Genomic Analysis of a Hybrid Enteroaggregative Hemorrhagic Escherichia coli O181:H4 Strain Causing Colitis with Hemolytic-Uremic Syndrome
Abstract
:1. Introduction
2. Results
2.1. Bacterial Strain, Pathotype Identification, and Susceptibility to Antimicrobials
2.2. Genomic Characteristics of EAHEC Strain SCPM-O-B-9427
2.3. Genotypic Profiling of the EAHEC Strain SCPM-O-B-9427
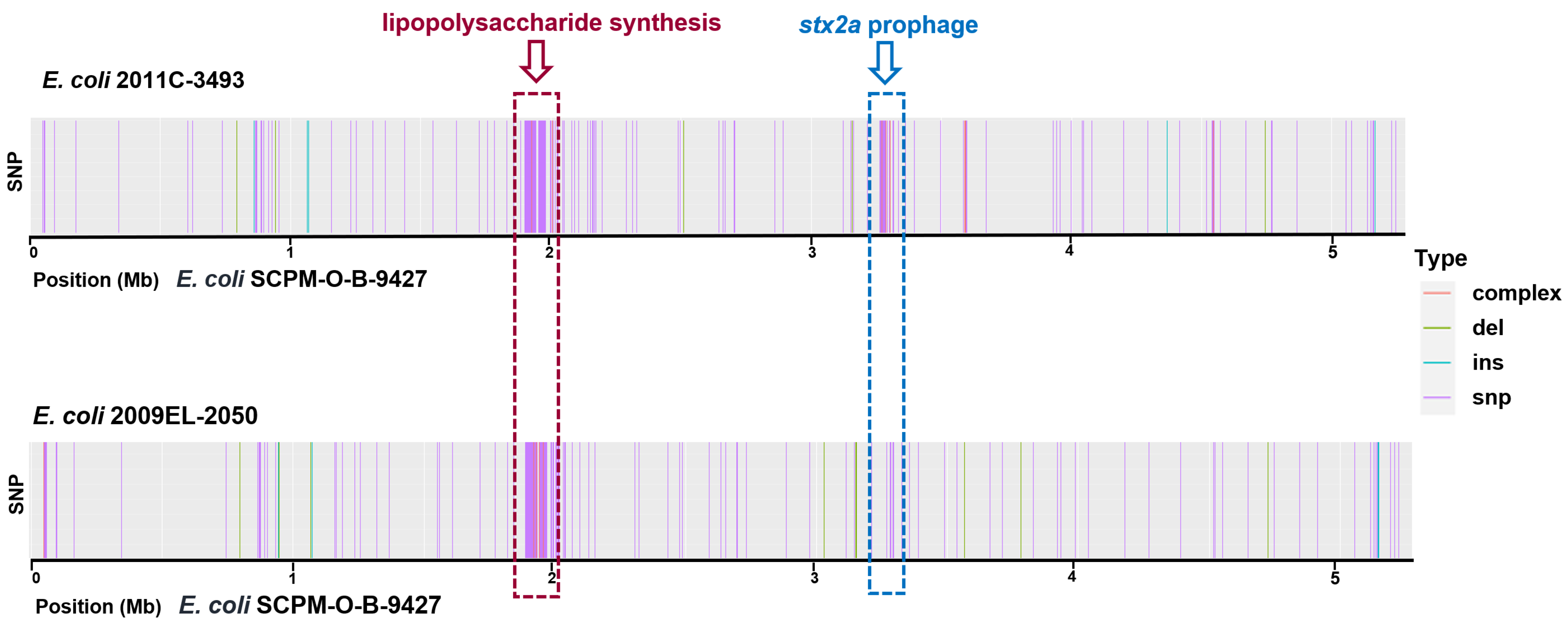
2.4. Chromosomal SNPs in the EAHEC Strains SCPM-O-B-9427, 2011C-3493, and 2009EL-2050
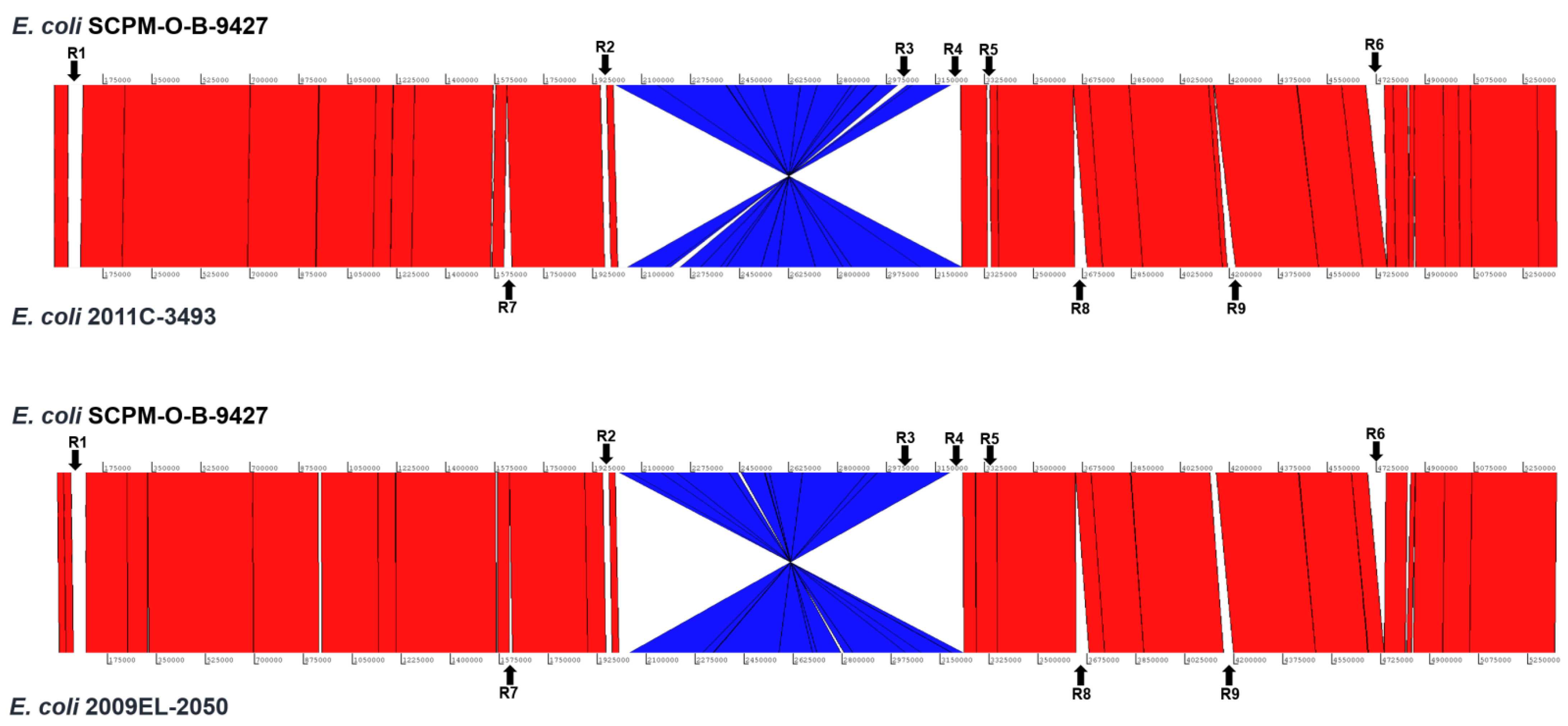
2.5. Chromosomal Structural Comparison of the Strains SCPM-O-B-9427, 2011C-3493, and 2009EL-2050
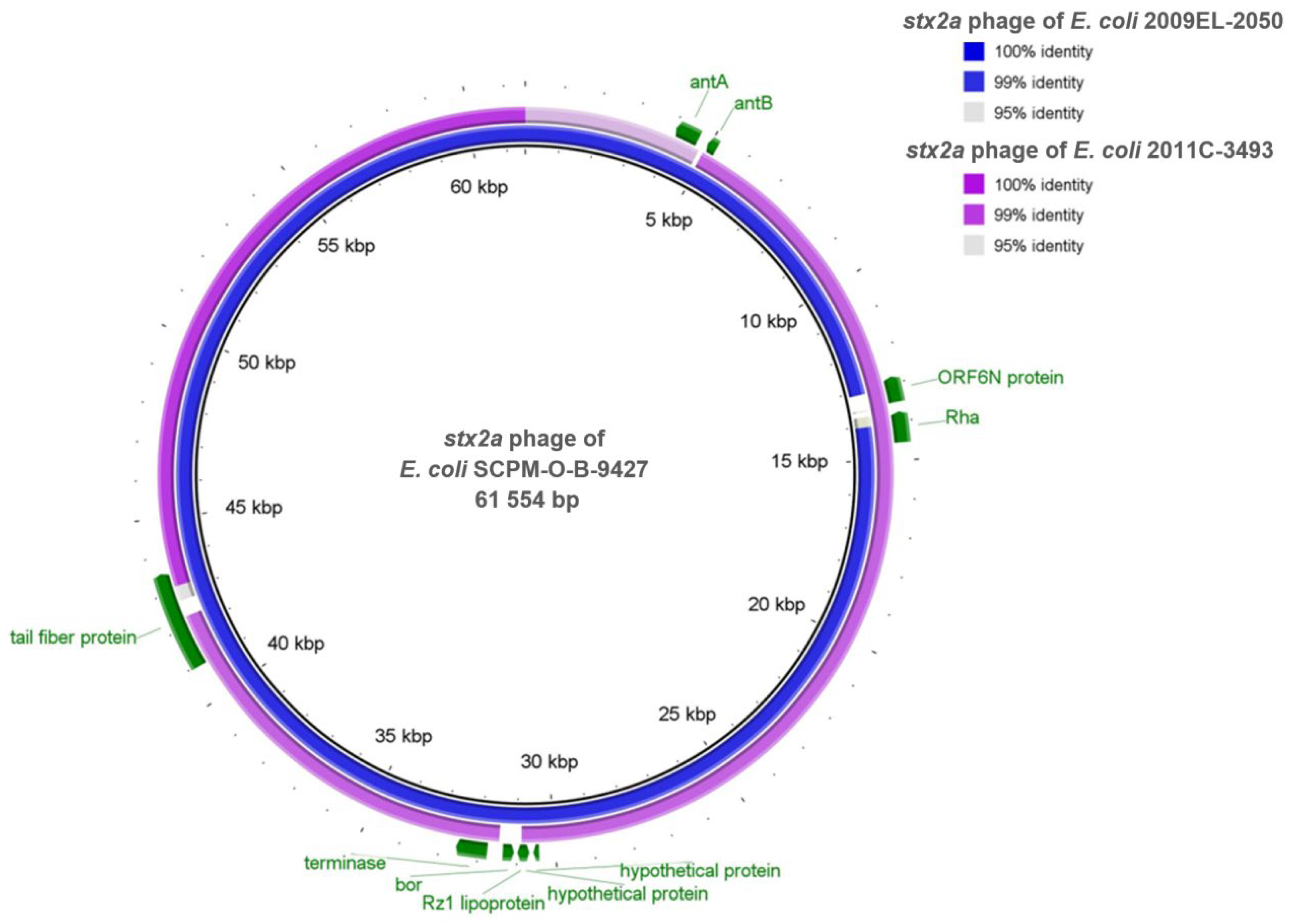
2.6. Comparison of the stx2a Prophages Located in the Genomes of the Strains SCPM-O-B-9427, 2011C-3493, and 2009EL-2050
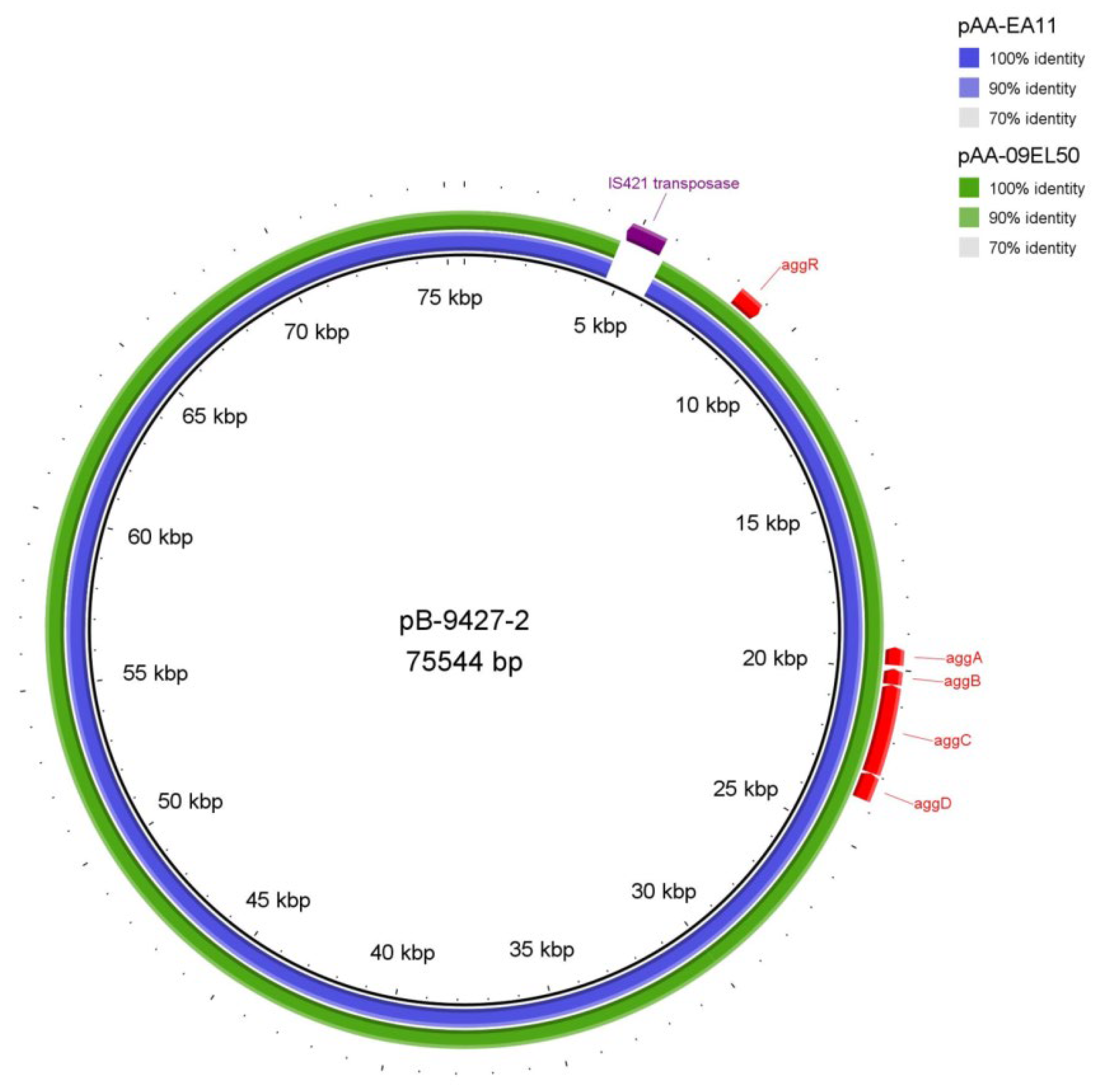
2.7. The Plasmids of the Strain SCPM-O-B-9427
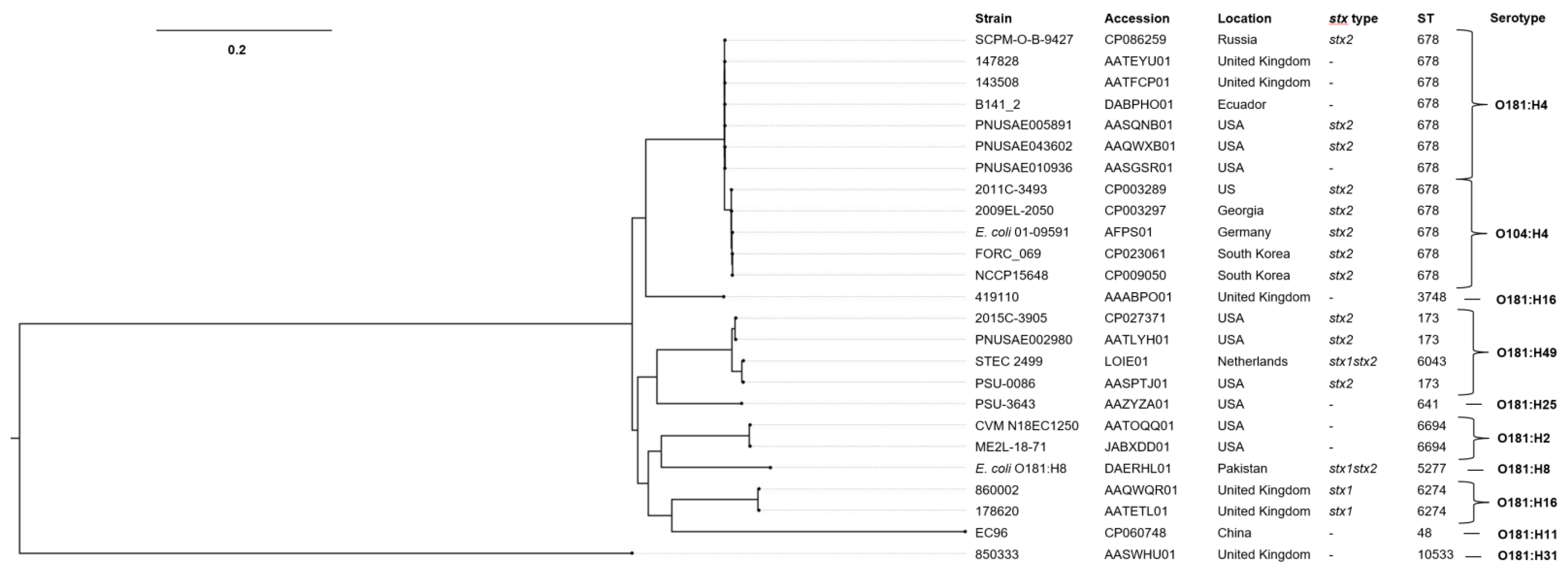
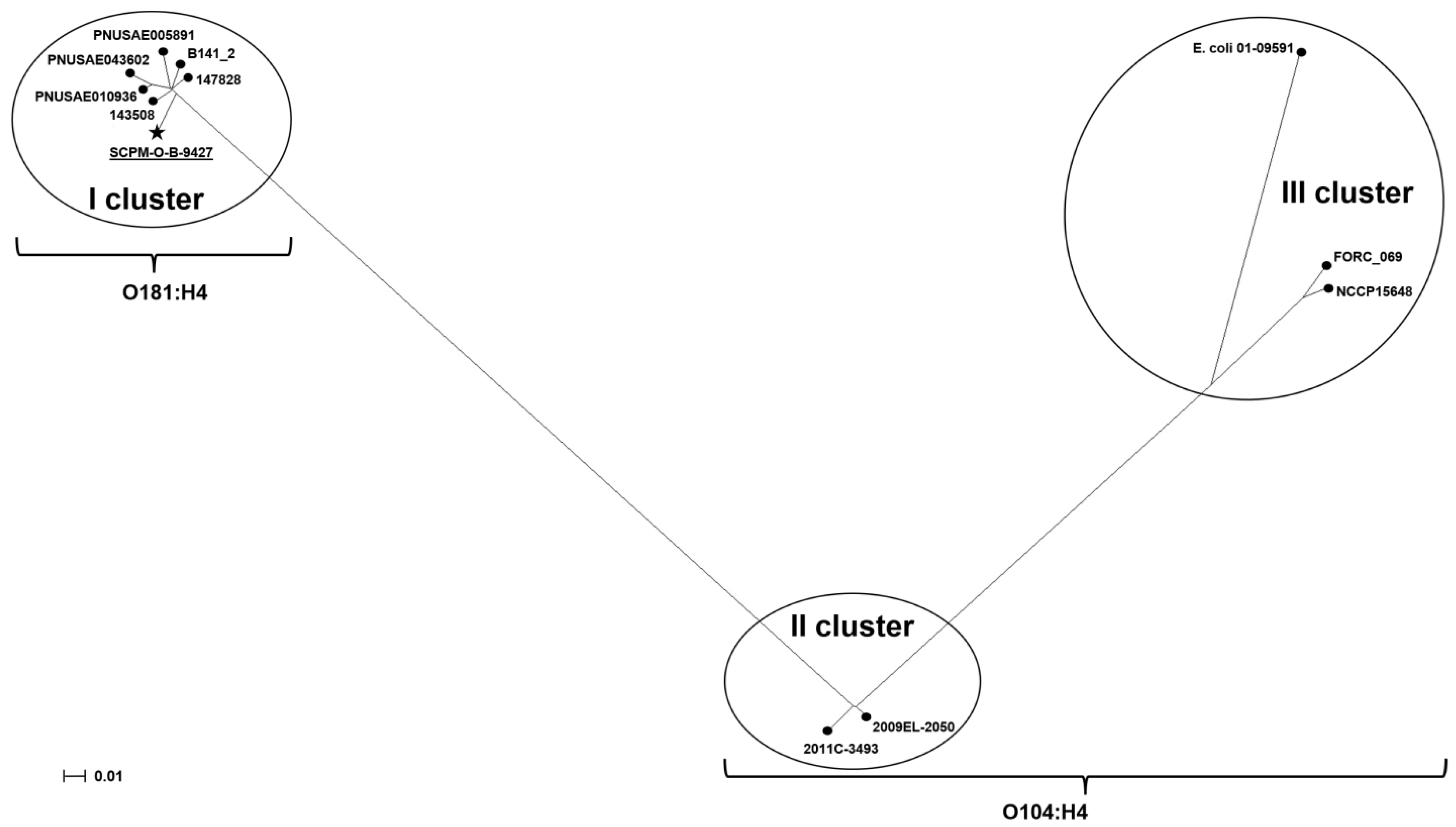
2.8. Phylogenetic Analysis for E. coli O181 and O104:H4
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains Used in This Study
4.2. Strain Isolation, Growing, Identification, and Susceptibility Testing
4.3. DNA Isolation and Pathotype and Phylogroup Identification
4.4. Whole Genome Sequencing, Assembly and Annotation
4.5. Whole Genome Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss, R.; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PLoS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Clements, A.; Young, J.C.; Constantinou, N.; Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3, 71–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyong, E.C.; Zaia, S.R.; Allué-Guardia, A.; Rodriguez, A.L.; Irion-Byrd, Z.; Koenig, S.; Feng, P.; Bono, J.L.; Eppinger, M. Pathogenomes of Atypical Non-shigatoxigenic Escherichia coli NSF/SF O157:H7/NM: Comprehensive Phylogenomic Analysis Using Closed Genomes. Front. Microbiol. 2020, 11, 619. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, R.R.; Henderson, I.R. The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 2012, 12, 214–226. [Google Scholar] [CrossRef]
- Dixit, P.D.; Pang, T.Y.; Studier, F.W.; Maslov, S. Recombinant transfer in the basic genome of Escherichia coli. Proc. Natl. Acad. Sci. USA 2015, 112, 9070–9075. [Google Scholar] [CrossRef] [Green Version]
- Desvaux, M.; Dalmasso, G.; Beyrouthy, R.; Barnich, N.; Delmas, J.; Bonnet, R. Pathogenicity Factors of Genomic Islands in Intestinal and Extraintestinal Escherichia coli. Front. Microbiol. 2020, 11, 2065. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Pakbin, B.; Brück, W.M.; Rossen, J. Virulence Factors of Enteric Pathogenic Escherichia coli: A Review. Int. J. Mol. Sci. 2021, 22, 9922. [Google Scholar] [CrossRef]
- Jesser, K.J.; Levy, K. Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. Dis. 2020, 33, 372–380. [Google Scholar] [CrossRef]
- Bolukaoto, J.Y.; Singh, A.; Alfinete, N.; Barnard, T.G. Occurrence of Hybrid Diarrhoeagenic Escherichia coli Associated with Multidrug Resistance in Environmental Water, Johannesburg, South Africa. Microorganisms 2021, 9, 2163. [Google Scholar] [CrossRef]
- Díaz-Jiménez, D.; García-Meniño, I.; Herrera, A.; García, V.; López-Beceiro, A.M.; Alonso, M.P.; Blanco, J.; Mora, A. Genomic Characterization of Escherichia coli Isolates Belonging to a New Hybrid aEPEC/ExPEC Pathotype O153:H10-A-ST10 eae-beta1 Occurred in Meat, Poultry, Wildlife and Human Diarrheagenic Samples. Antibiotics 2020, 9, 192. [Google Scholar] [CrossRef]
- Valiatti, T.B.; Santos, F.F.; Santos, A.; Nascimento, J.; Silva, R.M.; Carvalho, E.; Sinigaglia, R.; Gomes, T. Genetic and Virulence Characteristics of a Hybrid Atypical Enteropathogenic and Uropathogenic Escherichia coli (aEPEC/UPEC) Strain. Front. Cell. Infect. Microbiol. 2020, 10, 492. [Google Scholar] [CrossRef]
- Santos, A.; Santos, F.F.; Silva, R.M.; Gomes, T. Diversity of Hybrid- and Hetero-Pathogenic Escherichia coli and Their Potential Implication in More Severe Diseases. Front. Cell. Infect. Microbiol. 2020, 10, 339. [Google Scholar] [CrossRef]
- Rasko, D.A.; Webster, D.R.; Sahl, J.W.; Bashir, A.; Boisen, N.; Scheutz, F.; Paxinos, E.E.; Sebra, R.; Chin, C.S.; Iliopoulos, D.; et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 2011, 365, 709–717. [Google Scholar] [CrossRef] [Green Version]
- Tietze, E.; Dabrowski, P.W.; Prager, R.; Radonic, A.; Fruth, A.; Aura, P.; Nitsche, A.; Mielke, M.; Flieger, A. Comparative genomic analysis of two novel sporadic Shiga toxin-producing Escherichia coli O104:H4 strains isolated 2011 in Germany. PLoS ONE 2015, 10, e0122074. [Google Scholar] [CrossRef] [Green Version]
- Wald, M.; Rieck, T.; Nachtnebel, M.; Greute’laers, B.; an der Heiden, M.; Altmann, D.; Hellenbrand, W.; Faber, M.; Frank, C.; Schweickert, B.; et al. Enhanced surveillance during a large outbreak of bloody diarrhoea and haemolytic uraemic syndrome caused by Shiga toxin/verotoxin-producing Escherichia coli in Germany, May to June 2011. Euro Surveill. 2011, 16, 19893. [Google Scholar]
- Chokoshvili, O.; Lomashvili, K.; Malakmadze, N.; Geleishvil, M.; Brant, J.; Imnadze, P.; Chitadze, N.; Tevzadze, L.; Chanturia, G.; Tevdoradze, T.; et al. Investigation of an outbreak of bloody diarrhea complicated with hemolytic uremic syndrome. J. Epidemiol. Glob. Health 2014, 4, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.A.; Awosika, J.; Baldwin, C.; Bishop-Lilly, K.A.; Biswas, B.; Broomall, S.; Chain, P.S.; Chertkov, O.; Chokoshvili, O.; Coyne, S.; et al. Threat Characterization Consortium. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including shiga toxin encoding phage stx2. PLoS ONE 2012, 7, e48228. [Google Scholar] [CrossRef] [Green Version]
- Nowrouzian, F.L.; Wold, A.E.; Adlerberth, I. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 2005, 191, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Cheasty, T.; Woodward, D.; Smith, H.R. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include Verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS 2004, 112, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Ballem, A.; Gonçalves, S.; Garcia-Meniño, I.; Flament-Simon, S.C.; Blanco, J.E.; Fernandes, C.; Saavedra, M.J.; Pinto, C.; Oliveira, H.; Blanco, J.; et al. Prevalence and serotypes of Shiga toxin-producing Escherichia coli (STEC) in dairy cattle from Northern Portugal. PLoS ONE 2020, 15, e0244713. [Google Scholar] [CrossRef] [PubMed]
- Eklund, M.; Scheutz, F.; Siitonen, A. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: Serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J. Clin. Microbiol. 2001, 39, 2829–2834. [Google Scholar] [CrossRef] [Green Version]
- García-Aljaro, C.; Muniesa, M.; Blanco, J.E.; Blanco, M.; Blanco, J.; Jofre, J.; Blanch, A.R. Characterization of Shiga toxin-producing Escherichia coli isolated from aquatic environments. FEMS Microbiol. Lett. 2005, 246, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Ori, E.L.; Takagi, E.H.; Andrade, T.S.; Miguel, B.T.; Cergole-Novella, M.C.; Guth, B.; Hernandes, R.T.; Dias, R.; Pinheiro, S.; Camargo, C.H.; et al. Diarrhoeagenic Escherichia coli and Escherichia albertii in Brazil: Pathotypes and serotypes over a 6-year period of surveillance. Epidemiol. Infect. 2018, 147, e10. [Google Scholar] [CrossRef] [Green Version]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [Green Version]
- Berger, P.; Knödler, M.; Förstner, K.U.; Berger, M.; Bertling, C.; Sharma, C.M.; Vogel, J.; Karch, H.; Dobrindt, U.; Mellmann, A. The primary transcriptome of the Escherichia coli O104:H4 pAA plasmid and novel insights into its virulence gene expression and regulation. Sci. Rep. 2016, 6, 35307. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Garcia, F. Escherichia coli O104:H4 pathogenesis: An enteroaggregative E. coli/Shiga toxin-Producing E. coli explosive cocktail of high virulence. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef] [Green Version]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Wilson, K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001, 56, 2–4. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies. Available online: https://nanoporetech.com/ (accessed on 12 April 2021).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic. Acids. Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [Green Version]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Micobiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic. Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Kevin Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Genomics %G~C Content Calculator—Science Buddies. Available online: https://www.sciencebuddies.org/science-fair-projects/references/genomics-g-c-content-calculator (accessed on 9 November 2011).
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis Comparison Tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Wombac. Available online: https://github.com/tseemann/wombac (accessed on 15 November 2021).
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 November 2021).
- Snippy. Available online: https://github.com/tseemann/snippy (accessed on 15 November 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org (accessed on 15 February 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 15 February 2022).
| Antimicrobials | MIC, mg/L | Interpretation |
|---|---|---|
| Ampicillin | 16 | R 1 |
| Amoxicillin/clavulanic acid | 4 | S 2 |
| Cefotaxime | ≤1 | S |
| Ceftazidime | ≤1 | S |
| Cefoperazone/sulbactam | ≤8 | S |
| Cefepime | ≤1 | S |
| Aztreonam | ≤1 | S |
| Imipenem | ≤1 | S |
| Meropenem | ≤0.25 | S |
| Amikacin | ≤2 | S |
| Gentamicin | ≤1 | S |
| Netilmicin | 2 | S |
| Ciprofloxacin | 1 | R |
| Fosfomycin | ≤16 | S |
| Nitrofurantoin | ≤16 | S |
| Trimethoprim/sulfamethoxazole | ≤20 | S |
| Feature/Strain | SCPM-O-B-9427 | 2011C-3493 | 2009EL-2050 |
|---|---|---|---|
| GenBank chromosome | CP086259 | CP003289 | CP003297 |
| Region | Russian Federation | Germany | Georgia |
| Isolation year | 2018 | 2011 | 2009 |
| Serotype | O181:H4 | O104:H4 | O104:H4 |
| ST | ST678 | ST678 | ST678 |
| stx subtype | 2a | 2a | 2a |
| aggDCBA cluster | + | + | + |
| Chromosome size, bp | 5,268,110 | 5,273,097 | 5,253,138 |
| GC content of chromosome, % | 50.7 | 50.7 | 50.7 |
| Genes (total) | 5408 | 5314 | 5325 |
| CDS (total) | 5283 | 5191 | 5197 |
| Genes (RNA) | 125 | 123 | 128 |
| rRNAs | 8, 7, 7 (5S, 16S, 23S) | 8, 7, 7 (5S, 16S, 23S) | 8, 7, 7 (5S, 16S, 23S) |
| tRNAs | 95 | 92 | 97 |
| Pseudogenes (total) | 297 | 227 | 237 |
| Plasmids | - | - | p09EL50 (109,274 bp) |
| - | pESBL-EA11 (88,544 bp) | - | |
| pB-9427-1 (83,340 bp) | - | - | |
| pB-9427-2 (75,544 bp) | pAA-EA11 (74,213 bp) | pAA-09EL50 (74,213 bp) | |
| pB-9427-3 (51,013 bp) | - | ||
| pB-9427-4 (7939 bp) | - | ||
| pB-9427-5 (6728 bp) | - | ||
| - | pG-EA11 (1549 bp) | pG-09EL50 (1549 bp) |
| Region | Sequence, Kbp/GC Content. % | Note | ||
|---|---|---|---|---|
| SCPM-O-B-9427 | 2009EL-2050 | 2011C-3493 | ||
| R1 | 52.0/47.5 | 45.0/56.5 | 44.5/56.8 | Part of R1 are incomplete prophages and in the strain SCPM-O-B-9427 are related to Enterobacteria phage BP-4795 (NC_004813) and in other strains to Enterobacteria phage P1 (NC_005856). |
| R2 | 11.7/31.4 | 9.6/30.7 | 9.6/30.7 | The region contains the genes involved in lipopolysaccharide synthesis of O104 (the strains 2011C-3493 and 2009EL-2050) and O181 (the strain SCPM-O-B-9427) antigen gene clusters. |
| R3 | 33.9/52.6 | 33.9/52.6 | 32.6/53.0 | The region of the strain 2011C-3493 is related to Bacteriophage sp. isolate ctdA53 (BK038108) and the regions of the strains SCPM-O-B-9427 and 2009EL-2050 to Escherichia phage Lambda_ev017 (NC_049948). |
| R4 | 21.6/51.4 | 20.1/52.3 | 20.1/52.3 | The region contains the chromosomal genes, IS elements, and genes coding the hypothetical proteins. |
| R5 | 61.6/50.2 | 60.6/50.2 | 60.9/50.2 | The region is the stx2a prophage. |
| R6 | 65.6/49.2 | - | - | The region contains the chromosome genes, part of them coding the type II secretion system. |
| R7 | - | 11.0/43.3 | 27.8/45.7 | The region contains the genes related to Enterobacteria phage Sf6 (NC_005344). |
| R8 | - | 44.6/50.5 | 44.6/50.5 | The region contains the genes related to intact Enterobacteria phage lambda (NC_001416). |
| R9 | - | 14.8/53.1 | 30.2/49.0 | The region coding the chromosome genes that are partly missed in the strain 2009EL-2050. |
| Plasmid | Accession Number | GC Contain (%) | Homolog | Query Cover (%) | Percentage of Identity (%) | Note |
|---|---|---|---|---|---|---|
| pB-9427-1 | CP086260 | 53.3 | Escherichia coli FDAARGOS_1300 plasmid unnamed3 (CP069999)—82,270 bp | 98 | 99.53 | F plasmid |
| pB-9427-2 | CP086261 | 47.2 | Escherichia coli O104:H4 FDAARGOS_348 plasmid unnamed2 (CP022087)—75,559 bp | 100 | 99.91 | pAA plasmid |
| pB-9427-3 | CP086262 | 49.6 | Escherichia coli SJ7 plasmid pSJ7-2 (CP051658)—53,543 bp | 94 | 99.23 | Contains genes encoding the type IV conjugative transfer system |
| pB-9427-4 | CP086263 | 41.6 | Escherichia coli RHBSTW-00895 plasmid pRHBSTW-00895_4 (CP056266)—7939 bp | 100 | 99.67 | Contains genes related to mobilization proteins |
| pB-9427-5 | CP086264 | 51.3 | Shigella sonnei 500867 plasmid pSSE2 (KP979588)—6728 bp | 100 | 100 | Contains plasmid-borne E-type colicin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kislichkina, A.A.; Kartsev, N.N.; Skryabin, Y.P.; Sizova, A.A.; Kanashenko, M.E.; Teymurazov, M.G.; Kuzina, E.S.; Bogun, A.G.; Fursova, N.K.; Svetoch, E.A.; et al. Genomic Analysis of a Hybrid Enteroaggregative Hemorrhagic Escherichia coli O181:H4 Strain Causing Colitis with Hemolytic-Uremic Syndrome. Antibiotics 2022, 11, 1416. https://doi.org/10.3390/antibiotics11101416
Kislichkina AA, Kartsev NN, Skryabin YP, Sizova AA, Kanashenko ME, Teymurazov MG, Kuzina ES, Bogun AG, Fursova NK, Svetoch EA, et al. Genomic Analysis of a Hybrid Enteroaggregative Hemorrhagic Escherichia coli O181:H4 Strain Causing Colitis with Hemolytic-Uremic Syndrome. Antibiotics. 2022; 11(10):1416. https://doi.org/10.3390/antibiotics11101416
Chicago/Turabian StyleKislichkina, Angelina A., Nikolay N. Kartsev, Yury P. Skryabin, Angelika A. Sizova, Maria E. Kanashenko, Marat G. Teymurazov, Ekaterina S. Kuzina, Alexander G. Bogun, Nadezhda K. Fursova, Edward A. Svetoch, and et al. 2022. "Genomic Analysis of a Hybrid Enteroaggregative Hemorrhagic Escherichia coli O181:H4 Strain Causing Colitis with Hemolytic-Uremic Syndrome" Antibiotics 11, no. 10: 1416. https://doi.org/10.3390/antibiotics11101416
APA StyleKislichkina, A. A., Kartsev, N. N., Skryabin, Y. P., Sizova, A. A., Kanashenko, M. E., Teymurazov, M. G., Kuzina, E. S., Bogun, A. G., Fursova, N. K., Svetoch, E. A., & Dyatlov, I. A. (2022). Genomic Analysis of a Hybrid Enteroaggregative Hemorrhagic Escherichia coli O181:H4 Strain Causing Colitis with Hemolytic-Uremic Syndrome. Antibiotics, 11(10), 1416. https://doi.org/10.3390/antibiotics11101416






