Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021
Abstract
:1. Introduction
2. Results
2.1. E. coli Isolation
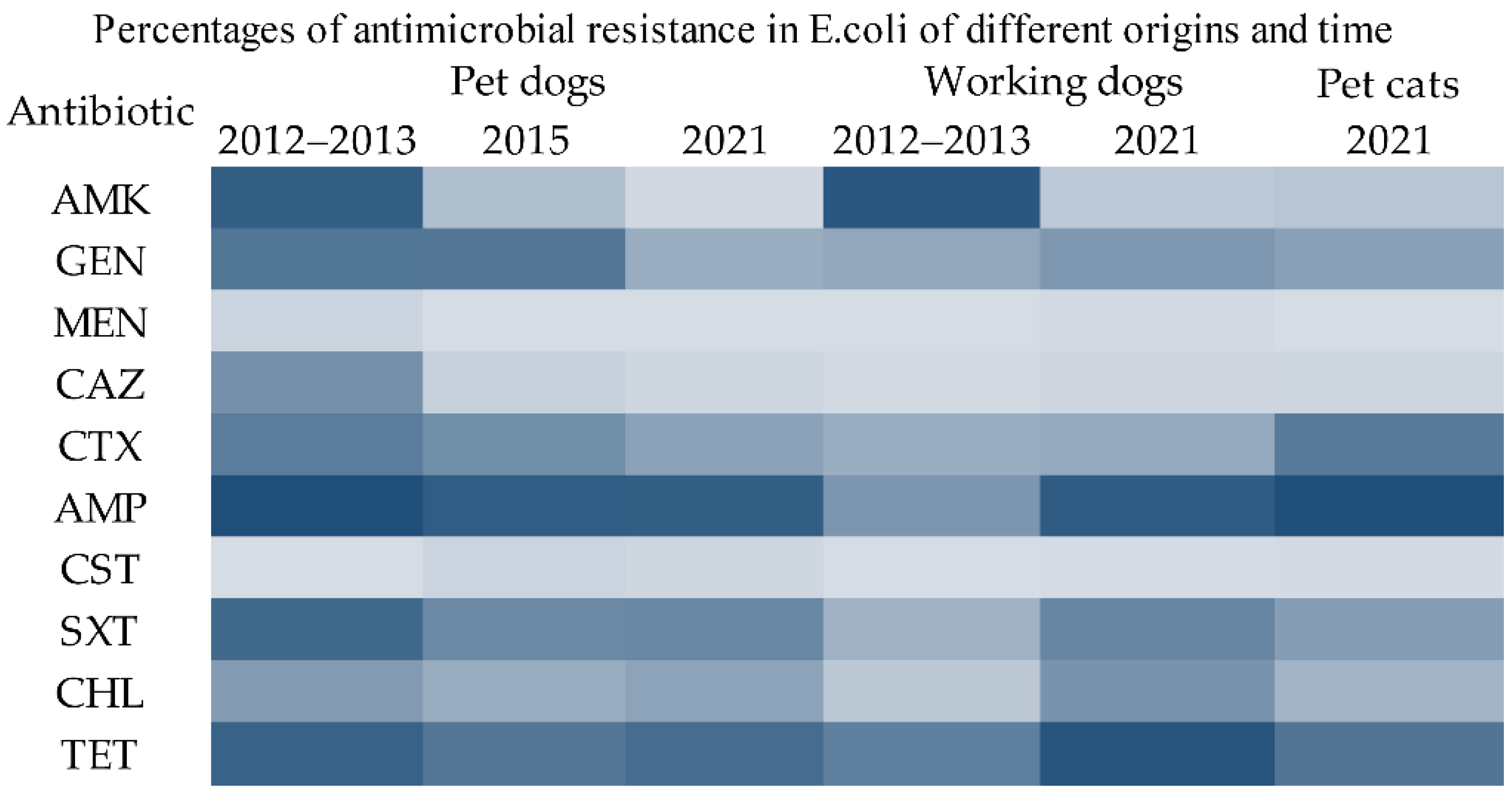
2.2. Drug Resistance Phenotypes
2.3. ESBL-Producing E. coli
2.4. Antimicrobial Resistance Genes
2.5. Phylogenetic Groups
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation and Identification of E. coli
4.3. Antimicrobial Susceptibility and Initial ESBL Identification
4.4. Detection of ARGs (Antibiotic Resistance Genes)
4.5. Phylogenetic Grouping
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pomba, C.; Rantala, M.; Greko, C.; Baptiste, K.E.; Catry, B.; van Duijkeren, E.; Mateus, A.; Moreno, M.A.; Pyorala, S.; Ruzauskas, M.; et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017, 72, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.T.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2018, 73, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Mannion, A.; Madden, C.M.; Swennes, A.G.; Townes, C.; Byrd, C.; Marini, R.P.; Fox, J.G. Cytotoxic Escherichia coli strains encoding colibactin and cytotoxic necrotizing factor (CNF) colonize laboratory macaques. Gut Pathog. 2017, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 2021, 19, 37–54. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Buldain, D.; Gortari Castillo, L.; Buchamer, A.; Chirino-Trejo, M.; Mestorino, N. Pet and stray dogs as reservoirs of antimicrobial-resistant Escherichia coli. Int. J. Microbiol. 2021, 2021, 6664557. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Cummins, E.A.; Snaith, A.E.; McNally, A.; Hall, R.J. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Ur Rahman, S.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The growing genetic and functional diversity of extended spectrum beta-lactamases. BioMed Res. Int. 2018, 2018, 9519718. [Google Scholar] [CrossRef]
- Derakhshandeh, A.; Eraghi, V.; Boroojeni, A.M.; Niaki, M.A.; Zare, S.; Naziri, Z. Virulence factors, antibiotic resistance genes and genetic relatedness of commensal Escherichia coli isolates from dogs and their owners. Microb. Pathog. 2018, 116, 241–245. [Google Scholar] [CrossRef]
- Alves, J.C.; Jorge, P.; Santos, A. A survey on the prevalence of diarrhea in a Portuguese population of police working dogs. BMC Vet. Res. 2021, 17, 211. [Google Scholar] [CrossRef]
- Bhat, A.H. Bacterial zoonoses transmitted by household pets and as reservoirs of antimicrobial resistant bacteria. Microb. Pathog. 2021, 155, 104891. [Google Scholar] [CrossRef] [PubMed]
- Wedley, A.L.; Dawson, S.; Maddox, T.W.; Coyne, K.P.; Pinchbeck, G.L.; Clegg, P.; Nuttall, T.; Kirchner, M.; Williams, N.J. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 2017, 199, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Thungrat, K.; Boothe, D.M. Multilocus sequence typing and virulence profiles in uropathogenic Escherichia coli isolated from cats in the United States. PLoS ONE 2015, 10, e0143335. [Google Scholar]
- Sevilla, E.; Mainar-Jaime, R.C.; Moreno, B.; Martin-Burriel, I.; Morales, M.; Andres-Lasheras, S.; Chirino-Trejo, M.; Badiola, J.J.; Bolea, R. Antimicrobial resistance among canine enteric Escherichia coli isolates and prevalence of attaching-effacing and extraintestinal pathogenic virulence factors in Spain. Acta Vet. Hung. 2020, 68, 1–7. [Google Scholar] [CrossRef]
- Leonard, E.K.; Pearl, D.L.; Finley, R.L.; Janecko, N.; Reid-Smith, R.J.; Peregrine, A.S.; Weese, J.S. Comparison of antimicrobial resistance patterns of Salmonella spp. and Escherichia coli recovered from pet dogs from volunteer households in Ontario (2005-06). J. Antimicrob. Chemother. 2012, 67, 174–181. [Google Scholar] [CrossRef]
- Saputra, S.; Jordan, D.; Mitchell, T.; Wong, H.S.; Abraham, R.J.; Kidsley, A.; Turnidge, J.; Trott, D.J.; Abraham, S. Antimicrobial resistance in clinical Escherichia coli isolated from companion animals in Australia. Vet. Microbiol. 2017, 211, 43–50. [Google Scholar] [CrossRef]
- Hata, A.; Fujitani, N.; Ono, F.; Yoshikawa, Y. Surveillance of antimicrobial-resistant Escherichia coli in Sheltered dogs in the Kanto Region of Japan. Sci. Rep. 2022, 12, 773. [Google Scholar] [CrossRef]
- Thongratsakul, S.; Poolkhet, C.; Amavisit, P.; Sato, T.; Fukuda, A.; Usui, M.; Tamura, Y. Antimicrobial resistance and Stec virulence genes of Escherichia coli isolated from non-diarrheic and diarrheic dogs at a veterinary teaching hospital in Thailand. Southeast Asian J. Trop. Med. Public Health 2019, 50, 708–714. [Google Scholar]
- Lei, T.; Tian, W.; He, L.; Huang, X.H.; Sun, Y.X.; Deng, Y.T.; Sun, Y.; Lv, D.H.; Wu, C.M.; Huang, L.Z.; et al. Antimicrobial resistance in Escherichia coli isolates from food animals, animal food products and companion animals in China. Vet. Microbiol. 2010, 146, 85–89. [Google Scholar] [CrossRef]
- Liu, X.Q.; Liu, H.X.; Li, Y.Q.; Hao, C.J. High prevalence of beta-lactamase and plasmid-mediated quinolone resistance genes in extended-spectrum cephalosporin-resistant Escherichia coli from dogs in Shaanxi, China. Front. Microbiol. 2016, 7, 1843. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Li, Y.; Hao, C. Association between virulence profile and fluoroquinolone resistance in Escherichia coli isolated from dogs and cats in China. J. Infect. Dev. Ctries. 2017, 11, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Z.; Zhang, Y.; Zhang, Z.; Lei, L.; Xia, Z. Increasing prevalence of ESBL-producing multidrug resistance Escherichia coli From diseased pets in Beijing, China from 2012 to 2017. Front. Microbiol. 2019, 10, 2852. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Y.; Chen, Y.; Huang, M.; Wang, Y.; Shen, Z.; Xia, Z.; Li, G. Antimicrobial resistance of bacterial pathogens isolated from canine urinary tract infections. Vet. Microbiol. 2020, 241, 108540. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.T.; Wang, Y.P.; Yuan, Y.W.; Xie, Y.J. A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.Q.; Lei, L.; Lv, Y.; Zhang, R.M.; Liu, X.Y.; Li, M.; Zhang, F.; Wang, Y. bla(NDM-1)-producing multidrug-resistant Escherichia coli isolated from a companion dog in China. J. Glob. Antimicrob. Resist. 2018, 13, 24–27. [Google Scholar] [CrossRef]
- Stolle, I.; Prenger-Berninghoff, E.; Stamm, I.; Scheufen, S.; Hassdenteufel, E.; Guenther, S.; Bethe, A.; Pfeifer, Y.; Ewers, C. Emergence of OXA-48 carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in dogs. J. Antimicrob. Chemother. 2013, 68, 2802–2808. [Google Scholar] [CrossRef]
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. Updat. 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Dickson, A.; Smith, M.; Smith, F.; Park, J.; King, C.; Currie, K.; Langdridge, D.; Davis, M.; Flowers, P. Understanding the relationship between pet owners and their companion animals as a key context for antimicrobial resistance-related behaviours: An interpretative phenomenological analysis. Health Psychol. Behav. Med. 2019, 7, 45–61. [Google Scholar] [CrossRef]
- Xu, J.; Wang, X.; Sun, K.S.; Lin, L.; Zhou, X. Parental self-medication with antibiotics for children promotes antibiotic over-prescribing in clinical settings in China. Antimicrob. Resist. Infect. Control 2020, 9, 150. [Google Scholar] [CrossRef]
- Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum beta-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health 2021, 12, 100236. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Wang, L.; Peng, Q.; Li, Y.; Zhou, H.; Li, Q. Molecular characterization of extended-spectrum beta-lactamase-producing multidrug resistant Escherichia coli from swine in Northwest China. Front. Microbiol. 2018, 9, 1756. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, J.; Li, X.; Ma, L.; Cao, X.; Hu, W.; Zhao, L.; Jing, W.; Lan, X.; Li, Y.; et al. Genetic diversity, antimicrobial resistance and extended-spectrum beta-lactamase type of Escherichia coli isolates from chicken, dog, pig and yak in Gansu and Qinghai Provinces, China. J. Glob. Antimicrob. Resist. 2020, 22, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Vega-Manriquez, X.D.; Ubiarco-Lopez, A.; Verdugo-Rodriguez, A.; Hernandez-Chinas, U.; Navarro-Ocana, A.; Ahumada-Cota, R.E.; Ramirez-Badillo, D.; Hernandez-Diaz de Leon, N.; Eslava, C.A. Pet dogs potential transmitters of pathogenic Escherichia coli with resistance to antimicrobials. Arch. Microbiol. 2020, 202, 1173–1179. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, Y.; Zhang, Q.; Jin, L.; Wang, Q.; Zhang, Y.; Wang, X.; Hu, M.; Li, L.; Qi, J.; et al. The prevalence of colistin resistance in Escherichia coli and Klebsiella pneumoniae isolated from food animals in China: Coexistence of mcr-1 and blaNDM with low fitness cost. Int. J. Antimicrob. Agents 2018, 51, 739–744. [Google Scholar] [CrossRef]
- Al-Bayssari, C.; Nawfal Dagher, T.; El Hamoui, S.; Fenianos, F.; Makdissy, N.; Rolain, J.M.; Nasreddine, N. Carbapenem and colistin-resistant bacteria in North Lebanon: Coexistence of mcr-1 and NDM-4 genes in Escherichia coli. J. Infect. Dev. Ctries. 2021, 15, 934-342. [Google Scholar] [CrossRef]
- Zogg, A.L.; Simmen, S.; Zurfluh, K.; Stephan, R.; Schmitt, S.N.; Nuesch-Inderbinen, M. High prevalence of extended-spectrum beta-lactamase producing Enterobacteriaceae among clinical isolates from cats and dogs admitted to a veterinary hospital in Switzerland. Front. Vet. Sci. 2018, 5, 62. [Google Scholar] [CrossRef]
- Hordijk, J.; Schoormans, A.; Kwakernaak, M.; Duim, B.; Broens, E.; Dierikx, C.; Mevius, D.; Wagenaar, J.A. High prevalence of fecal carriage of extended spectrum beta-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013, 4, 242. [Google Scholar] [CrossRef]
- Hardefeldt, L.Y.; Holloway, S.; Trott, D.J.; Shipstone, M.; Barrs, V.R.; Malik, R.; Burrows, M.; Armstrong, S.; Browning, G.F.; Stevenson, M. Antimicrobial prescribing in dogs and cats in Australia: Results of the Australasian infectious disease advisory panel survey. J. Vet. Intern. Med. 2017, 31, 1100–1107. [Google Scholar] [CrossRef]
- Gonzalez-Ramirez, M.T.; Landero-Hernandez, R. Pet-human relationships: Dogs versus cats. Animals 2021, 11, 2745. [Google Scholar] [CrossRef]
- Yasugi, M.; Hatoya, S.; Motooka, D.; Matsumoto, Y.; Shimamura, S.; Tani, H.; Furuya, M.; Mie, K.; Miyake, M.; Nakamura, S.; et al. Whole-genome analyses of extended-spectrum or AmpC beta-lactamase-producing Escherichia coli isolates from companion dogs in Japan. PLoS ONE 2021, 16, e0246482. [Google Scholar] [CrossRef]
- Xiao, Y.; Hu, Y. The major aminoglycoside-modifying enzyme AAC(3)-II found in Escherichia coli determines a significant disparity in its resistance to gentamicin and amikacin in China. Microb. Drug Resist. 2012, 18, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Mohanam, L.; Menon, T. Emergence of rmtC and rmtF 16S rRNA methyltransferase in clinical isolates of Pseudomonas aeruginosa. Indian J. Med. Microbiol. 2017, 35, 282–285. [Google Scholar] [CrossRef]
- Yousefi, A.; Torkan, S. Uropathogenic Escherichia coli in the urine samples of Iranian dogs: Antimicrobial resistance pattern and distribution of antibiotic resistance genes. BioMed Res. Int. 2017, 2017, 4180490. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.Y.; Lupo, A.; Schink, A.K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Enne, V.I.; Bennett, P.M.; Livermore, D.M.; Hall, L.M.C. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 2004, 53, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Kotb, D.N.; Mahdy, W.K.; Mahmoud, M.S.; Khairy, R.M.M. Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis. 2019, 19, 979. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.Y.; Wang, H.N.; Tian, G.B.; Zhang, Y.; Yang, X.; Xia, Q.Q.; Tang, J.N.; Zou, L.K. Phenotypic and genotypic characterisation of antimicrobial resistance in faecal bacteria from 30 Giant pandas. Int. J. Antimicrob. Agents 2009, 33, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Infections with extended-spectrum beta-lactamase-producing Enterobacteriaceae changing epidemiology and drug treatment choices. Drugs 2010, 70, 313–333. [Google Scholar] [CrossRef]
- Karahutova, L.; Mandelik, R.; Bujnakova, D. Antibiotic resistant and biofilm-associated Escherichia coli isolates from diarrheic and healthy dogs. Microorganisms 2021, 9, 1334. [Google Scholar] [CrossRef]
- Hutton, T.A.; Innes, G.K.; Harel, J.; Garneau, P.; Cucchiara, A.; Schifferli, D.M.; Rankin, S.C. Phylogroup and virulence gene association with clinical characteristics of Escherichia coli urinary tract infections from dogs and cats. J. Vet. Diagn. Investig. 2018, 30, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Skurnik, D.; Le Menac’h, A.; Zurakowski, D.; Mazel, D.; Courvalin, P.; Denamur, E.; Andremont, A.; Ruimy, R. Integron-associated antibiotic resistance and phylogenetic grouping of Escherichia coli isolates from healthy subjects free of recent antibiotic exposure. Antimicrob. Agents Chemother. 2005, 49, 3062–3065. [Google Scholar] [CrossRef] [PubMed]
- Shahi, S.K.; Singh, V.K.; Kumar, A. Detection of Escherichia coli and associated beta-lactamases genes from diabetic foot ulcers by multiplex PCR and molecular modeling and docking of SHV-1, TEM-1, and OXA-1 beta-lactamases with clindamycin and piperacillin-tazobactam. PLoS ONE 2013, 8, e68234. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Askari, A.; Ghanbarpour, R.; Akhtardanesh, B.; Aflatoonian, M.R.; Sharifi, H.; Jajarmi, M.; Molaei, R. Detection of zoonotic diarrheagenic pathotypes of Escherichia coli in healthy household dogs. Iran. J. Microbiol. 2020, 12, 522–530. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coliphylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Jemima, S.A.; Verghese, S. Multiplex PCR for bla(CTX-M) & bla(SHV) in the extended spectrum beta lactamase (ESBL) producing Gram-negative isolates. Indian J Med Res 2008, 128, 313–317. [Google Scholar]
- Lin, C.F.; Hsu, S.K.; Chen, C.H.; Huang, J.R.; Lo, H.H. Genotypic detection and molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a regional hospital in central Taiwan. J Med Microbiol 2010, 59, 665–671. [Google Scholar] [CrossRef]
- Al-Agamy, M.H.; Aljallal, A.; Radwan, H.H.; Shibl, A.M. Characterization of carbapenemases, ESBLs, and plasmid-mediated quinolone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J. Infect. Public Health 2018, 11, 64–68. [Google Scholar] [CrossRef]
- Sengeløv, G.; Agersø, Y.; Halling-Sørensen, B.; Baloda, S.B.; Andersen, J.S.; Jensen, L.B. Bacterial antibiotic resistance levels in Danish farmland as a result of treatment with pig manure slurry. Environ. Int. 2003, 28, 587–595. [Google Scholar] [CrossRef]
- Ng, L.K.; Martin, I.; Alfa, M.; Mulvey, M. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell Probe. 2001, 15, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Xu, Y.C.; Sun, J.; Huang, M.; Jia, X.; Jiang, C.J.; Feng, Y.J. Harnessing efficient multiplex PCR methods to detect the expanding Tet(X) family of tigecycline resistance genes. Virulence 2020, 11, 49–56. [Google Scholar] [CrossRef]
- Luo, Y.; Mao, D.Q.; Rysz, M.; Zhou, D.X.; Zhang, H.J.; Xu, L.; Alvarez, P.J.J. Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci. Technol. 2010, 44, 7220–7225. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, A.; Strahilevitz, J.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 2006, 50, 2872–2874. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Hsueh, P.R.; Jacoby, G.A.; Hooper, D.C. Risk factors and clinical characteristics of patients with qnr-positive Klebsiella pneumoniae bacteraemia. J. Antimicrob. Chemother. 2013, 68, 2907–2914. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Z.L.; Liu, J.H.; Zeng, Z.L.; Ma, J.Y.; Jiang, H.X. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 2007, 59, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.T.; Zhou, Y.; Yang, L.; Xu, Y. Multidrug-resistant genes of aminoglycoside-modifying enzymes and 16S rRNA methylases in Acinetobacter baumannii strains. Genet. Mol. Res. 2014, 13, 3842–3849. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, N.; Rezaee, M.A.; Kafil, H.S.; Hasani, A.; Barhaghi, M.H.S.; Milani, M.; Sefidan, F.Y.; Memar, M.Y.; Lalehzadeh, A.; Ghotaslou, R. Evaluation of Resistance Mechanisms in Carbapenem-Resistant Enterobacteriaceae. Infect. Drug Resist. 2020, 13, 1377–1385. [Google Scholar] [CrossRef]
- Warburg, G.; Hidalgo-Grass, C.; Partridge, S.R.; Tolmasky, M.E.; Temper, V.; Moses, A.E.; Block, C.; Strahilevitz, J. A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: Sequence type 512 carrying a plasmid encoding aac(6)-Ib. J. Antimicrob. Chemoth. 2012, 67, 898–901. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Liu, L.; Lin, J.; Ma, K.; Long, H.Y.; Wei, L.; Xie, Y.; McNally, A.; Zong, Z.Y. Key evolutionary events in the emergence of a globally disseminated, carbapenem resistant clone in the Escherichia coli ST410 lineage. Commun. Biol. 2019, 2, 322. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 29–39. [Google Scholar] [CrossRef] [PubMed]


| blaESBL | 2012–2013 | 2015 | 2021 | 2021 | 2021 |
|---|---|---|---|---|---|
| Pet Dogs | Pet Dogs | Pet Dogs | Pet Cats | Working Dogs | |
| blaCTX-M | 20 | 15 | 29 | 18 | 17 |
| blaTEM | 0 | 0 | 0 | 0 | 2 |
| blaCTX-M+TEM | 13 | 10 | 30 | 20 | 28 |
| blaCTX-M+OXA-1 | 4 | 1 | 0 | 4 | 0 |
| blaCTX-M+TEM+OXA-1 | 4 | 1 | 1 | 1 | 1 |
| Antibiotic | blaCTX-M Positive (n = 149) | blaCTX-M Negative (n = 102) | p-Value |
|---|---|---|---|
| Amikacin | 13.42% | 9.80% | 0.385 |
| Gentamicin | 61.74% | 32.35% | <0.001 ** |
| Imipenem | 2.01% | 3.92% | 0.447 |
| Meropenem | 1.34% | 0.98% | 1.000 |
| Cefazolin | 100.00% | 12.75% | <0.001 ** |
| Ceftazidime | 7.38% | 2.94% | 0.132 |
| Cefotaxime | 91.28% | 0.98% | <0.001 ** |
| Cefepime | 86.58% | 0.98% | <0.001 ** |
| Aztreonam | 60.40% | 0.00% | <0.001 ** |
| Ampicillin | 100.00% | 95.10% | 0.010 * |
| Piperacillin | 100.00% | 81.37% | <0.001 ** |
| Amoxicillin–Clavulanate | 4.03% | 10.78% | 0.036 * |
| Ampicillin–Sulbactam | 41.61% | 33.33% | 0.185 |
| Piperacillin–Tazobactam | 3.36% | 0.98% | 0.406 |
| Colistin | 4.03% | 1.96% | 0.479 |
| Trimethoprim–Sulfamethoxazole | 60.40% | 72.55% | 0.047 * |
| Chloramphenicol | 53.69% | 47.06% | 0.302 |
| Ciprofloxacin | 47.65% | 32.35% | 0.016 * |
| Levofloxacin | 46.31% | 30.39% | 0.011 * |
| Moxifloxacin | 52.35% | 33.33% | 0.003 * |
| Tetracycline | 85.91% | 91.18% | 0.206 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Ji, X.; Liang, B.; Jiang, B.; Li, Y.; Yuan, T.; Zhu, L.; Liu, J.; Guo, X.; Sun, Y. Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021. Antibiotics 2022, 11, 1506. https://doi.org/10.3390/antibiotics11111506
Zhou Y, Ji X, Liang B, Jiang B, Li Y, Yuan T, Zhu L, Liu J, Guo X, Sun Y. Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021. Antibiotics. 2022; 11(11):1506. https://doi.org/10.3390/antibiotics11111506
Chicago/Turabian StyleZhou, Yifan, Xue Ji, Bing Liang, Bowen Jiang, Yan Li, Tingyv Yuan, Lingwei Zhu, Jun Liu, Xuejun Guo, and Yang Sun. 2022. "Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021" Antibiotics 11, no. 11: 1506. https://doi.org/10.3390/antibiotics11111506
APA StyleZhou, Y., Ji, X., Liang, B., Jiang, B., Li, Y., Yuan, T., Zhu, L., Liu, J., Guo, X., & Sun, Y. (2022). Antimicrobial Resistance and Prevalence of Extended Spectrum β-Lactamase-Producing Escherichia coli from Dogs and Cats in Northeastern China from 2012 to 2021. Antibiotics, 11(11), 1506. https://doi.org/10.3390/antibiotics11111506





