1. Introduction
Candida albicans (
C. albicans) is the most common human fungal pathogen and is responsible for notorious candidiasis ranging from superficial mucosal to life-threatening systemic infection. In regard to contemporary antifungals,
C. albicans infections carry an alarming mortality in the United States, as high as 40%, in systemic candidiasis attributed to the incomplete eradication of the yeast and the emergence of resistant phenotypes [
1]. The fungal pathogenicity is due to its virulence factors such as the ability to adhere to host tissues, the formation of hyphae, and the development of a complex and robust biofilm.
C. albicans readily forms biofilms on the surface of the host or medical devices. It is known that biofilm formation poses a crucial cause for more drug resistance than its free yeast to conventional antifungal agents and that it is less affected by host immune responses. It has been estimated that microbes in biofilms can tolerate 100–1000 times higher concentrations of antibiotics than their counterpart planktonic microbes, making them harder to eradicate [
2]. Even worse, mature biofilms continuously release
Candida cells into the environment; these then colonize new sites, resulting in chronic infections [
3]. Yeast and filamentous fungi biofilm-associated infections have been increasingly studied, and in 2021, this compelled the WHO to compile the first fungal priority pathogen list (FPPL) [
4]. However, only a handful of antifungal drugs are currently approved for the treatment of fungal infections (e.g., azoles, polyenes, and echinocandins). Among these, the most broadly used azoles are ineffective against
C. albicans biofilm due to the upregulation of efflux pumps in the biofilm, while amphotericin B, a representative of polyenes, failed to reach biofilms because of the enhanced extracellular matrix or beta-glucan synthesis during biofilm growth. Nevertheless, some
Candida species with intrinsic resistance to fluconazole also have a decreased susceptibility to echinocandins, which are occasionally associated with serious bloodstream infections [
5]. Furthermore,
Candida spp. with an
fks mutation can form resistance to echinocandins, reducing antibiofilm activity [
6]. Consequently, the emergence of azole-resistant yeasts in many parts of the world draws grave concern. Even worse, the treatment of
C. albicans biofilm-related infections is limited since the biofilm extracellular matrix acts as a physical barrier, thus preventing the diffusion of antimycotic drugs [
7,
8]. For these reasons, there is an urgent need to explore alternative therapeutic strategies for developing new antifungal drugs with efficient antibiofilm activity and non-toxicity, and novel drug targets are waiting to be discovered to solve the problem of
C. albicans biofilm formation.
Recently, antimicrobial peptides (AMPs) have garnered attention for their broad-spectrum antimicrobial activities against bacteria, fungi, and some enveloped viruses [
9,
10,
11,
12], as well as their strong antibiofilm ability [
13]. AMPs are natural, small, and highly conserved effector molecules that play a key role in innate immunity, with the antimicrobial mode of action mainly focused on the integrity of microbial wall cells by pore formation or related activities. AMPs are small polypeptide molecules generated by all living organisms to protect their host from pathogens, with properties of multiple cellular targets, low toxicity, and functional diversity. These characteristics enable the AMPs to exhibit potential ability as treatment options against organisms resistant to the conventional antifungal drugs and as a new treatment for biofilm-related infections [
14,
15]. So far, several natural peptides, including nisin, daptomycin, polymyxin B, and colistin, have been approved by the US Food and Drug Administration (FDA) for the clinical treatment of microbial infections [
16].
In previous work, our research group discovered AMP-17, a novel antimicrobial peptide with a significant antifungal effect against
C. albicans,
C. neoformans, et al. [
17,
18], as well as exhibiting a low toxicity towards human erythrocytes and stable physicochemical characteristics [
19]. Further studies showed that the antifungal activity of the peptide was closely associated with cell wall destruction, an increase in membrane permeability, filamentous growth delay, and the abnormal metabolism of macromolecules [
17,
20]. Given that filamentation as the structural basis of the robust biofilm acts an essential role in the
C. albicans biofilm development, the inhibitory effect of AMP-17 on hyphae undoubtedly implies that it has potential antibiofilm ability. Therefore, it is necessary and valuable to investigate whether AMP-17 can inhibit
C. albicans biofilm and classify the potential mode of the antibiofilm action.
According to the literature, the severity of
C. albican biofilm has been linked to the increased cellular density associated with the biofilm community,
C. albicans morphological changes such as yeast-hyphae transition, the protective effect of the extracellular matrix, and the production of drug efflux pumps [
21]. Antifungal agents could play a crucial role in preventing and controlling biofilm-related infections if they target the aforementioned causes of biofilm formation. At present, the mechanisms of AMPs are referred to in the following ways: disruption of the cell signaling system, avoidance of excessive microbial reaction, suppression of binding protein transport, or degradation of the extracellular polymeric matrix [
13]. Furthermore, the same AMPs can exert their biological activity in a corresponding way in different biofilm-forming stages [
22]. In this work, we evaluate the antibiofilm activity of AMP-17 in the biofilm-forming and mature phase and summarize the specific antifungal characteristic of the peptide in different stages of the
C. albicans biofilm.
Finally, to shed light on the antifungal mechanisms of AMP-17 against C. albicans biofilm and the putative targets, and considering that yeast germination and the filamentous growth in C. albicans play a significant role in biofilm formation, we performed morphological techniques such as SEM and CLSM to observe the structural features of C. albicans cells. RNA-seq was applied to study the gene expression profile of AMP-17-treated biofilms to find out the closely related response genes and signaling pathways, which could facilitate elucidating the molecular mechanisms of the peptide’s action on the C. albicans biofilm and open up new therapeutic alternatives for the treatment of Candida-related biofilm infection.
3. Discussion
C. albicans is one of the leading causes of hospital-acquired bloodstream infections. When patients undergo immunosuppressive therapies, organ transplantation, and chemotherapy, they are susceptible to systemic
C. albicans bloodstream infections, also known as candidemia and tissue colonization [
36]. Nevertheless, the presence of
C. albicans biofilms makes the current problems more serious because the biofilm form is less sensitive to common antifungal drugs, such as azoles and conventional amphotericin B, than planktonic cells [
37,
38]. Furthermore,
C. albicans biofilms are dynamic and highly structured three-dimensional networks composed of a large number of hyphae and an extracellular matrix, which worsen the problem. Given the severity of biofilm-related diseases, it is urgent to find alternative antifungal agents that can efficiently control biofilms. Interestingly, some antimicrobial peptides (AMPs) have been found to have a broad-spectrum antimicrobial ability and excellent antibiofilm capacity, according to previous reports [
39,
40]. In our previous study, AMP-17 showed a strong in vitro inhibitory effect toward multiple fungi of reference strains and clinical isolates, including
C. albicans,
C. neoformans,
C. glabrata, and
C. tropicals, while it was not toxic towards human erythrocyte cells [
17,
18,
19]. In the current study, AMP-17 showed strong antibiofilm activity against early-forming biofilm and mature biofilm. In addition, the CV assays also displayed a similar inhibitory effect of the peptide against
C. albicans in both the biofilm-forming and mature biofilm phases (
Figure 1). Interestingly, the antibiofilm activity of AMP-17 was stronger than that of FLC at the same concentration. This effect was further confirmed through morphological studies using the SEM and CLSM assays (
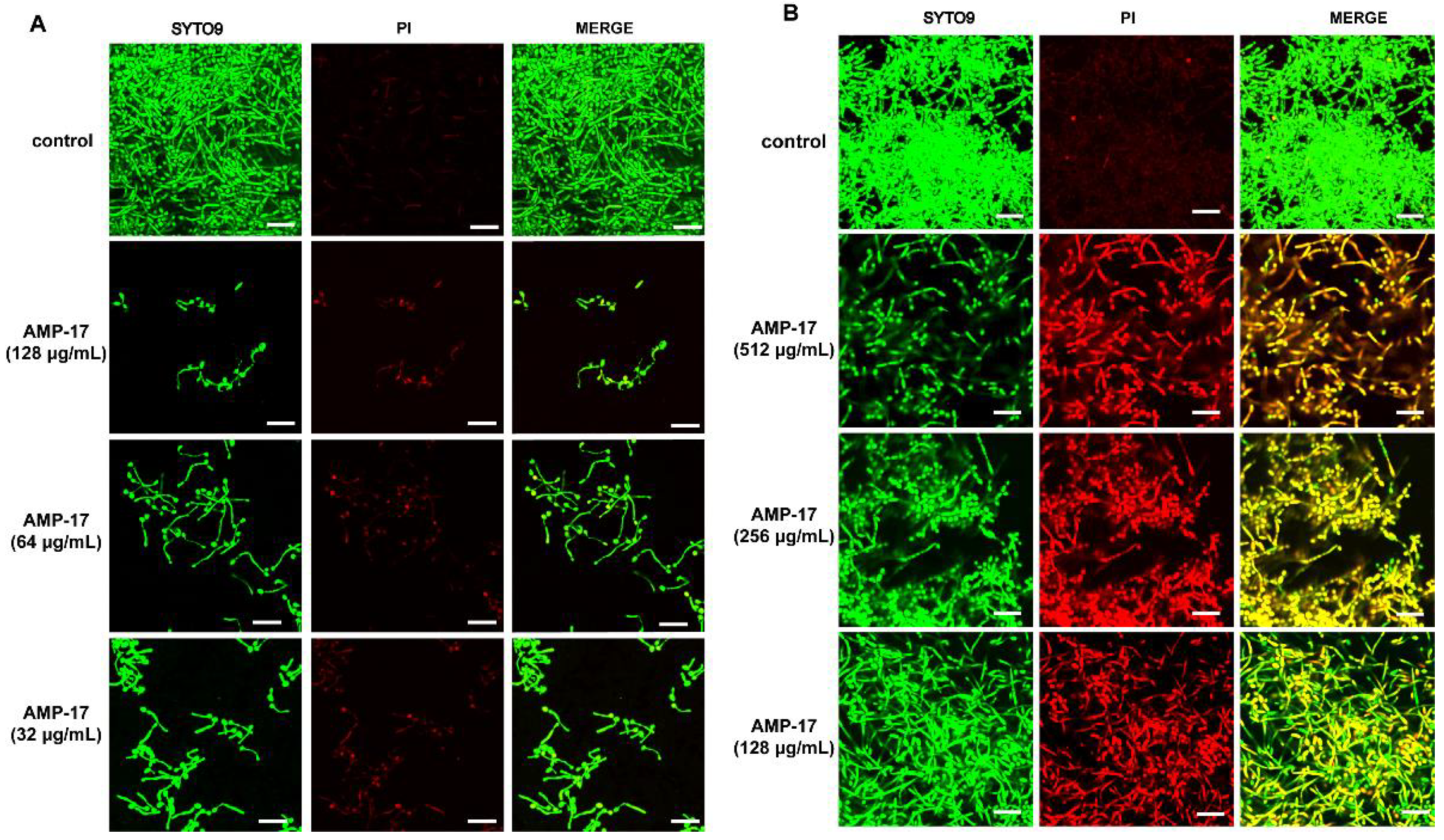
Figure 2 and
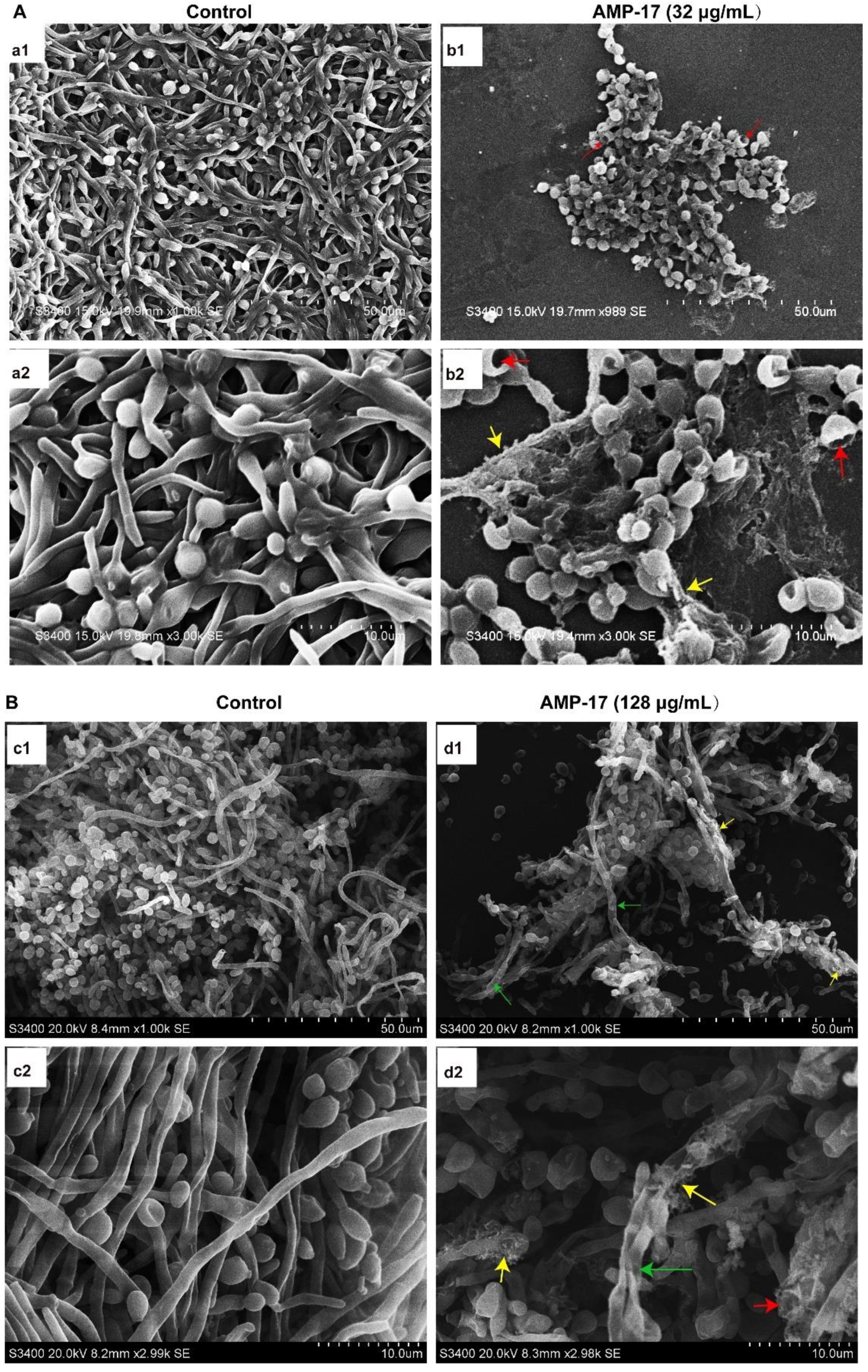
Figure 3). Morphological analysis validated the specific 3D network was hardly formed and the structure was damaged in response to AMP-17. As biofilm infections are too stubborn to control effectively in clinical practice, we were inspired to elucidate the potential molecular mechanisms of AMP-17 and to provide an alternative candidate agent for candidiasis therapy.
The formation of
C. albicans biofilms involves several specific stages, including the early, developmental, and mature phases. Cell adherence is the first step in biofilm formation, followed by the
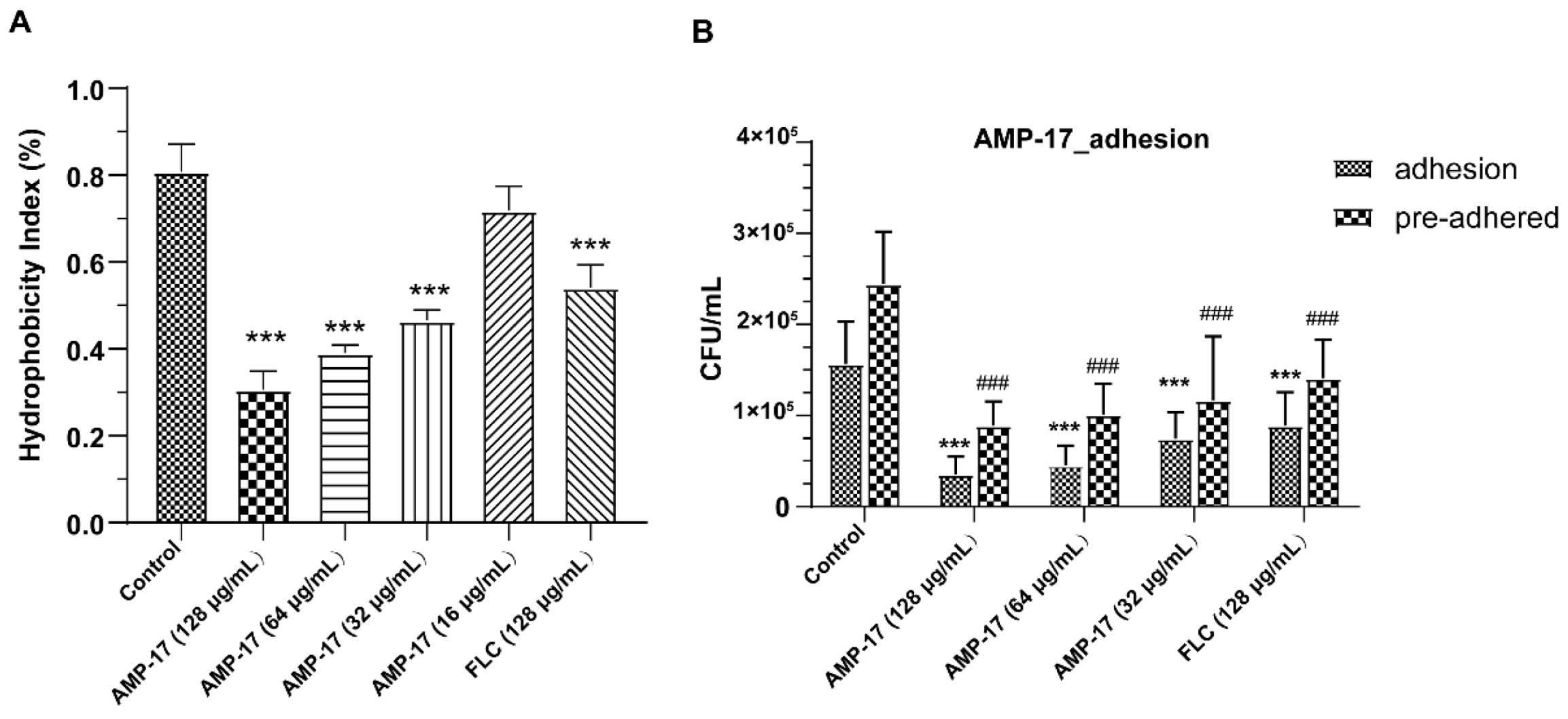
C. albicans yeast cell gradually transforming into hyphal forms, which is essential not only for biofilm growth but also for biofilm dissemination. Repression of adhesion or the yeast-to-hyphae transition can cause biofilm formation defects, making a novel target for biofilm-specific treatment. Our data demonstrated that AMP-17 not only suppressed adhesion effectively but also inhibited the yeast-to-hyphae transition. In terms of inhibitory adhesion, our data showed that 32 μg/mL AMP-17 remarkably removed more than 50% of adhering cells or adhered cells from the microplate surface (
Figure 4). To the best of our knowledge, relative hydrophobicity is considered a pathogenic factor contributing to
Candida spp. adherence on the host surface [
24]. We also observed that 64 μg/mL AMP-17 significantly reduced the hydrophobicity index. Therefore, the inhibition of adhesion in AMP-17 treatment was likely due to the decrease in hydrophobicity induced by the peptide, contributing to the earliest antibiofilm effect in the
C. albicans biofilm life cycle. While, in terms of the inhibitory yeast-to-hyphae transition, the filamentation assay showed that AMP-17 obviously suppressed the morphological transition (
Figure 5). Starting at 8 μg/mL of AMP-17, only smooth-edged colonies were found on solid Spider medium. At 32 μg/mL, filamentous cells were hardly seen in the RPMI 1640—10% FBS medium, and yeast-like cells were locked in the hyphae-induced medium. Nevertheless, the inhibition of adhesion and yeast-to-hyphae in the AMP-17 treatment was stronger than that produced by FLC treatment at the same concentration. The antibiofilm action of AMP-17 seemed to be the inhibitory adhesion or yeast-to-hyphae transition.
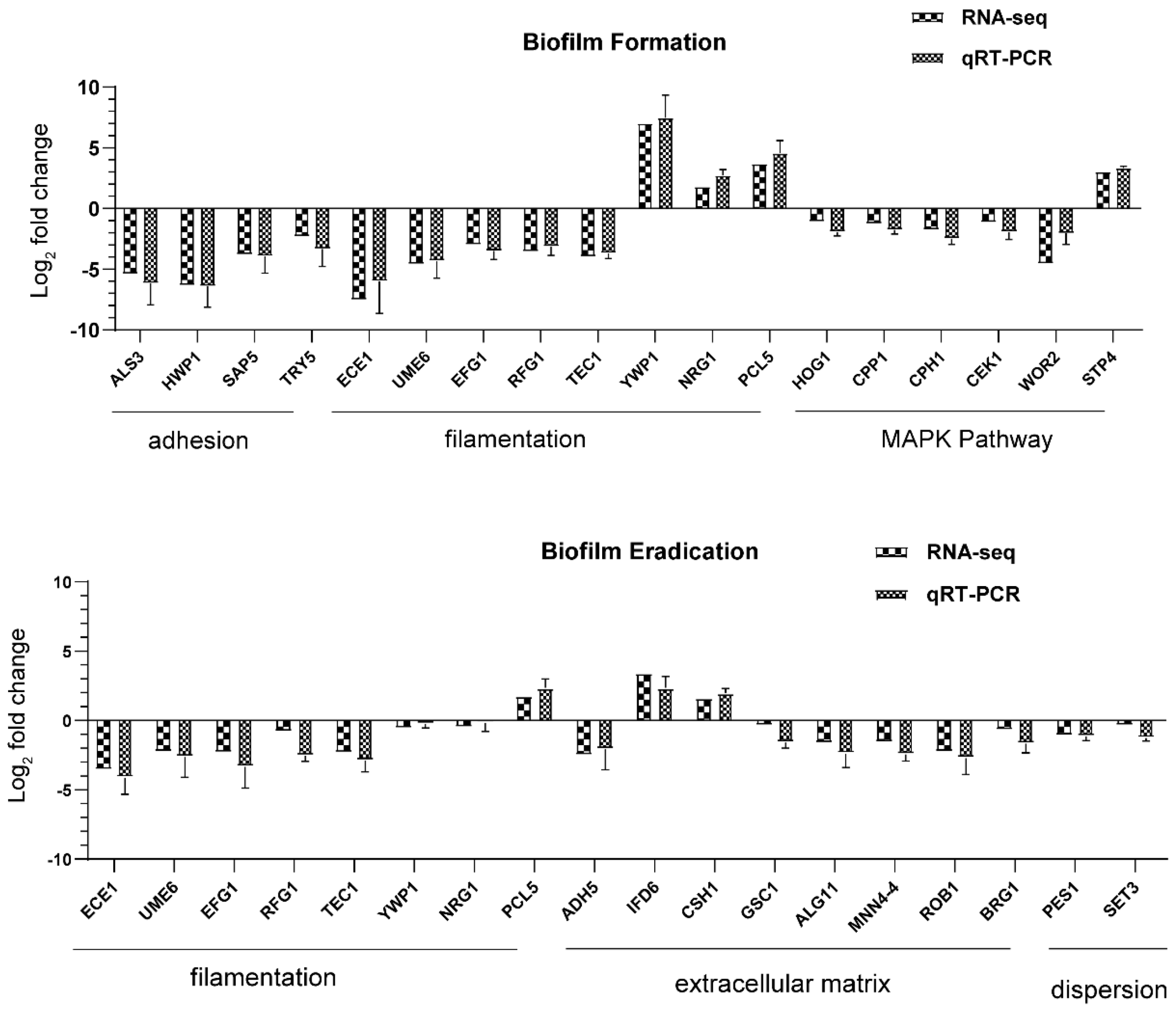
Adhesion is thought to be the first event in hyphal growth, and a family of cell surface proteins known as agglutinin-like sequence (Als) proteins are associated with the cell adhesion and aggregation of yeast cells and are an essential component of biofilm formation [
41]. RNA-seq results showed that the relevant Als family genes including ALS1, ALS2, ALS3, ALS4, and ALS7 were downregulated, as expected (
Table S4). Among these genes, ALS3 reduction was greater than others, with a downregulation of 5.35-fold. Furthermore, the downregulation of other adhesion-related typical genes such as HWP1, SAP5, and TRY5 [
42,
43] was also evidenced to reduce the cell adhesion and virulence ability, contributing to the detachment of biofilms from an abiotic substrate (
Table S1,
Figure 9). On the other hand, hyphal elements contribute substantially to the
C. albicans biofilm volume, and the lack of this morphological form in AMP-17 treated biofilms was evidenced by a sparse, preliminary biofilm with scattered blastopores shown in SEM and CLSM assays (
Figure 2 and
Figure 3). In addition, the expression of hyphal initiation (EFG1, CPH1, and TEC1) and long-term maintenance (UME6, HGC1, and ECE1) stages were downregulated after AMP-17 treatment. Reportedly, Efg1 and Cph1, the first identified regulators of hyphal development, synergistically regulate virulence genes, while Tec1 has been shown to regulate hyphal development and virulence in
C. albicans [
42]. Furthermore, genes negatively regulating biofilm formation were upregulated when exposed to AMP-17 in the current study, such as the yeast form-specific gene (YWP1) and genes encoding a repressor of the filamentation program (NRG1) [
44,
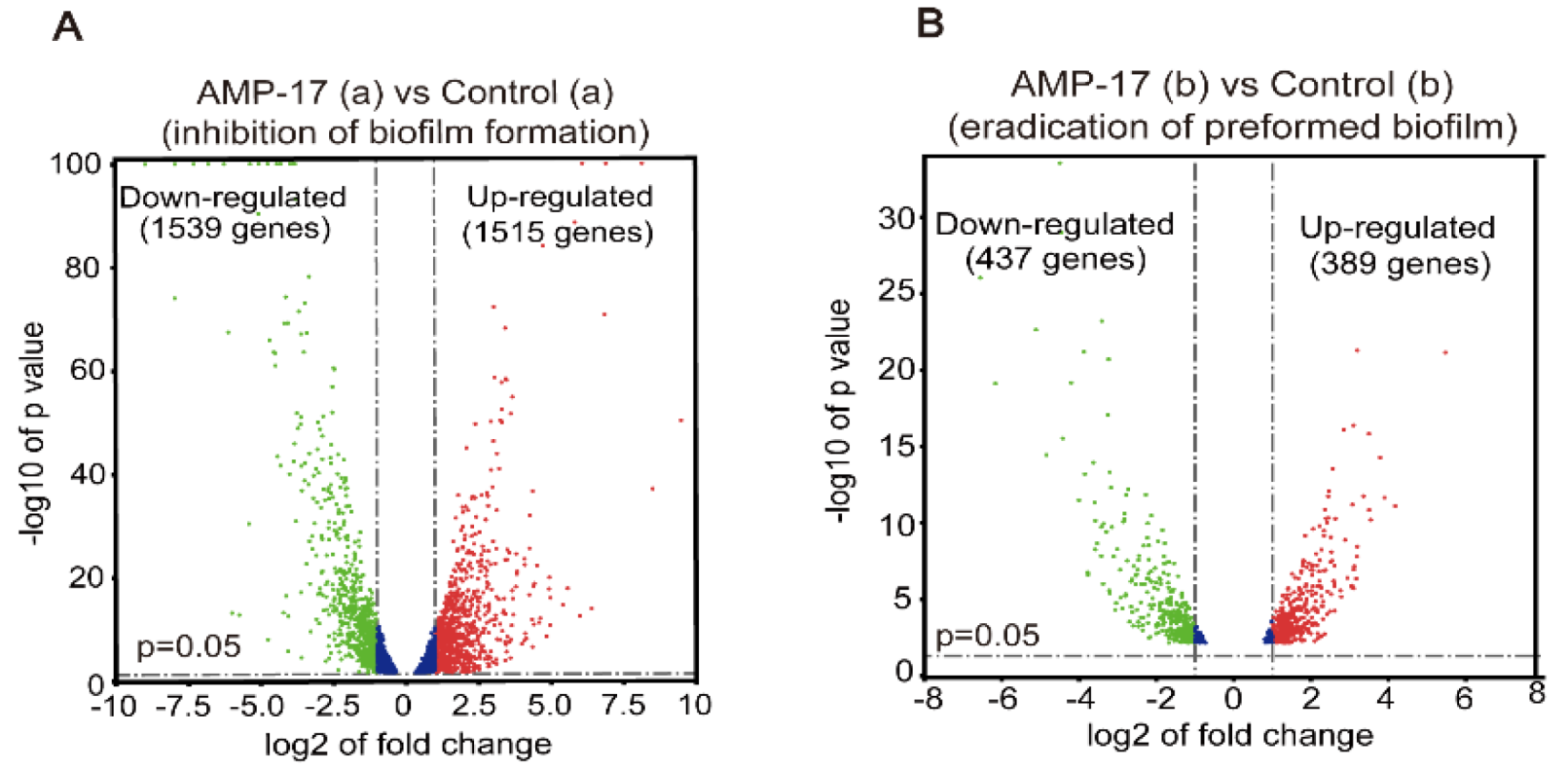
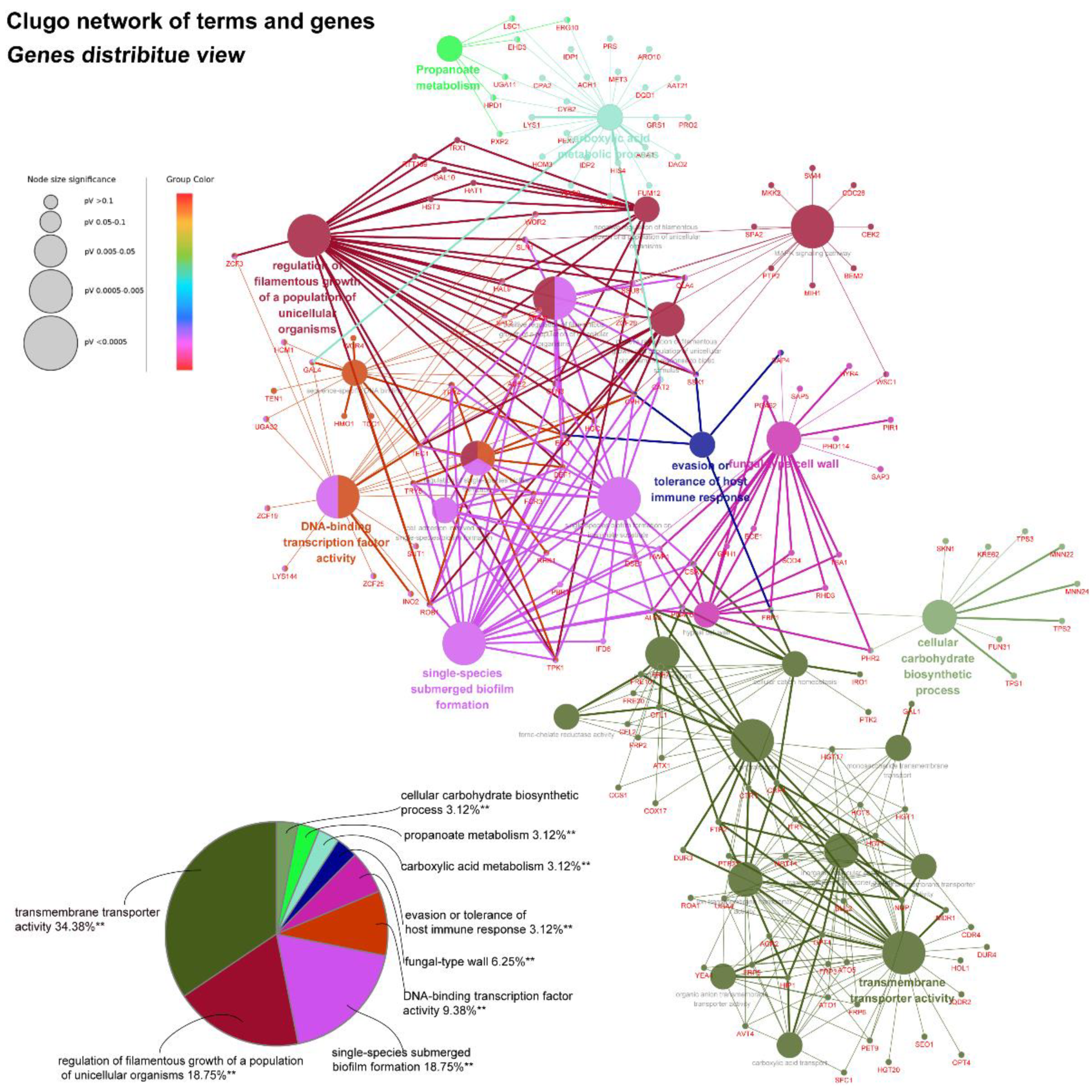
45]. As yeast-to-hyphae and filamentous development are necessary for the structure of 3D biofilms, these filamentation-regulated genes in the mature biofilm phase are also altered similarly to the biofilm-forming phase when treated with AMP-17. Consistently, 18.75% of genes participate in the regulation of filamentous growth in the co-expressed DEGs of both AMP-17-treated forming biofilms and mature biofilms (
Figure 8A). This phenomenon suggests that the repression of AMP-17 on hyphae took place during the whole biofilm developmental period. Above all, the downregulation of adhesion-related or filamentous-associated genes or upregulation of the negative regulators contributed to defective
C. albicans biofilm or its inability to form a biofilm, which powerfully explains the inhibitory effect of AMP-17 on biofilms. Importantly, the hyphae-related antibiofilm activity of AMP-17 in C. albicans is likely to produce a profound effect on the drug-resistant
Candida spp. Due to drug resistance, echinocandin occasionally exhibits reduced antibiofilm activity in
Candida spp. with an
fks mutation [
6], while AMP-17 might overcome the difficulties in treating echinocandin-resistant
Candida spp., leading to an increased interest in a future investigation into the synergist effect of the peptide against
C. albicans biofilm in combination with an echinocandin.
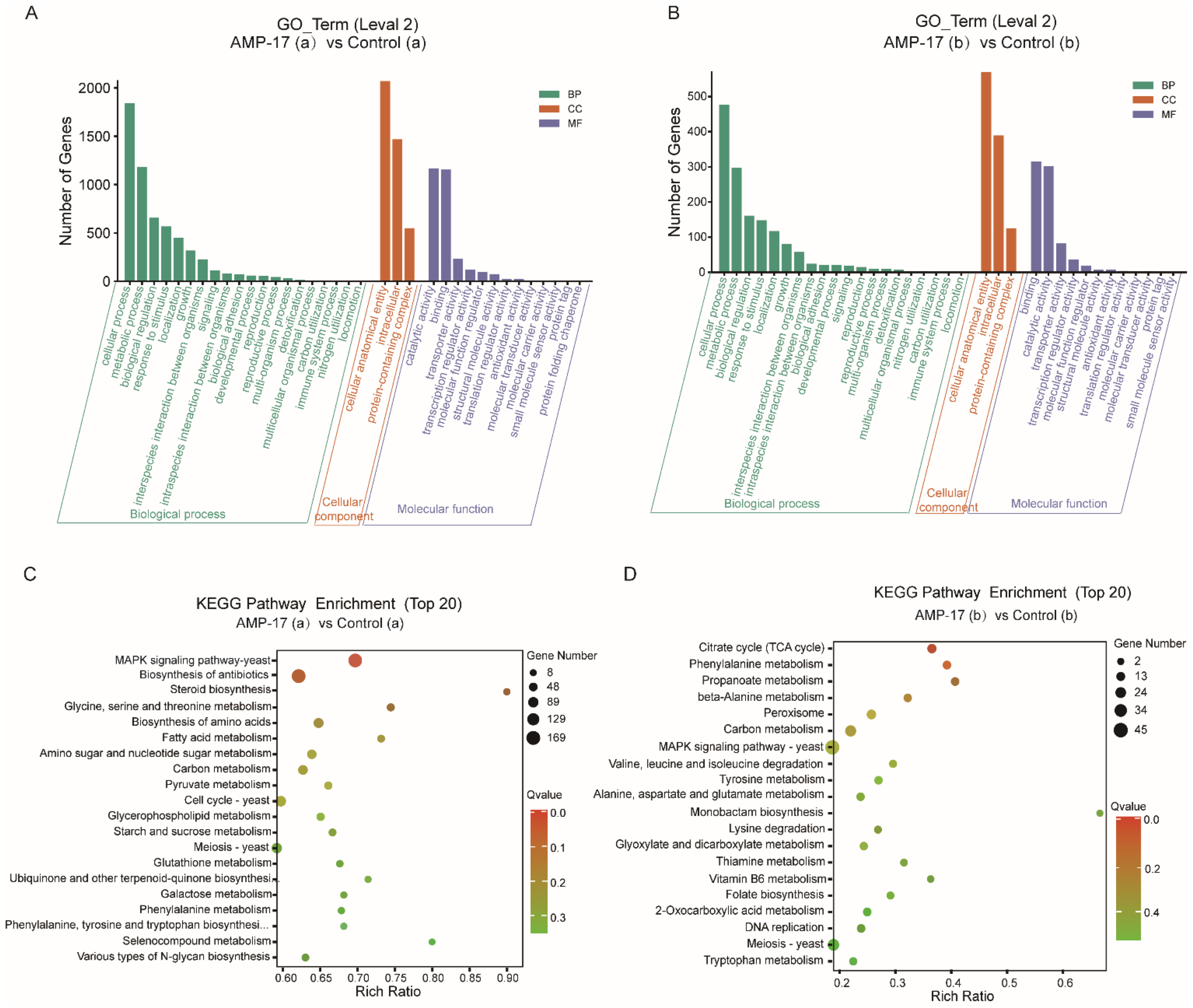
From the regulatory-pathway perspective, since the key transcriptional factor regulating hyphal growth (EFG1) involved in the RAS/cyclic AMP (cAMP) pathway was downregulated after AMP-17 treatment, as the RNA-seq and qRT-PCR results showed, we further verified the expression of the main genes in this pathway by RNA-seq, including RAS1, CYR1, PDE2, and EFG1. The results showed that, except for EFG1, all these genes remained unchanged. Therefore, AMP-17 treatment exerted a minor influence on the Ras/cAMP pathway. Nevertheless, KEGG enrichment showed that a large number of genes in the MAPK pathway were altered greatly in response to AMP-17 (
Figure S4). Previous reports suggested that MAPK participates in cell wall forming under vegetative and filamentous growth, playing an important role in morphogenesis [
25,
46]. The gene expression level of HOG1, CEK1, CCP1, CPH1, and WOR3 was found to be downregulated after AMP-17 treatment. As one of the MAPKs, Cek1 has a role in the biofilm formation and filamentous growth of
C. albicans. While another MAPK, Mkc1, has a function in the formation and maintenance of cell wall structure [
25]. Our data showed that CEK1 was downregulated and MKC1 nearly unchanged, indicative of hyphae inhibition mediated by the Cek1-MAPK pathway other than Mkc1 under AMP-17 treatment. Furthermore, some antifungal agents exert antibiofilm effects by targeting cell walls or membrane components, such as FLC, which exerts its antifungal activity by interfering with the ergosterol biosynthesis pathway [
47]. Hence, the inhibition of ergosterol biosynthesis by AMP-17 may also account for its antibiofilm effect. As we found, five ergosterol biosynthetic genes (ERG1, ERG27, ERG251, ERG4, and ERG6) were collectively upregulated after AMP-17 treatment, which directly resulted in the reduction in ergosterol biosynthesis (
Figure S4). Blocking ergosterol biosynthesis could be effective in both yeasts and biofilms, and the depletion of ergosterol directly changes cell membrane functions, which eventually leads to cell death. This explained the increased numbers of dead hyphae and cells under AMP-17 treatment, illustrated by the CLSM images (
Figure 2). Collectively, we postulate that the antibiofilm effect of AMP-17 is due to its regulation of the MAPK pathway and suppression of ergosterol biosynthesis metabolism, which in turn led to abnormal morphogenesis and cell death, and finally, inhibited biofilm development.
As is well known, a salient feature of the
C. albicans biofilm is the presence of the biofilm matrix composed of major macromolecules including polysaccharides, proteins, and nucleic acids, which plays a preponderant role in protecting the biofilm cells from antifungal drug treatment [
8]. The AMP-17 treatment inhibited the expression of several genes required for encoding enzymes that produce polysaccharide components (e.g., ALG11 and MNN4) in the mature biofilm phase, indicative of the repression of biofilm matrix production [
48]. In the AMP-17-treated preformed biofilm group, there was a reduction in GSC1 involved in the β-1,3-glucan synthesis, resulting in broad defects in the matrix synthesis. Interestingly, a few genes encoding alcohol dehydrogenases, such as Adh5, Csh1, and Ifd6, play roles in matrix production: Adh5 acts positively, while Csh1 and Ifd6 act negatively [
49]. In the current study, the AMP-17-treated preformed biofilms exhibited a higher gene expression of CSH1 and IFD6, and lower expression of ADH5, compared to the untreated samples. ADH5 encodes alcohol dehydrogenases that may promote the entry of ethanol into the TCA cycle for energy or via the glyoxylate shunt to provide hexose for β-1,3 glucan synthesis, while CSH1 and IFD6 act preferentially to yield a matrix inhibitory signal, although the roles of such genes may be too complex to reveal how AMP-17 regulates the
C. albicans biofilm matrix production or degradation. Indeed, the changes in these matrix-regulated genes gave us a glimpse that the inhibition of matrix production with AMP-17 treatment plays a critical role in antibiofilm activity, particularly in mature biofilm, due to their impact on carbon metabolism. Sarah et al. studied that the TCA cycle plays a vital part in mediating biofilm formation, especially by influencing the matrix composition in
S. aureus [
50]. As a result, the pathways of the TCA cycle, phenylamine metabolism, propanoate metabolism, and beta-Alanine metabolism, enriched in the AMP-17 treated preformed biofilms (
Figure 7) evidenced the interference of the peptide with matrix production and biofilm development. On the other hand, the dispersal of
C. albicans into the surrounding environment (primarily as round, yeast-form cells) occurs throughout biofilm formation, with great numbers of cells being dispersed once the biofilm reaches maturity. Therefore, it is necessary for antifungal agents to prevent the
C. albicans cells within biofilms from dispersing or disseminating. Set3, a component of a chromatin-modifying complex, is also required for dispersal, probably by being recruited to specific genes by the transcriptional regulator Nrg1. In response to AMP-17, some genes linked to dispersals, such as NRG1, PES1, and SET3, were suppressed in the mature biofilm phase (
Figure 9). Overexpression of either NRG1 or PES1 increased the number of cells released from the biofilm. Therefore, the inhibition of filamentous growth, biofilm matrix production, and cell dispersal may account for the elimination activity of AMP-17 on preformed biofilm.
In addition, AMP-17’s antibiofilm activity might also be achieved through a multitarget action, as considerable DEGs are involved in carbohydrate metabolism, fatty acid metabolism, nucleotide metabolisms, and amino acid metabolism. For example, genes encoding carboxylic acid metabolic proteins (CDC28, LSC1, CYB2, ACH1, and GLA4) [
51] and metal ion transport (CTR1, CFL1, CFL2, and CRP1) [
52], required for various metabolic or catalytic activities, were altered greatly after AMP-17 treatment. Furthermore, as the necessary components of the cell membrane and cell skeleton, the abnormal metabolism of carbohydrates, glycerophospholipid, and various amino acids certainly affects the organism’s life cycle, including cell integrity and biofilm growth. Previous studies have revealed a link between the abnormal cell cycle and cell death of AMP-17-treated planktonic cells [
20]. Hence, the significant antibiofilm effect of AMP-17 not only depends on the repression of adhesion, yeast-to-hyphal, matrix production, and dispersion but also on the damage to yeast cells in the biofilm.
4. Materials and Methods
4.1. Antimicrobial Peptide
AMP-17 was obtained from the recombinant expression plasmid pET-28a (+)—(AMP-17) using a prokaryotic expression system and purified with Ni-NTA beads (Novagen, Germany) [
17,
20]. Briefly,
E. coli BL21 (DE3) harboring the plasmid was incubated at 37 °C in liquid luria broth (LB) medium containing 100 μg/mL kanamycin with shaking at 200 rpm and until its OD
600 reached 0.5. Then, a final concentration of 0.05 mmol/L IPTG was added, and the culture was incubated for 24 h at 32 °C. Finally, bacterial cells were collected by centrifugation (12,000 rpm for 10 min) at 4 °C, resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, and 100 mM NaCl), and disrupted by sonication (160 W, ultrasonic for 1 s, pause for 2 s, 3 min). Subsequently, the lysate was dissolved in urea, the prepared protein was purified through His Tag’s Ni-NTA affinity chromatography, and the imidazole was removed by ultrafiltration using an ultrafiltration tube. AMP-17 was granted a Chinese patent license in 2019 (patent number: ZL 2016 1 0428119. 8) and the amino acid sequence was shown in our previous report [
20].
4.2. Strains and Growth Condition
A C. albicans reference strain SC5314 was preserved by the Key Laboratory of Modern Pathogenic Biology of Guizhou Medical University. C. albicans were grown at 35 °C in YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) and stored on YPD plates solidified with 1.5% agar at 4 °C. A single C. albicans colony was inoculated into YPD broth and incubated at 35 °C for 12 h with shaking at 200 rpm and grown to the exponential phase for further experiments. To stimulate the hyphal growth of C. albicans, RPMI-1640 (Invitrogen, Carlslad, CA, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) was used as the culture medium.
4.3. Minimal Inhibitory Concentration (MIC) of Planktonic C. albicans
The minimum inhibitory concentration (MIC) for AMP-17 against
C. albicans was tested by a broth micro-dilution assay in 96-well microtiter, according to the procedures of the Clinical and Laboratory Standards Institute (CLSI) standard, with slight modifications [
53]. Briefly, the exponential fungal cells incubated in YPD medium overnight were diluted to 1–2 × 10
3 CFU/mL and co-incubated with various concentrations of AMP-17 in a Sabouraud-dextrose broth (SDB) at 35 °C for 24 h. The lowest concentration at which no visual growth was observed by the naked eye was considered the MIC value. Specifically, FLC was used as a positive control drug and
Candida.
parapsilosis ATCC22019 as the quality control strain. When the MIC of FLC against
C. parapsilosis ATCC22019 was between 0.5 μg/mL and 4.0 μg/mL, the experimental data were reliable. The experiments were repeated in triplicate and three multiple wells were set up in each experiment.
4.4. Biofilm Formation
As fetal bovine serum (FBS) is needed for filamentous growth during C. albicans biofilm formation, FBS was added to RPMI-1640 (RPMI 1640-FBS) at a final concentration of 10% to guarantee biofilm development. Briefly, yeast cells at a density of 1 × 106 CFU/mL were seeded into a flat-bottomed 96-well microplate of polypropylene at 37 °C for 90 min. Then, non-adhered cells were removed by washing the walls twice using sterile PBS, followed by RPMI 1640-FBS. Next, the adherent yeast was co-incubated with AMP-17 for 12 h at 37 °C to detect the effect of the peptide on biofilm formation. In order to observe the eradication ability of AMP-17 on mature biofilm, the culture needed to be incubated for 24 h at 37 °C to grow mature, and then was co-incubated with AMP-17.
4.5. XTT Reduction Assays
The XTT/Menadione assay was applied to assess the biofilm mass of
C. albicans and determine the antibiofilm efficacy of AMP-17 on biofilm formation and mature biofilm, measured by the microtiter plate reader at 490 nm [
29]. Before each assay, the XTT salt [2,3-bis (2-methoxy-4-nitro-5-sulfophenyl) 2H-tetrazolium-5-carboxanilide sodium salt] was dissolved in Ringer’s solution at a final concentration of 0.5 mg/mL. Notably, a menadione solution at a final concentration of 0.4 nM was prepared and filter-sterilized. Finally, the XTT solution was mixed with the menadione solution at a ratio of 20:1 (
v/
v). At the end of the incubation period, the biofilm was washed with sterile PBS to remove planktonic cells, followed by 100 μL of freshly prepared XTT solution added to each wall and incubated for 30 min in the dark at 37 °C. The supernatants were transferred to a new flat bottom plate and an OD490 nm, which represents the metabolic activity of the
C. albicans biofilm, was detected at 490 nm.
4.6. Crystal Violet (CV) Assays
In addition to the XTT reduction method, CV assays that represent biofilm biomass are also usually used to evaluate biofilm viability. Once the samples were finished, they were washed twice using sterile PBS to remove non-adhered cells, and then stained with 5% crystal violet (Solarbio, Beijing, China) for 10 min. Next, the stained biofilms were washed gently with sterile PBS, and destained with 95% ethanol; then, 100 μL PBS was added to every microplate wall. Finally, the absorbance of each well was measured at a wavelength of 560 nm.
4.7. Detection of Biofilm Inhibition and Biofilm Eradication
C. albicans biofilm formation in the presence of AMP-17 was studied in accordance with the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI) for antifungal susceptibility testing. For the biofilm inhibition of AMP-17,
C. albicans suspension on logarithmic phase was diluted to 2 × 10
6 CFU/mL in RPMI medium and incubated with 2-fold serial dilutions of AMP-17 in a 96-wall flat-bottomed microplate at 37 °C for 12 h. After incubation, the supernatant was removed without disturbing the intact biofilms and washed twice with sterile PBS buffer to remove any planktonic cells. Finally, the biofilm biomass was measured by XTT reduction assays, and the biofilm viability was analyzed with the following equation:
For biofilm eradication, mature biofilms needed to be prepared as previously described. Then, the preformed biofilm was washed without disturbing the formed biofilm and incubated with various concentrations of AMP-17 for 12 h at 37 °C. The subsequent processing of AMP-17-treated preformed biofilm was the same as the above biofilm formation.
4.8. Cell Surface Hydrophobicity Test
A cell surface hydrophobicity (CSH) assay was performed to assess the cell adhesion of
C. albicans. The logarithmic phase of
C. albicans was diluted to 1.0 × 10
7 CFU/mL in YPD medium and treated with AMP-17 at 35 °C for 24 h. The cells were harvested, resuspended using sterile PBS, and the optical density (OD) values were measured at 600 nm (
A0). Chloroform was added to the suspension, vortexed for 1 min, and settled at room temperature for 30 min. The upper aqueous phase was collected and the OD
600 (
A1) was determined. The CHS of
C. albicans cells was quantified by the hydrophobicity index using the following equation:
4.9. Cell Adhesion Assay
Efficient adhesion is the first step to attaching closely to the host surface and plays an important role in biofilm formation. In vitro cell adherence was performed as previously reported, with some modifications [
29,
54]. Briefly,
C. albians cells cultured on the exponential phase were diluted to 2 × 10
6 CFU/mL and co-incubated with AMP-17 in an RPMI medium in a 6-well flat-bottomed microplate at 37 °C for 90 min. Subsequently, the walls were washed using sterile PBS to remove the non-adherent cells and scraped using a cell scraper. Next, the suspension containing adhered cells for each group was serially diluted, and the diluted suspension (5 μL) was spot-seeded on a YPD plate, and the colonies were counted after 24 h incubation. For the inhibitory effect of AMP-17 on preformed adhesion, a
C. albicans suspension was previously cultured for 90 min until the cells adhered to the surface of the microplate, co-incubated with AMP-17 for another 90 min, and finally washed twice. Finally, the number of adhered cells was counted as per the previous description.
4.10. Yeast-to-Hypha Transition Assay
The effect of AMP-17 on the transition of yeast-to-hyphal in
C. albicans was performed according to the previous report with minor modifications [
29,
42].
C. albicans cells were diluted to 2 × 10
5 CFU/mL and co-incubated with AMP-17 in an RPMI-FBS medium in a 12-well flat-bottomed microplate at 37 °C for 2, 4, and 6 h. Images of cells were captured using an inverted microscope (IX51, Olympus, Tokyo, Japan). To test the inhibitory effect of AMP-17 on hyphal formation,
C. albicans cells were treated with AMP-17 for 2 h and it was observed whether the yeast cells sprouted. At least 200
C. albicans cells were counted to quantify the hyphal formation rate, and Image J was applied to measure the length of the hyphae. In addition, the
C. albicans filamentation assay was carried out on a solid Spider agar plate. A total of 10 μL of
C. albicans suspension (1 × 10
6 CFU/mL) was added to the plate containing AMP-17 or FLC and incubated at 37 °C for 3 days. Images of the colony edges were obtained using a microscope.
4.11. Confocal Laser Scanning Microscope (CLSM) Assays
A confocal laser scanning microscope (CLSM) was applied to visualize the C. albicans biofilm. Briefly, sterile polylysine-treated cover slips with an area of 1 cm2 were used as the carrier and placed in a 12-well plate. Following the method described above, the C. albicans biofilm was incubated for 30 min at 37 °C in the dark with 10mM PI (Sigma, St. Louis, MO, USA) and 10 mM SYTO9 (Invitrogen, USA). Next, the slips were washed, fixed with paraformaldehyde, and stuck on a sterile polylysine-treated glass slide. Finally, images were taken with an Olympus CLSM (Markham, ON, Canada) at an excitation wavelength of 488 nm for SYTO9 and 525 nm for PI. SYTO9 could move through the membrane of viable cells, resulting in the accumulation of green fluorescence, while PI stained the dead cells, emitting red fluorescence.
4.12. Scanning Electron Microscopy (SEM)
Scanning electron microscopy (SEM) was applied to evaluate the morphological surface changes of the biofilms. A sterile polylysine-treated cover glass as a biofilm carrier was placed in a 6-well plate and C. albicans biofilms were cultured according to the above method. After incubation, the samples were washed twice in PBS and then fixed with 2.5% glutaraldehyde at 4 °C overnight. Next, the fixed samples were washed with PBS, followed by dehydration using a tertiary butyl alcohol series (50, 75, 95, and 100%) for 10 min. Subsequently, the samples were dried in a high vacuum evaporator and coated with a thin layer of gold-palladium. Finally, the prepared sample was placed on the Hitachi H-7650 Scanning Electron Microscope (Tokyo, Japan) for observation and to obtain images.
4.13. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
qRT-PCR assays were performed to verify the accuracy of RNA-seq. Biofilm samples were collected in a 1.5 mL EP tub (Axygen, Sigma-Aldrich, St. Louis, MO, USA) of free-RNase with a cell spatula. Total RNA was extracted using Trizol and quantified using a NanoDrop Spectrophotometer (ND-200, Thermo Scientific, Waltham, MA, USA). Then, the RNA was reversed to cDNA in a 20 μL reaction mixture using a PrimeScript RT reagent Kit with gDNA Remover (Takara, Maebashi, Japan), with procedures as follows: gDNA removal reaction at 42 °C for 2 min, followed by a reverse transcriptional reaction at 42 °C for 2 min, and 85 °C for 5 sec to generate cDNA. Primers of
C. albicans for qRT-PCR were designed using Primer Premier 5.0 and synthesized by Sangon Biotech. According to the SYBR Premix Ex Taq TM Kit (Takara, Japan) protocol, the qRT-PCR mixture (20 μL) was freshly prepared, containing SYBR green fluorescent dyes, PCR reverse primer, cDNA, and RNase-free water. The qRT-PCR process was performed using an ABI7300 fluorescence quantitative PCR system with the following cycles: 95 °C for 30 s for pre-denaturation, along with 95 °C for 5 s and 60 °C for 30 s for a total of 40 cycles. A 20 μL reaction system was used in the qRCR system. All data were normalized to the internal reference gene ACT1. The relative quantification of gene expression was computed using the 2
−ΔΔCt method, in which ΔCt = Ct target gene-Ct internal reference genes. The primer sequences are provided in
Table S2.
4.14. RNA-Seq
4.14.1. RNA Isolation, Library Construction, and Sequencing
RNA-Seq was performed using an Illumine HisSeq System by BGI Tech Company (Shenzhen, China). For the inhibitory effect of AMP-17 on biofilm formation in RNA-seq analysis, the exponential C. albicans (2 × 106 CFU/mL) was co-incubated with 32 μg/mL AMP-17 using RPMI medium in a 6-well flat-bottomed plate at 37 °C for 12 h, harvested in a 1.5 mL EP tub, and stored at −70 °C after liquid nitrogen freezing. In order to test the eradication effect of AMP-17 on preformed biofilm, the exponential C. albicans (2 × 106 CFU/mL) was incubated in RPMI medium in a 6-well flat-bottomed plate at 37 °C for 24 h to grow mature before being exposed to 128 μg/mL AMP-17 for 12 h. Subsequently, biofilm samples were collected as in the previous step. Meanwhile, untreated biofilm or untreated preformed biofilm were set as controls and each group was prepared with three parallel replicates. Later, all the samples were sent to BGI Corporation for further RNA-seq detection and analysis via the BGISEQ-500 sequencer. The total RNA extraction, quantification, qualification, cDNA library construction, and transcriptome sequencing were performed by Beijing Genomics Institute (BGI) Co., Ltd. (Shenzhen, China). Finally, the platform converted the sequenced image signals into textual signals and stored them as raw data in the FASTQ format.
4.14.2. RNA-Seq Data Analysis
The raw reads generated from sequencing were cleaned by removing adaptor-polluted reads, reads with unknown sequence “N” accounting for more than 50%, and low-quality reads (more than 20% of bases with a Phred threshold score < 15). The clean reads were mapped to the
C. albicans SC5314 genome (
https://www.ncbi.nlm.nih.gov/gene/?term=candida+albicans+sc5314 accessed on 12 March 2021) using the Hierarchical indexing for spiced alignment of transcripts (HISAT) v2.1.0 software [
55]. Reads per kilobase million mapped reads (RPKM) were used to access gene expression levels to eliminate the bias caused by the effects of sequencing depth and gene length. Differentially expressed genes (DEGs) were identified with a threshold of Q-value (adjust
p-value) ≤ 0.001 and log2(fold change) ≥ 1. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional classifications were carried out using Blast2GO and KOBAS for annotation of the functions of DEGs (
http://kobas.cbi.pku.edu.cn accessed on 18 May 2022). The
p-value was corrected by the false discovery rate (FDR). The pathway with FDR < 0.01 was considered to be significantly enriched. Data mining and figure presentation processes were analyzed by BGI’s in-house customized data mining system called Dr. Tom (
http://biosys.bgi.com accessed on 22 April 2021). The gene interaction network including selected DEGs and neighbors was created by STRING (version 10) and visualized by Cytoscape (version 3.6.1). Default parameters (score value above two and at least four nodes) were used as the cutoff criteria for network module screening. The enrichment analysis of clusters was qualified with a threshold of
p < 0.05, FDR-adjusted. Finally, DEGs owing to specific functions were selected for qRT-PCR analysis to validate the data from RNA-Seq.
4.15. Statistical Analysis
Figures depicting the differential gene expression between AMP-17 treated and untreated groups were created using the DESeq2 package in RStudio. Gene interaction networks were created using the ClueGo application in Cytoscape (
http://cytoscape.org, accessed on 13 December 2021).
All other graphs and analyses were carried out in GraphPad Prism (version 7, La Jolla, CA, USA). Every experiment was independently performed at least three times, and the data were expressed as the mean ± standard deviation (SD) of three independent experiments. Differences between experimental groups were assessed for significance using one-way ANOVA. The * p < 0.05, ** p < 0.01, and *** p < 0.001 levels were considered to indicate statistical significance.