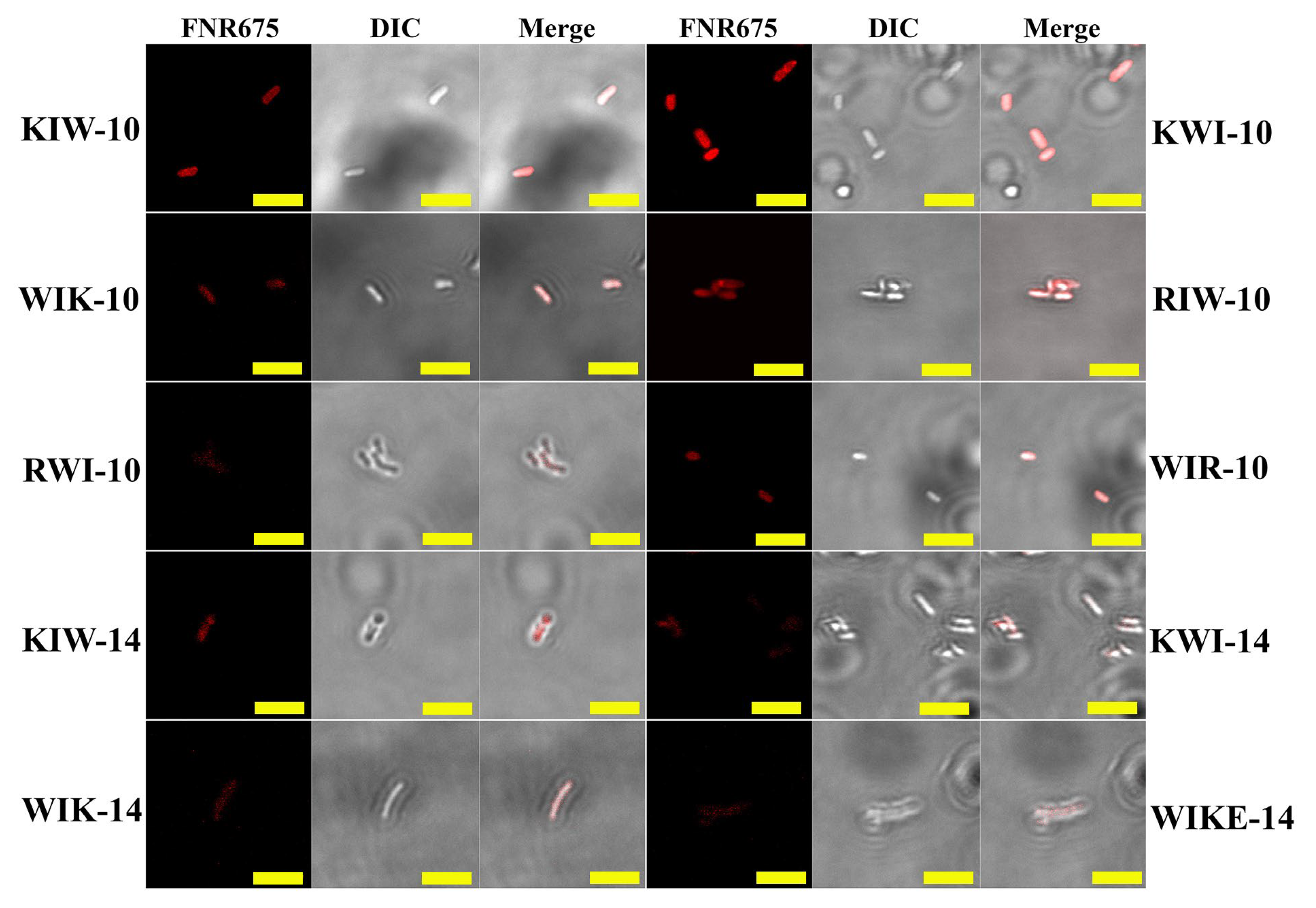
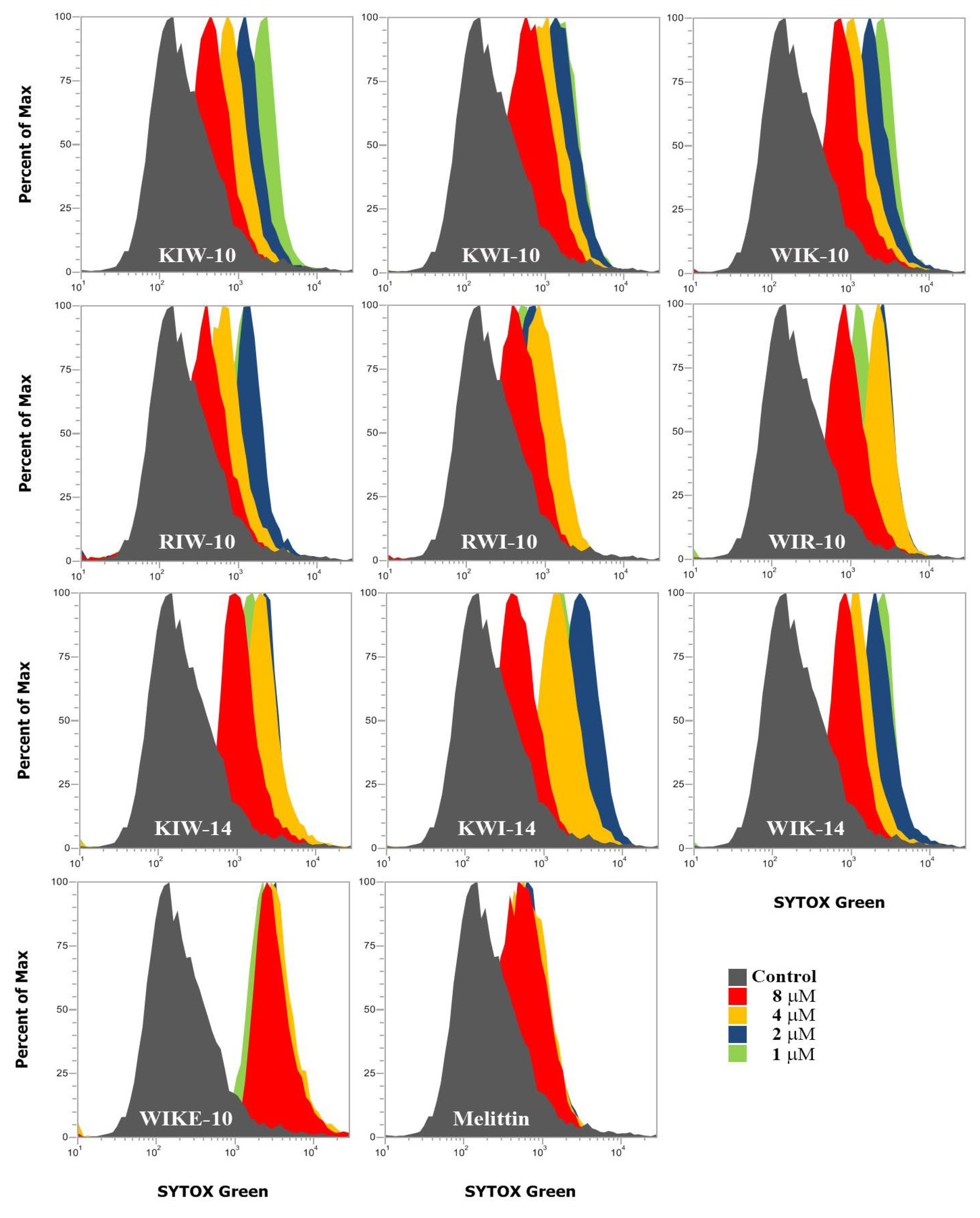
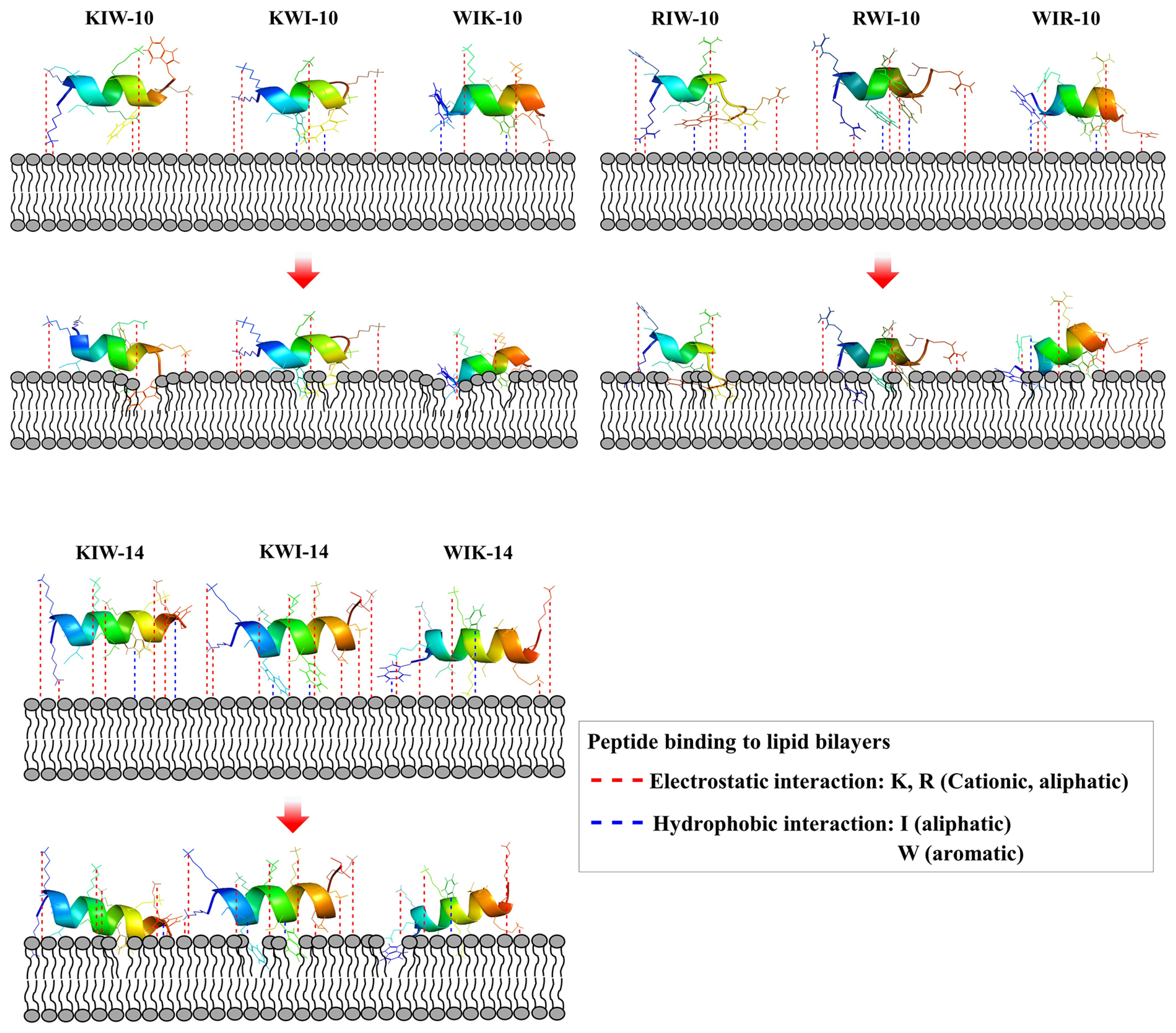
2.3. Mode of Action of Peptides
2.4. In Vivo Antibacterial Effects of the Selected Peptides
3. Materials and Methods
3.1. Materials
3.2. Solid-Phase Peptide Synthesis by Microwave
3.3. Antibacterial Assay by a Microdilution Method
3.4. Hemolysis and Cytotoxicity
3.5. Inhibition Zone Assay
3.6. Membrane Depolarization
3.7. Confocal Laser-Scanning Microscopy (CLSM)
3.8. SYTOX Green Uptake Assay
3.9. Small Unilamellar Vesicles Preparation
3.10. Tryptophan Fluorescence Blue Shift
3.11. Calcein Leakage Assay
3.12. Scanning Electron Microscopy (SEM) Preparation
3.13. Murine Infection Model
3.14. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, X.; Lü, Y.; Yue, C. Development and Research Progress of Anti-Drug Resistant Bacteria Drugs. Infect. Drug Resist. 2021, 14, 5575–5593. [Google Scholar] [CrossRef] [PubMed]
- Mohd Asri, N.A.; Ahmad, S.; Mohamud, R.; Mohd Hanafi, N.; Mohd Zaidi, N.F.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Mohd Noor, N.; Mustafa, F.H.; et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella Pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef] [PubMed]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic Resistance in Microbes: History, Mechanisms, Therapeutic Strategies and Future Prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Elmaidomy, A.H.; Shady, N.H.; Abdeljawad, K.M.; Elzamkan, M.B.; Helmy, H.H.; Tarshan, E.A.; Adly, A.N.; Hussien, Y.H.; Sayed, N.G.; Zayed, A.; et al. Antimicrobial potentials of natural products against multidrug resistance pathogens: A comprehensive review. RSC Adv. 2022, 12, 29078–29102. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wang, Y.; Tian, W.; Cui, X.; Tu, P.; Li, J.; Shi, S.; Liu, X. Biosynthesis Investigations of Terpenoid, Alkaloid, and Flavonoid Antimicrobial Agents Derived from Medicinal Plants. Antibiotics 2022, 11, 1380. [Google Scholar] [CrossRef]
- Jiménez, M.C.; Kowalski, L.; Souto, R.B.; Alves, I.A.; Viana, M.D.; Aragón, D.M. New drugs against multidrug-resistant Gram-negative bacteria: A systematic review of patents. Future Microbiol. 2022, 17, 1393–1408. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, Y.; Hahm, K.-S. The Role of Antimicrobial Peptides in Preventing Multidrug-Resistant Bacterial Infections and Biofilm Formation. Int. J. Mol. Sci. 2011, 12, 5971–5992. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349. [Google Scholar] [CrossRef]
- Yu, L.; Li, K.; Zhang, J.; Jin, H.; Saleem, A.; Song, Q.; Jia, Q.; Li, P. Antimicrobial Peptides and Macromolecules for Combating Microbial Infections: From Agents to Interfaces. ACS Appl. Bio Mater. 2022, 5, 366–393. [Google Scholar] [CrossRef]
- Drayton, M.; Deisinger, J.P.; Ludwig, K.C.; Raheem, N.; Müller, A.; Schneider, T.; Straus, S.K. Host Defense Peptides: Dual Antimicrobial and Immunomodulatory Action. Int. J. Mol. Sci. 2021, 22, 11172. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Cell Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef] [PubMed]
- El Shazely, B.; Yu, G.; Johnston, P.R.; Rolff, J. Resistance Evolution Against Antimicrobial Peptides in Staphylococcus aureus Alters Pharmacodynamics Beyond the MIC. Front. Microbiol. 2020, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, L.; Dong, C. Antimicrobial Peptides: An Overview of their Structure, Function and Mechanism of Action. Protein Pept. Lett. 2022, 29, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.; Sharma, A.; Alajangi, H.K.; Jaiswal, P.K.; Lim, Y.B.; Singh, G.; Barnwal, R.P. In pursuit of next-generation therapeutics: Antimicrobial peptides against superbugs, their sources, mechanism of action, nanotechnology-based delivery, and clinical applications. Int. J. Biol. Macromol. 2022, 218, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Boparai, J.K.; Sharma, P.K. Mini Review on Antimicrobial Peptides, Sources, Mechanism and Recent Applications. Protein Pept. Lett. 2020, 27, 4–16. [Google Scholar] [CrossRef]
- Benfield, A.H.; Henriques, S.T. Mode-of-Action of Antimicrobial Peptides: Membrane Disruption vs. Intracellular Mechanisms. Front. Med. Technol. 2020, 2, 610997. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial Peptides: Application Informed by Evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Park, S.-C.; Kim, J.-Y.; Shin, S.-O.; Jeong, C.-Y.; Kim, M.-H.; Shin, S.Y.; Cheong, G.-W.; Park, Y.; Hahm, K.-S. Investigation of Toroidal Pore and Oligomerization by Melittin Using Transmission Electron Microscopy. Biochem. Biophys. Res. Commun. 2006, 343, 222–228. [Google Scholar] [CrossRef]
- Scocchi, M.; Mardirossian, M.; Runti, G.; Benincasa, M. Non-Membrane Permeabilizing Modes of Action of Antimicrobial Peptides on Bacteria. Curr. Top. Med. Chem. 2016, 16, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Moghal, M.M.R.; Saha, S.K.; Yamazaki, M. The Role of Membrane Tension in the Action of Antimicrobial Peptides and Cell-Penetrating Peptides in Biomembranes. Biophys. Rev. 2019, 11, 431–448. [Google Scholar] [CrossRef] [PubMed]
- Javadpour, M.M.; Juban, M.M.; Lo, W.C.; Bishop, S.M.; Alberty, J.B.; Cowell, S.M.; Becker, C.L.; McLaughlin, M.L. De novo antimicrobial peptides with low mammalian cell toxicity. J. Med. Chem. 1996, 39, 3107–3113. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Tarantino, C.; Romeo, D. Design of synthetic antimicrobial peptides based on sequence analogy and amphipathicity. Eur. J. Biochem. 1997, 250, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Sothiselvam, S.; Lu, T.K.; de la Fuente-Nunez, C. Peptide Design Principles for Antimicrobial Applications. J. Mol. Biol. 2019, 431, 3547–3567. [Google Scholar] [CrossRef]
- Puentes, P.R.; Henao, M.C.; Torres, C.E.; Gómez, S.C.; Gómez, L.A.; Burgos, J.C.; Arbeláez, P.; Osma, J.F.; Muñoz-Camargo, C.; Reyes, L.H.; et al. Design, Screening, and Testing of Non-Rational Peptide Libraries with Antimicrobial Activity: In Silico and Experimental Approaches. Antibiotics 2020, 9, 854. [Google Scholar] [CrossRef]
- Chan, D.I.; Prenner, E.J.; Vogel, H.J. Tryptophan- and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. Biochim. Biophys. Acta 2006, 1758, 1184–1202. [Google Scholar] [CrossRef]
- Brogden, K.A.; De Lucca, A.J.; Bland, J.; Elliott, S. Isolation of an Ovine Pulmonary Surfactant-Associated Anionic Peptide Bactericidal for Pasteurella haemolytica. Proc. Natl. Acad. Sci. USA 1996, 93, 412–416. [Google Scholar] [CrossRef]
- Subbalakshmi, C.; Sitaram, N. Mechanism of Antimicrobial Action of Indolicidin. FEMS Microbiol. Lett. 1998, 160, 91–96. [Google Scholar] [CrossRef]
- Brötz, H.; Bierbaum, G.; Leopold, K.; Reynolds, P.E.; Sahl, H.G. The Lantibiotic Mersacidin Inhibits Peptidoglycan Synthesis by Targeting Lipid II. Antimicrob. Agents Chemother. 1998, 42, 154–160. [Google Scholar] [CrossRef]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E.W. Sublethal Concentrations of Pleurocidin-Derived Antimicrobial Peptides Inhibit Macromolecular Synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G.; Agerberth, B.; Boman, A. Mechanisms of Action on Escherichia coli of Cecropin P1 and PR-39, Two Antibacterial Peptides from Pig Intestine. Infect. Immun. 1993, 61, 2978–2984. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L.; Insug, O.; Rogers, M.E.; Consolvo, P.J.; Condie, B.A.; Lovas, S.; Bulet, P.; Blaszczyk-Thurin, M. Interaction between Heat Shock Proteins and Antimicrobial Peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Kabelka, I.; Vácha, R. Advances in Molecular Understanding of α-Helical Membrane-Active Peptides. Acc. Chem. Res. 2021, 54, 2196–2204. [Google Scholar] [CrossRef]
- Matsuzaki, K. Membrane Permeabilization Mechanisms. Adv. Exp. Med. Biol. 2019, 1117, 9–16. [Google Scholar] [CrossRef]
- de Planque, M.R.; Kruijtzer, J.A.; Liskamp, R.M.; Marsh, D.; Greathouse, D.V.; Koeppe, R.E.; de Kruijff, B.; Killian, J.A. Different Membrane Anchoring Positions of Tryptophan and Lysine in Synthetic Transmembrane Alpha-Helical Peptides. J. Biol. Chem. 1999, 274, 20839–20846. [Google Scholar] [CrossRef]
- Persson, S.; Killian, J.A.; Lindblom, G. Molecular Ordering of Interfacially Localized Tryptophan Analogs in Ester- and Ether-Lipid Bilayers Studied by 2H-NMR. Biophys. J. 1998, 75, 1365–1371. [Google Scholar] [CrossRef]
- Monné, M.; Nilsson, I.; Johansson, M.; Elmhed, N.; von Heijne, G. Positively and Negatively Charged Residues Have Different Effects on the Position in the Membrane of a Model Transmembrane Helix. J. Mol. Biol. 1998, 284, 1177–1183. [Google Scholar] [CrossRef][Green Version]
- Braun, P.; von Heijne, G. The Aromatic Residues Trp and Phe Have Different Effects on the Positioning of a Transmembrane Helix in the Microsomal Membrane. Biochemistry 1999, 38, 9778–9782. [Google Scholar] [CrossRef]
- Fennell, J.F.; Shipman, W.H.; Cole, L.J. Antibacterial action of a bee venom fraction (melittin) against a penicillin-resistant staphylococcus and other microorganisms. USNRDL-TR-67-101. Res. Dev. Tech. Rep. 1967, 5, 1–13. [Google Scholar] [CrossRef]
- Approved Standard: NCCLS M7-A3; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. National Committee for Clinical Laboratory Standards Antimicrobial Susceptibility Testing. NCCLS: Villanova, PA, USA, 1993.
- Park, S.-C.; Kim, M.-H.; Hossain, M.A.; Shin, S.Y.; Kim, Y.; Stella, L.; Wade, J.D.; Park, Y.; Hahm, K.-S. Amphipathic Alpha-Helical Peptide, HP (2-20), and Its Analogues Derived from Helicobacter Pylori: Pore Formation Mechanism in Various Lipid Compositions. Biochim. Biophys. Acta 2008, 1778, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-K.; Park, S.-C.; Hahm, K.-S.; Park, Y. A Helix-PXXP-Helix Peptide with Antibacterial Activity without Cytotoxicity against MDRPA-Infected Mice. Biomaterials 2014, 35, 1025–1039. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Kim, J.-Y.; Jeong, C.; Yoo, S.; Hahm, K.-S.; Park, Y. A Plausible Mode of Action of Pseudin-2, an Antimicrobial Peptide from Pseudis Paradoxa. Biochim. Biophys. Acta 2011, 1808, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-C.; Kim, H.; Kim, J.-Y.; Kim, H.; Cheong, G.-W.; Lee, J.R.; Jang, M.-K. Improved Cell Selectivity of Pseudin-2 via Substitution in the Leucine-Zipper Motif: In Vitro and In Vivo Antifungal Activity. Antibiotics 2020, 9, 921. [Google Scholar] [CrossRef] [PubMed]




| Name | MIC (μM) | ||||||
|---|---|---|---|---|---|---|---|
| Sequence | Molecular Mass (Da) | Ec * | Pa * | Sa * | Bs * | Lm * | |
| KIW-10 | KKIIKKIWKW-NH2 | 1369 | 8(64) | 16(64) | 16(128) | 4(32) | 4(128) |
| KWI-10 | KKIWKKWIKI-NH2 | 1369 | 8(64) | 16(64) | 16(128) | 2(32) | 2(64) |
| WIK-10 | WIKKIWKKIK-NH2 | 1369 | 8(32) | 8(32) | 8(64) | 2(8) | 2(16) |
| RIW-10 | RRIIRRIWRW-NH2 | 1509 | 8(16) | 16(32) | 8(16) | 2(4) | 2(8) |
| RWI-10 | RRIWRRWIRI-NH2 | 1509 | 2(4) | 16(32) | 8(16) | 1(4) | 2(8) |
| WIR-10 | WIRRIWRRIR-NH2 | 1509 | 2(4) | 4(8) | 4(8) | 1(1) | 2(2) |
| KIW-14 | KKIIKKIIKKIWKW-NH2 | 1852 | 4(4) | 2(2) | 4(8) | 1(2) | 0.5(1) |
| KWI-14 | KKIWKKWIKKIIKI-NH2 | 1852 | 4(4) | 2(2) | 4(8) | 1(2) | 0.5(1) |
| WIK-14 | WIKKIWKKIIKKIK-NH2 | 1852 | 4(4) | 2(2) | 2((2) | 1(1) | 0.5(1) |
| WIKE-14 | WIKKIWKKIIKEIK-NH2 | 1853 | 4(64) | 8(32) | 16(128) | 2(8) | 4(8) |
| Melittin | GIGAVLKVLTTGLPALISWIKRKRQQ-NH2 | 2845 | 2(2) | 2(16) | 2(2) | 2(2) | 1(2) |
| Peptide | Aqueous Condition | PE/PG Vesicle (7:3, w/w) c | PG/CL Vesicle (1:1, w/w) c | PC/CH/SM Vesicle (1:1:1, w/w/w) | ||||
|---|---|---|---|---|---|---|---|---|
| Buffer I a | Buffer II b | Buffer I | Buffer II | Buffer I | Buffer II | Buffer II c | Buffer II d | |
| KIW-10 | 352 | 356 | 343 | 346 | 340 | 341 | 355 | 351 |
| KWI-10 | 354 | 354 | 338 | 345 | 340 | 341 | 354 | 352 |
| WIK-10 | 354 | 354 | 334 | 339 | 331 | 332 | 354 | 354 |
| RIW-10 | 352 | 356 | 339 | 341 | 339 | 339 | 356 | 353 |
| RWI-10 | 353 | 355 | 338 | 338 | 337 | 338 | 355 | 353 |
| WIR-10 | 352 | 355 | 337 | 337 | 337 | 338 | 354 | 354 |
| KIW-14 | 353 | 357 | 340 | 340 | 339 | 340 | 347 | 343 |
| KWI-14 | 353 | 357 | 332 | 332 | 331 | 332 | 349 | 352 |
| WIK-14 | 356 | 356 | 318 | 318 | 321 | 321 | 354 | 350 |
| WIKE-14 | 353 | 356 | 332 | 337 | 322 | 328 | 353 | 352 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.-C.; Son, H.; Kim, Y.-M.; Lee, J.-K.; Park, S.; Lim, H.S.; Lee, J.R.; Jang, M.-K. Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria. Antibiotics 2022, 11, 1619. https://doi.org/10.3390/antibiotics11111619
Park S-C, Son H, Kim Y-M, Lee J-K, Park S, Lim HS, Lee JR, Jang M-K. Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria. Antibiotics. 2022; 11(11):1619. https://doi.org/10.3390/antibiotics11111619
Chicago/Turabian StylePark, Seong-Cheol, Hyosuk Son, Young-Min Kim, Jong-Kook Lee, Soyoung Park, Hye Song Lim, Jung Ro Lee, and Mi-Kyeong Jang. 2022. "Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria" Antibiotics 11, no. 11: 1619. https://doi.org/10.3390/antibiotics11111619
APA StylePark, S.-C., Son, H., Kim, Y.-M., Lee, J.-K., Park, S., Lim, H. S., Lee, J. R., & Jang, M.-K. (2022). Design of Antimicrobial Peptides with Cell-Selective Activity and Membrane-Acting Mechanism against Drug-Resistant Bacteria. Antibiotics, 11(11), 1619. https://doi.org/10.3390/antibiotics11111619








