Insights into the Vibrio Genus: A One Health Perspective from Host Adaptability and Antibiotic Resistance to In Silico Identification of Drug Targets
Abstract
1. Introduction
2. Results
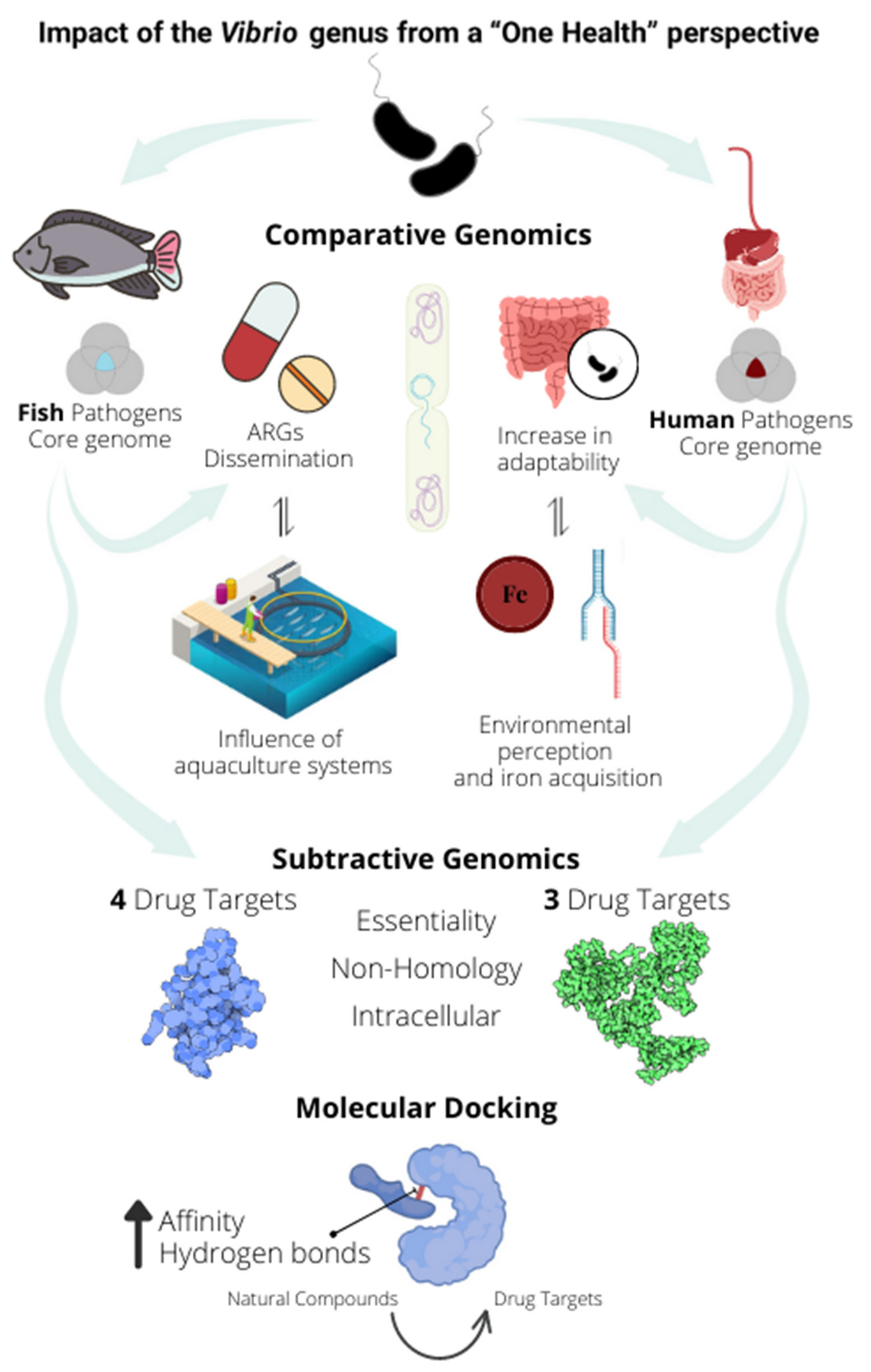
2.1. Human Pathogens Have More Genes Associated with Host Adaptation Than Fish Pathogens
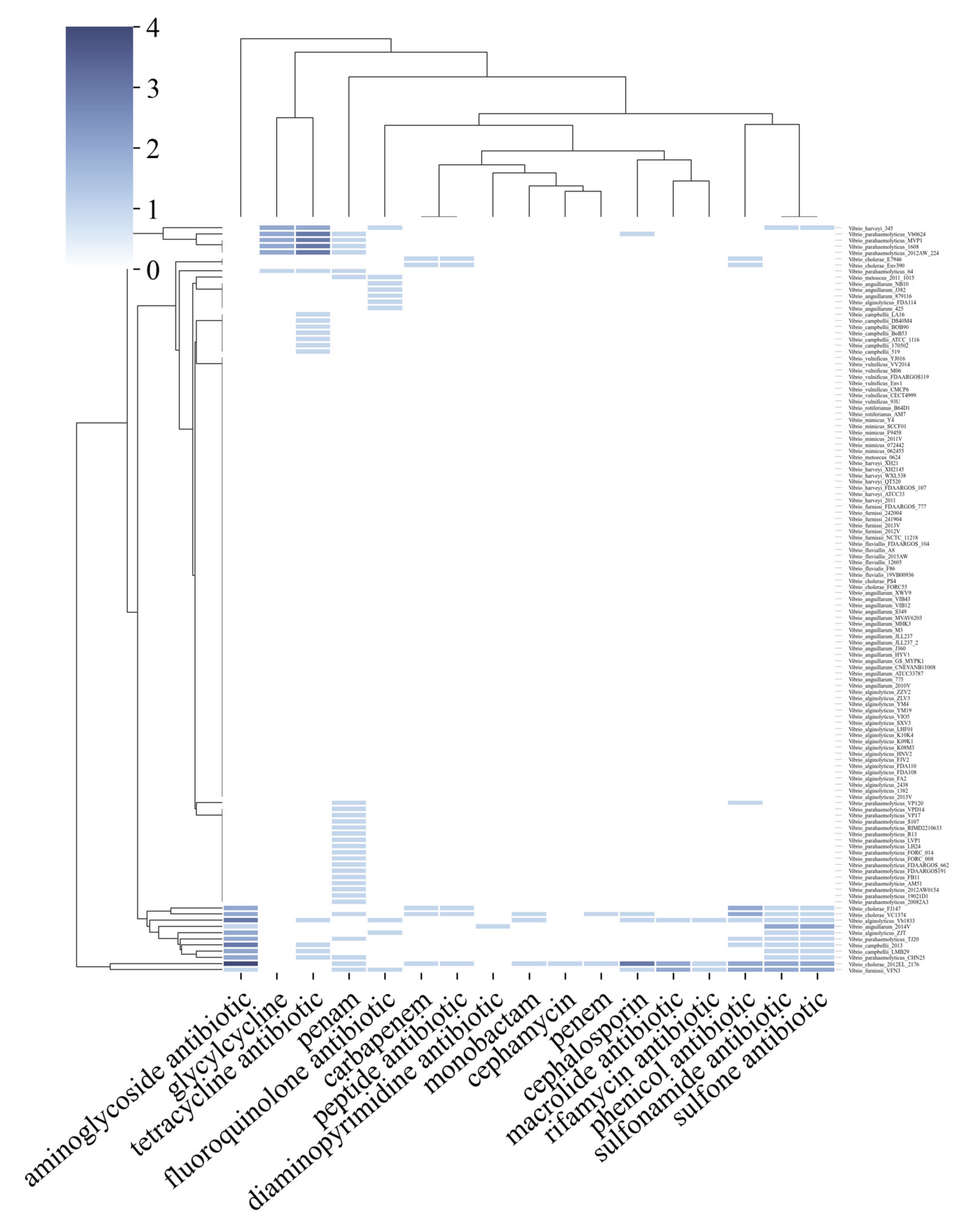
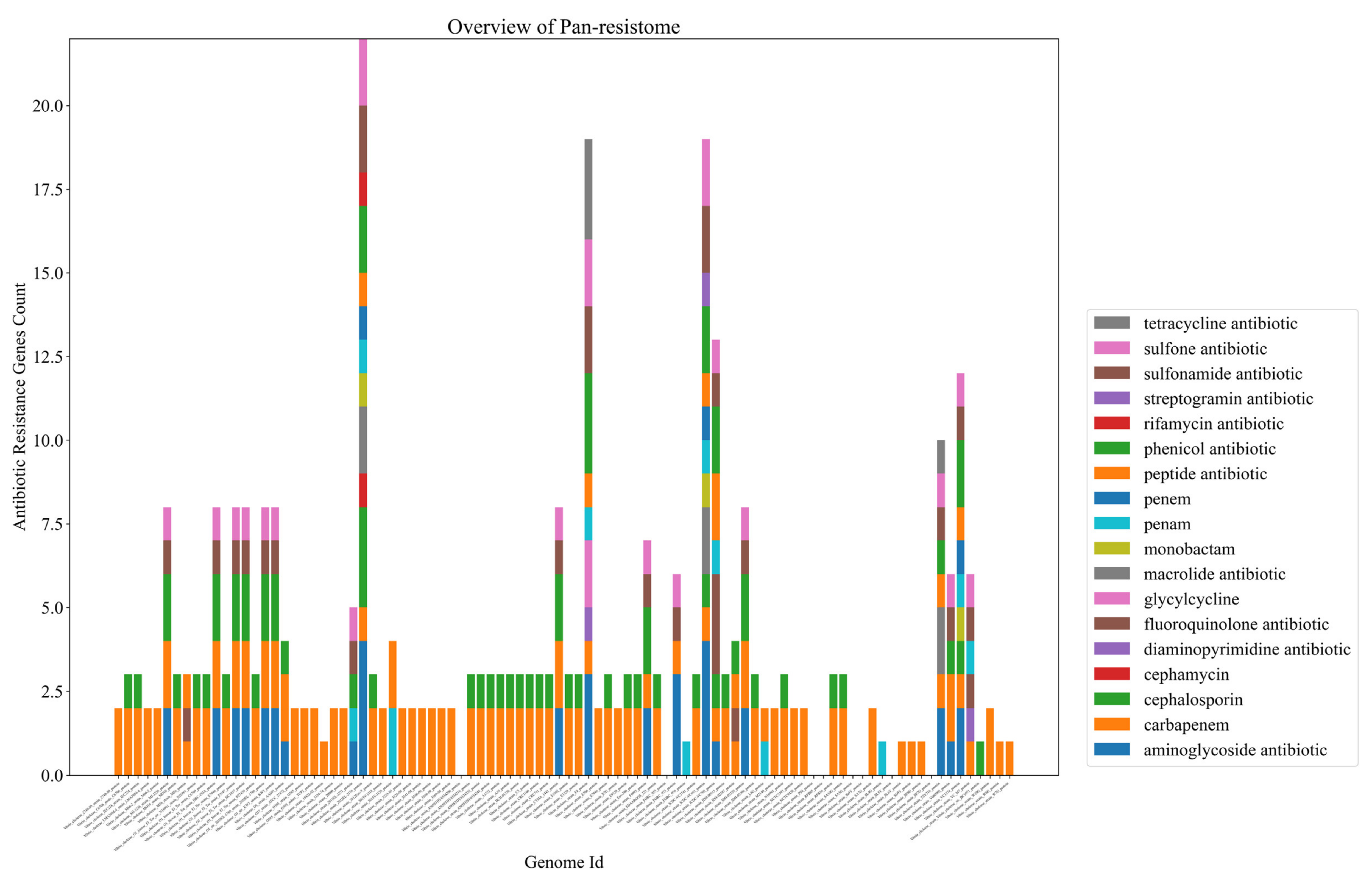
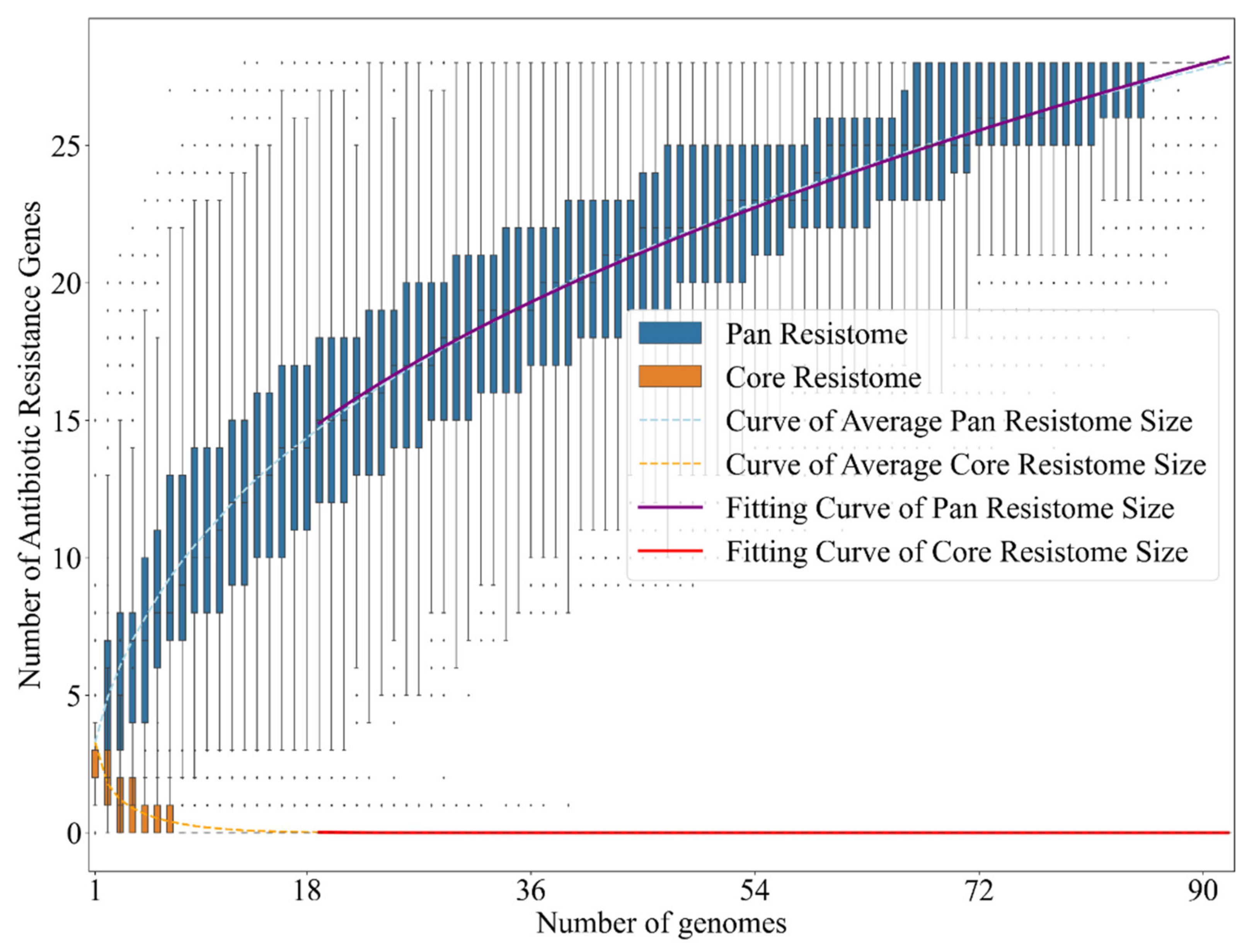
2.2. Bacteria of the Vibrio Genus Associated with Fish Infection Have a Much Higher Amount of Resistance-Associated Genes
2.3. Subtractive Genomics Reveals Seven Potential Drug Targets to Fight against Bacteria of the Vibrio Genus
3. Discussion
4. Materials and Methods
4.1. Data Acquisition
4.2. Adaptation and Resistance
4.3. Drug Target Identification
4.4. Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brumfield, K.D.; Usmani, M.; Chen, K.M.; Gangwar, M.; Jutla, A.S.; Huq, A.; Colwell, R.R. Environmental Parameters Associated with Incidence and Transmission of Pathogenic Vibrio spp. Environ. Microbiol. 2021, 23, 7314–7340. [Google Scholar] [CrossRef] [PubMed]
- Vezzulli, L.; Pezzati, E.; Brettar, I.; Höfle, M.; Pruzzo, C. Effects of Global Warming on Vibrio Ecology. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.D.; Lemos, L.S.; Roges, E.M.; de Moura, J.F.; Tavares, D.C.; Matias, C.A.R.; Rodrigues, D.P.; Siciliano, S. A Comprehensive Survey of Aeromonas Sp. and Vibrio Sp. in Seabirds from Southeastern Brazil: Outcomes for Public Health. J. Appl. Microbiol. 2018, 124, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Canesi, L.; Oyanedel, D.; Travers, M.A.; Charrière, G.M.; Pruzzo, C.; Vezzulli, L. Vibrio–Bivalve Interactions in Health and Disease. Environ. Microbiol. 2020, 22, 4323–4341. [Google Scholar] [CrossRef]
- Guardiola-Avila, I.; Martínez-Vázquez, V.; Juárez-Rendón, K.; Alvarez-Ainza, M.; Paz-González, A.; Rivera, G. Prevalence and Virulence of Vibrio Species Isolated from Raw Shrimp from Retail Markets in Reynosa, Mexico. Lett. Appl. Microbiol. 2020, 71, 280–286. [Google Scholar] [CrossRef]
- Montánchez, I.; Kaberdin, V.R. Vibrio harveyi: A Brief Survey of General Characteristics and Recent Epidemiological Traits Associated with Climate Change. Mar. Environ. Res. 2020, 154, 104850. [Google Scholar] [CrossRef]
- Zhang, X.H.; He, X.; Austin, B. Vibrio harveyi: A Serious Pathogen of Fish and Invertebrates in Mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef]
- Hundenborn, J.; Thurig, S.; Kommerell, M.; Haag, H.; Nolte, O. Severe Wound Infection with Photobacterium damselae ssp. damselae and Vibrio harveyi, Following a Laceration Injury in Marine Environment: A Case Report and Review of the Literature. Case Rep. Med. 2013, 2013, 610632. [Google Scholar] [CrossRef]
- Pavia, A.T.; Bryan, J.A.; Maher, K.L.; Roderick Hester, T.; Farmer, J.J. Brief Reports Vibrio carchariae Infection after a Shark Bite. Available online: https://www.acpjournals.org/doi/abs/10.7326/0003-4819-111-1-85 (accessed on 18 May 2022).
- Brehm, T.T.; Dupke, S.; Hauk, G.; Fickenscher, H.; Rohde, H.; Berneking, L. Non-Cholera Vibrio Species—Currently Still Rare but Growing Danger of Infection in the North Sea and the Baltic Sea. Internist 2021, 62, 876–886. [Google Scholar] [CrossRef]
- Newton, A.; Kendall, M.; Vugia, D.J.; Henao, O.L.; Mahon, B.E. Increasing Rates of Vibriosis in the United States, 1996–2010: Review of Surveillance Data from 2 Systems. Clin. Infect. Dis. 2012, 54 (Suppl. S5), S391–S395. [Google Scholar] [CrossRef]
- Jacobs Slifka, K.M.; Newton, A.E.; Mahon, B.E. Vibrio alginolyticus Infections in the USA, 1988–2012. Epidemiol. Infect. 2017, 145, 1491–1499. [Google Scholar] [CrossRef]
- Hwang, S.; Kim, Y.; Jung, H.; Chang, H.H.; Kim, S.J.; Park, H.K.; Lee, J.M.; Kim, H.I.; Kim, S.W. A Fatal Case of Bacteremia Caused by Vibrio cholerae Non-O1/O139. Infect. Chemother. 2021, 53, 384–390. [Google Scholar] [CrossRef]
- Yang, A.; Yassin, M.; Phan, T. Vibrio mimicus Wound Infection in a Burn Patient. Radiol. Case Rep. 2021, 16, 1348–1351. [Google Scholar] [CrossRef]
- Mcmeeking, A.A.; Codd, W.J.; Hanna, B.A. Report of a Wound Infection Caused by Vibrio parahaemolyticus and Vibrio vulnificus. Diagn. Microbiol. Infect. Dis. 1986, 5, 221–223. [Google Scholar] [CrossRef]
- Brayton, P.R.; Bode, R.B.; Colwell, R.R.; Macdonell, M.T.; Hall, H.L.; Grimes, D.J.; West, P.A.; Bryant, A.T.N. Vibrio cincinnatiensis Sp. Nov., a New Human Pathogen. J. Clin. Microbiol. 1986, 23, 104–108. [Google Scholar] [CrossRef]
- Kitaura, S.; Okamoto, K.; Wakabayashi, Y.; Okada, Y.; Okazaki, A.; Ikeda, M.; Hakuta, R.; Nakai, Y.; Okugawa, S.; Koike, K.; et al. Vibrio fluvialis Liver Abscess and Bacteremia in a Sashimi Lover: A Case Report and Review of the Literature. Open Forum Infect. Dis. 2020, 7, ofaa212. [Google Scholar] [CrossRef]
- Ballal, M.; Shetty, V.; Bangera, S.R.; Prabhu, M.; Umakanth, S. Vibrio furnissii, an Emerging Pathogen Causing Acute Gastroenteritis: A Case Report. JMM Case Rep. 2017, 4, e005111. [Google Scholar] [CrossRef]
- Li, J.; Woo, N.Y.S. Pathogenicity of Vibrios in Fish: An Overview. J. Ocean. Univ. Qingdao 2003, 2, 117–128. [Google Scholar]
- Dong, X.; Wang, H.; Zou, P.; Chen, J.; Liu, Z.; Wang, X.; Huang, J. Complete genome sequence of Vibrio campbellii strain 20130629003S01 isolated from shrimp with acute hepatopancreatic necrosis disease. Gut Pathog. 2019, 9, 31. [Google Scholar] [CrossRef]
- Mohamad, N.; Mohd Roseli, F.A.; Azmai, M.N.A.; Saad, M.Z.; Md Yasin, I.S.; Zulkiply, N.A.; Nasruddin, N.S. Natural Concurrent Infection of Vibrio harveyi and V. alginolyticus in Cultured Hybrid Groupers in Malaysia. J. Aquat. Anim. Health 2019, 31, 88–96. [Google Scholar] [CrossRef]
- Haenen, O.L.M.; van Zanten, E.; Jansen, R.; Roozenburg, I.; Engelsma, M.Y.; Dijkstra, A.; Boers, S.A.; Voorbergen-Laarman, M.; Möller, A.V.M. Vibrio vulnificus Outbreaks in Dutch Eel Farms since 1996: Strain Diversity and Impact. Dis. Aquat. Organ. 2014, 108, 201–209. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, Y.-X.; Jiang, Y.; Wang, Y.-G.; Liao, M.-J.; Rong, X.-J.; Wang, K.; Zhang, H.; Chen, J. First Report of Isolation and Complete Genome of Vibrio rotiferianus Strain SSVR1601 from Cage-Cultured Black Rockfish (Sebastes schlegelii) Associated with Skin Ulcer. J. Fish Dis. 2019, 42, 623–630. [Google Scholar] [CrossRef]
- Ali, S.; Hossain, M.; Azad, A.B.; Siddique, A.B.; Moniruzzaman, M.; Ahmed, M.A.; Amin, M.B.; Islam, M.S.; Rahman, M.M.; Mondal, D.; et al. Diversity of Vibrio parahaemolyticus in Marine Fishes of Bangladesh. J. Appl. Microbiol. 2021, 131, 2539–2551. [Google Scholar] [CrossRef]
- Frans, I.; Michiels, C.W.; Bossier, P.; Willems, K.A.; Lievens, B.; Rediers, H. Vibrio anguillarum as a Fish Pathogen: Virulence Factors, Diagnosis and Prevention. J. Fish Dis. 2011, 34, 643–661. [Google Scholar] [CrossRef]
- Lages, M.A.; Balado, M.; Lemos, M.L. The Expression of Virulence Factors in Vibrio anguillarum Is Dually Regulated by Iron Levels and Temperature. Front. Microbiol. 2019, 10, 2335. [Google Scholar] [CrossRef]
- Leung, K.Y.; Wang, Q.; Zheng, X.; Zhuang, M.; Yang, Z.; Shao, S.; Achmon, Y.; Siame, B.A. Versatile Lifestyles of Edwardsiella: Free-Living, Pathogen, and Core Bacterium of the Aquatic Resistome. Virulence 2022, 13, 5–18. [Google Scholar] [CrossRef]
- Hurtado, R.; Carhuaricra, D.; Soares, S.; Viana, M.V.C.; Azevedo, V.; Maturrano, L.; Aburjaile, F. Pan-Genomic Approach Shows Insight of Genetic Divergence and Pathogenic-Adaptation of Pasteurella multocida. Gene 2018, 670, 193–206. [Google Scholar] [CrossRef]
- Vannucci, F.A.; Kelley, M.R.; Gebhart, C.J. Comparative Genome Sequencing Identifies a Prophage-Associated Genomic Island Linked to Host Adaptation of Lawsonia intracellularis Infections. Vet. Res. 2013, 44, 49. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Das, B.; Kumar, N. Vibrio Pathogenicity Island-1: The Master Determinant of Cholera Pathogenesis. Front. Cell. Infect. Microbiol. 2020, 10, 561296. [Google Scholar] [CrossRef] [PubMed]
- Thiang, E.L.; Lee, C.W.; Takada, H.; Seki, K.; Takei, A.; Suzuki, S.; Wang, A.; Bong, C.W. Antibiotic Residues from Aquaculture Farms and Their Ecological Risks in Southeast Asia: A Case Study from Malaysia. Ecosyst. Health Sustain. 2021, 7, 1926337. [Google Scholar] [CrossRef]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic Resistance of Vibrio parahaemolyticus and Vibrio vulnificus in Various Countries: A Review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef]
- Onohuean, H.; Agwu, E.; Nwodo, U.U. Systematic Review and Meta-Analysis of Environmental Vibrio Species—Antibiotic Resistance. Heliyon 2022, 8, e08845. [Google Scholar] [CrossRef]
- Mok, J.S.; Cho, S.R.; Park, Y.J.; Jo, M.R.; Ha, K.S.; Kim, P.H.; Kim, M.J. Distribution and Antimicrobial Resistance of Vibrio parahaemolyticus Isolated from Fish and Shrimp Aquaculture Farms along the Korean Coast. Mar. Pollut. Bull. 2021, 171, 112785. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-Cholera Vibrios: The Microbial Barometer of Climate Change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef]
- Hackbusch, S.; Wichels, A.; Gimenez, L.; Döpke, H.; Gerdts, G. Potentially Human Pathogenic Vibrio Spp. in a Coastal Transect: Occurrence and Multiple Virulence Factors. Sci. Total Environ. 2020, 707, 136113. [Google Scholar] [CrossRef]
- Laverty, A.L.; Primpke, S.; Lorenz, C.; Gerdts, G.; Dobbs, F.C. Bacterial Biofilms Colonizing Plastics in Estuarine Waters, with an Emphasis on Vibrio Spp. And Their Antibacterial Resistance. PLoS ONE 2020, 15, e0237704. [Google Scholar] [CrossRef]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, Antibiotic-Resistant Bacteria, and Resistance Genes in Aquaculture: Risks, Current Concern, and Future Thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef]
- Shee, E.L.; Soo, Y.K.; Choon, M.K.; Kim, M.K.; Young, R.K.; Jeong, K.; Ryu, H.J.; Youn, S.L.; Sun, S.C.; Choy, H.E.; et al. The PyrH Gene of Vibrio vulnificus Is an Essential in Vivo Survival Factor. Infect. Immun. 2007, 75, 2795–2801. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D. Vibrio vulnificus: New Insights into a Deadly Opportunistic Pathogen. Environ. Microbiol. 2018, 20, 423–430. [Google Scholar] [CrossRef]
- Mohd Yazid, S.H.; Mohd Daud, H.; Azmai, M.N.A.; Mohamad, N.; Mohd Nor, N. Estimating the Economic Loss Due to Vibriosis in Net-Cage Cultured Asian Seabass (Lates calcarifer): Evidence from the East Coast of Peninsular Malaysia. Front. Vet. Sci. 2021, 8, 1111. [Google Scholar] [CrossRef]
- Defoirdt, T. Amino Acid–Derived Quorum Sensing Molecules Controlling the Virulence of Vibrios (and Beyond). PLoS Pathog. 2019, 15, e1007815. [Google Scholar] [CrossRef] [PubMed]
- Aujoulat, F.; Roger, F.; Bourdier, A.; Lotthé, A.; Lamy, B.; Marchandin, H.; Jumas-Bilak, E. From Environment to Man: Genome Evolution and Adaptation of Human Opportunistic Bacterial Pathogens. Genes 2012, 3, 191–232. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, G.D.; Hiller, N.L.; Hu, F.Z. What Makes Pathogens Pathogenic. Genome Biol. 2008, 9, 225–227. [Google Scholar] [CrossRef] [PubMed]
- Pazhani, G.P.; Chowdhury, G.; Ramamurthy, T. Adaptations of Vibrio parahaemolyticus to Stress During Environmental Survival, Host Colonization, and Infection. Front. Microbiol. 2021, 12, 737299. [Google Scholar] [CrossRef]
- Scott Merrell, D.; Bailey, C.; Kaper, J.B.; Camilli, A. The ToxR-Mediated Organic Acid Tolerance Response of Vibrio cholerae Requires OmpU. J. Bacteriol. 2001, 183, 2746–2754. [Google Scholar] [CrossRef]
- Minato, Y.; Fassio, S.R.; Kirkwood, J.S.; Halang, P.; Quinn, M.J.; Faulkner, W.J.; Aagesen, A.M.; Steuber, J.; Stevens, J.F.; Häse, C.C. Roles of the Sodium-Translocating NADH:Quinone Oxidoreductase (Na+-NQR) on Vibrio cholerae Metabolism, Motility and Osmotic Stress Resistance. PLoS ONE 2014, 9, e97083. [Google Scholar] [CrossRef]
- Shikuma, N.J.; Yildiz, F.H. Identification and Characterization of OscR, a Transcriptional Regulator Involved in Osmolarity Adaptation in Vibrio cholerae. J. Bacteriol. 2009, 191, 4082–4096. [Google Scholar] [CrossRef]
- Payne, S.M.; Mey, A.R.; Wyckoff, E.E. Vibrio Iron Transport: Evolutionary Adaptation to Life in Multiple Environments. Microbiol. Mol. Biol. Rev. 2016, 80, 69–90. [Google Scholar] [CrossRef]
- Wyckoff, E.E.; Schmitt, M.; Wilks, A.; Payne, S.M. HutZ Is Required for Efficient Heme Utilization in Vibrio cholerae. J. Bacteriol. 2004, 186, 4142–4151. [Google Scholar] [CrossRef]
- Sekine, Y.; Tanzawa, T.; Tanaka, Y.; Ishimori, K.; Uchida, T. Cytoplasmic Heme-Binding Protein (HutX) from Vibrio cholerae Is an Intracellular Heme Transport Protein for the Heme-Degrading Enzyme, HutZ. Biochemistry 2016, 55, 884–893. [Google Scholar] [CrossRef]
- Qin, W.; Li, D.; Xu, L.; Lin, W.; Tong, Y. Complete Genome Analysis of an Active Prophage of Vibrio Alginolyticus. Arch. Virol. 2021, 166, 891–896. [Google Scholar] [CrossRef]
- Rajpara, N.; Nair, M.; Bhardwaj, A.K. A Highly Promiscuous Integron, Plasmids, Extended Spectrum Beta Lactamases and Efflux Pumps as Factors Governing Multidrug Resistance in a Highly Drug Resistant Vibrio fluvialis Isolate BD146 from Kolkata, India. Indian J. Microbiol. 2018, 58, 60–67. [Google Scholar] [CrossRef]
- Helena Rebouças, R.; Viana de Sousa, O.; Sousa Lima, A.; Roger Vasconcelos, F.; de Carvalho, P.B.; dos Fernandes Vieira, R.H.S. Antimicrobial Resistance Profile of Vibrio Species Isolated from Marine Shrimp Farming Environments (Litopenaeus vannamei) at Ceará, Brazil. Environ. Res. 2011, 111, 21–24. [Google Scholar] [CrossRef]
- Jang, H.M.; Kim, Y.B.; Choi, S.; Lee, Y.; Shin, S.G.; Unno, T.; Kim, Y.M. Prevalence of Antibiotic Resistance Genes from Effluent of Coastal Aquaculture, South Korea. Environ. Pollut. 2018, 233, 1049–1057. [Google Scholar] [CrossRef]
- Kim, S.R.; Nonaka, L.; Suzuki, S. Occurrence of Tetracycline Resistance Genes Tet(M) and Tet(S) in Bacteria from Marine Aquaculture Sites. FEMS Microbiol. Lett. 2004, 237, 147–156. [Google Scholar] [CrossRef]
- Zago, V.; Veschetti, L.; Patuzzo, C.; Malerba, G.; Lleo, M.M. Resistome, Mobilome and Virulome Analysis of Shewanella algae and Vibrio Spp. Strains Isolated in Italian Aquaculture Centers. Microorganisms 2020, 8, 572. [Google Scholar] [CrossRef]
- Martins, A.F.M.; Fontana, H.; Moreira, B.M.; Bonelli, R.R. Draft Genome Sequence of a Tetracycline-Resistant Plesiomonas Shigelloides Strain Isolated from Aquaculture-Reared Tilapia. Microbiol. Resour. Announc. 2018, 7, e00832-18. [Google Scholar] [CrossRef]
- Tamminen, M.; Karkman, A.; Lõhmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline Resistance Genes Persist at Aquaculture Farms in the Absence of Selection Pressure. Environ. Sci. Technol. 2011, 45, 386–391. [Google Scholar] [CrossRef]
- Teicher, C.L.; Ronat, J.B.; Fakhri, R.M.; Basel, M.; Labar, A.S.; Herard, P.; Murphy, R.A. Antimicrobial Drug–Resistant Bacteria Isolated from Syrian War–Injured Patients, August 2011–March 2013. Emerg. Infect. Dis. 2014, 20, 1949–1951. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, Z.; Deng, Y.; Shi, Y.; Liu, Y.; Wu, C.; Luo, P.; Hu, C. Complete Genome Sequence of Vibrio campbellii LMB 29 Isolated from Red Drum with Four Native Megaplasmids. Front. Microbiol. 2017, 8, 2035. [Google Scholar] [CrossRef]
- Kong, W.; Huang, S.; Yang, Z.; Shi, F.; Feng, Y.; Khatoon, Z. Fish Feed Quality Is a Key Factor in Impacting Aquaculture Water Environment: Evidence from Incubator Experiments. Sci. Rep. 2020, 10, 187. [Google Scholar] [CrossRef]
- Qiu, H.; Jin, M.; Li, Y.; Lu, Y.; Hou, Y.; Zhou, Q. Dietary Lipid Sources Influence Fatty Acid Composition in Tissue of Large Yellow Croaker (Larmichthys crocea) by Regulating Triacylglycerol Synthesis and Catabolism at the Transcriptional Level. PLoS ONE 2017, 12, e0169985. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hern̗andez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Barakat, M.; Ortet, P.; Whitworth, D.E. P2RP: A Web-Based Framework for the Identification and Analysis of Regulatory Proteins in Prokaryotic Genomes. BMC Genom. 2013, 14, 269. [Google Scholar] [CrossRef]
- Soares, S.C.; Geyik, H.; Ramos, R.T.J.; de Sá, P.H.C.G.; Barbosa, E.G.V.; Baumbach, J.; Figueiredo, H.C.P.; Miyoshi, A.; Tauch, A.; Silva, A.; et al. GIPSy: Genomic Island Prediction Software. J. Biotechnol. 2016, 232, 2–11. [Google Scholar] [CrossRef]
- Hoff, J.; Daniel, B.; Stukenberg, D.; Thuronyi, B.W.; Waldminghaus, T.; Fritz, G. Vibrio natriegens: An Ultrafast-Growing Marine Bacterium as Emerging Synthetic Biology Chassis. Environ. Microbiol. 2020, 22, 4394–4408. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- He, Y.; Zhou, X.; Chen, Z.; Deng, X.; Gehring, A.; Ou, H.; Zhang, L.; Shi, X. PRAP: Pan Resistome Analysis Pipeline. BMC Bioinform. 2020, 21, 20–28. [Google Scholar] [CrossRef]
- Barh, D.; Tiwari, S.; Jain, N.; Ali, A.; Santos, A.R.; Misra, A.N.; Azevedo, V.; Kumar, A. In Silico Subtractive Genomics for Target Identification in Human Bacterial Pathogens. Drug Dev. Res. 2011, 72, 162–177. [Google Scholar] [CrossRef]
- Barinov, A.; Loux, V.; Hammani, A.; Nicolas, P.; Langella, P.; Ehrlichh, D.; Maguin, E.; van de Guchte, M. Prediction of Surface Exposed Proteins in Streptococcus Pyogenes, with a Potential Application to Other Gram-Positive Bacteria. Proteomics 2009, 9, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.I.; Ferdous, S.; Jewel, N.A.; Akter, A.; Mahmud, Z.; Islam, M.M.; Afrin, T.; Karim, N. Identification of Potential Drug Targets by Subtractive Genome Analysis of Escherichia Coli O157:H7: An in Silico Approach. Adv. Appl. Bioinform. Chem. 2015, 8, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.D.; Oliveira, P.H.E.; Siqueira, D.G.; Reis, V.C.C.; Dardenne, L.E.; Goliatt, P.V.Z.C. MHOLline 2.0: Workflow for Automatic Large-Scale Modeling and Analysis of Proteins. Rev. Mundi Eng. Tecnol. Gestão 2020, 5, 1–14. [Google Scholar] [CrossRef]
- Luo, H.; Lin, Y.; Liu, T.; Lai, F.L.; Zhang, C.T.; Gao, F.; Zhang, R. DEG 15, an Update of the Database of Essential Genes That Includes Built-in Analysis Tools. Nucleic Acids Res. 2021, 49, D677–D686. [Google Scholar] [CrossRef]
- Volkamer, A.; Kuhn, D.; Rippmann, F.; Rarey, M. Dogsitescorer: A Web Server for Automatic Binding Site Prediction, Analysis and Druggability Assessment. Bioinformatics 2012, 28, 2074–2075. [Google Scholar] [CrossRef]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15-Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Rodrigues, T.C.V.; Jaiswal, A.K.; de Sarom, A.; Oliveira, L.D.C.; Oliveira, C.J.F.; Ghosh, P.; Tiwari, S.; Miranda, F.M.; Benevides, L.D.J.; Azevedo, V.A.D.C.; et al. Reverse Vaccinology and Subtractive Genomics Reveal New Therapeutic Targets against Mycoplasma Pneumoniae: A Causative Agent of Pneumonia. R. Soc. Open Sci. 2019, 6, 190907. [Google Scholar] [CrossRef]
- Francoeur, P.G.; Koes, D.R. SolTranNet-A Machine Learning Tool for Fast Aqueous Solubility Prediction. J. Chem. Inf. Model. 2021, 61, 2530–2536. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Nogueira, W.G.; Jaiswal, A.K.; Tiwari, S.; Ramos, R.T.J.; Ghosh, P.; Barh, D.; Azevedo, V.; Soares, S.C. Computational Identification of Putative Common Genomic Drug and Vaccine Targets in Mycoplasma Genitalium. Genomics 2021, 113, 2730–2743. [Google Scholar] [CrossRef]
- Guillod, C.; Ghitti, F.; Mainetti, C. Vibrio parahaemolyticus Induced Cellulitis and Septic Shock after a Sea Beach Holiday in a Patient with Leg Ulcers. Case Rep. Dermatol. 2019, 11, 94–100. [Google Scholar] [CrossRef]
- Liang, J.H.; Liang, W.H.; Deng, Y.Q.; Fu, Z.G.; Deng, J.L.; Chen, Y.H. Vibrio vulnificus Infection Attributed to Bee Sting: A Case Report. Emerg. Microbes Infect. 2021, 10, 1890–1895. [Google Scholar] [CrossRef]
- Siddique, A.B.; Moniruzzaman, M.; Ali, S.; Dewan, M.N.; Islam, M.R.; Islam, M.S.; Amin, M.B.; Mondal, D.; Parvez, A.K.; Mahmud, Z.H. Characterization of Pathogenic Vibrio parahaemolyticus Isolated From Fish Aquaculture of the Southwest Coastal Area of Bangladesh. Front. Microbiol. 2021, 12, 635539. [Google Scholar] [CrossRef]
- Chen, J.; Chiu, C.; Gopalkrishnan, S.; Chen, A.Y.; Olinares, P.D.B.; Saecker, R.M.; Winkelman, J.T.; Maloney, M.F.; Chait, B.T.; Ross, W.; et al. Stepwise Promoter Melting by Bacterial RNA Polymerase. Mol. Cell 2020, 78, 275–288.e6. [Google Scholar] [CrossRef]
- Taylor, P.L.; Blakely, K.M.; de Leon, G.P.; Walker, J.R.; McArthur, F.; Evdokimova, E.; Zhang, K.; Valvano, M.A.; Wright, G.D.; Junop, M.S. Structure and Function of Sedoheptulose-7-Phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants. J. Biol. Chem. 2008, 283, 2835–2845. [Google Scholar] [CrossRef]
- Keyamura, K.; Fujikawa, N.; Ishida, T.; Ozaki, S.; Su’etsugu, M.; Fujimitsu, K.; Kagawa, W.; Yokoyama, S.; Kurumizaka, H.; Katayama, T. The Interaction of DiaA and DnaA Regulates the Replication Cycle in E. Coli by Directly Promoting ATP-DnaA-Specific Initiation Complexes. Genes Dev. 2007, 21, 2083–2099. [Google Scholar] [CrossRef]
- Hatzopoulos, G.N.; Mueller-Dieckmann, J. Structure of Translation Initiation Factor 1 from Mycobacterium Tuberculosis and Inferred Binding to the 30S Ribosomal Subunit. FEBS Lett. 2010, 584, 1011–1015. [Google Scholar] [CrossRef]
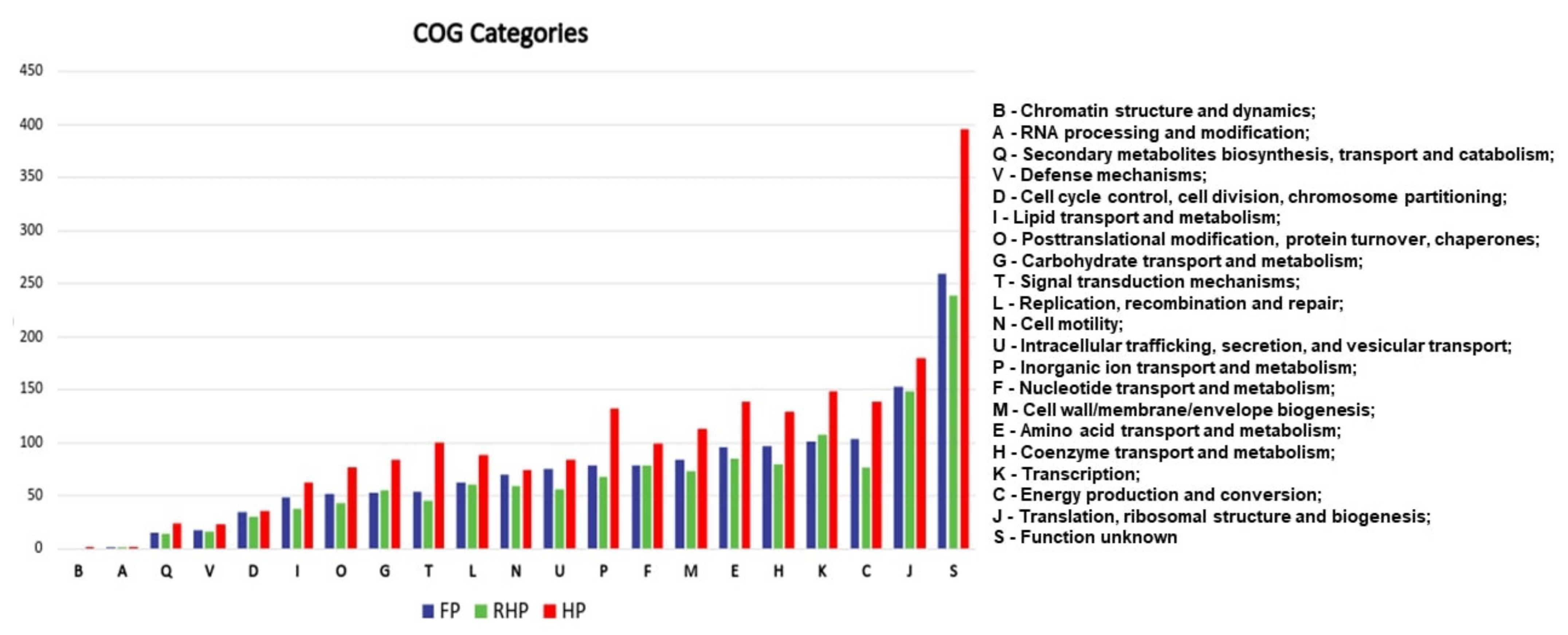


| (COGs: K) | HP | FP | RHP |
|---|---|---|---|
| RR | 12 | 9 | 7 |
| TR | 42 | 24 | 34 |
| OCS | 35 | 21 | 20 |
| SF | 7 | 5 | 5 |
| Genome/Strain | CHR 1 | CHR 2 | GI’s |
|---|---|---|---|
| V. parahaemolyticus RIMD | 11 | 9 | PAI’s |
| 4 | 2 | SI’s | |
| 7 | 4 | MI’s |
| Protein ID | Gene | Biological Process |
|---|---|---|
| WP_005491112.1 | hmuV | Part of the ABC transporter complex HmuTUV involved in hemin import; Responsible for energy coupling to the transport system. |
| WP_005491087.1 | VPA0422 | Transmembrane transporter activity; Putative hemin ABC transporter, permease protein |
| WP_021451074.1 | HutX | Heme utilization; cytosolic carrier protein |
| WP_005457554.1 | hutZ | Heme utilization protein |
| Protein ID | Product | Gene | Length (aa) | Structural Quality MHOLline | Biological Process | |
|---|---|---|---|---|---|---|
| FP | WP_005380329.1 | MULTISPECIES: UMP Kinase | pyrH | 241 | VERY HIGH | Catalyzes the reversible Phosphorylation of UMP to UDP. CTP biosynthesis via de novo pathway |
| WP_001040192.1 | MULTISPECIES: Translation initiation factor IF-1 | infA | 75 | HIGH | Stabilizes the binding of IF-2 and IF-3 on the 30 S subunit to which N-formylmethionyl-tRNA(fMet) subsequently binds | |
| WP_005372901.1 | MULTISPECIES: Phosphoheptose isomerase | gmhA | 196 | HIGH | D-glycero-D-manno-heptose 7-phosphate biosynthesis | |
| WP_005390081.1 | MULTISPECIES: bifunctional 3-hydroxydecanoyl- ACP dehydratase/trans-2-decenoyl-ACP isomerase | fabA | 172 | HIGH | Fatty acid biosynthesis | |
| HP | WP_005380329.1 | MULTISPECIES: UMP kinase [Vibrio] | pyrH | 241 | VERY HIGH | Catalyzes the reversible phosphorylation of UMP to UDP. CTP biosynthesis via de novo pathway |
| WP_005380392.1 | MULTISPECIES: D-sedoheptulose 7-phosphate isomerase | gmhA | 191 | VERY HIGH | D-glycero-D-manno-heptose 7-phosphate biosynthesis | |
| WP_005395986.1 | RNA polymerase sigma factor RpoD | rpoD | 620 | VERY HIGH | Promote the attachment of RNA polymerase |
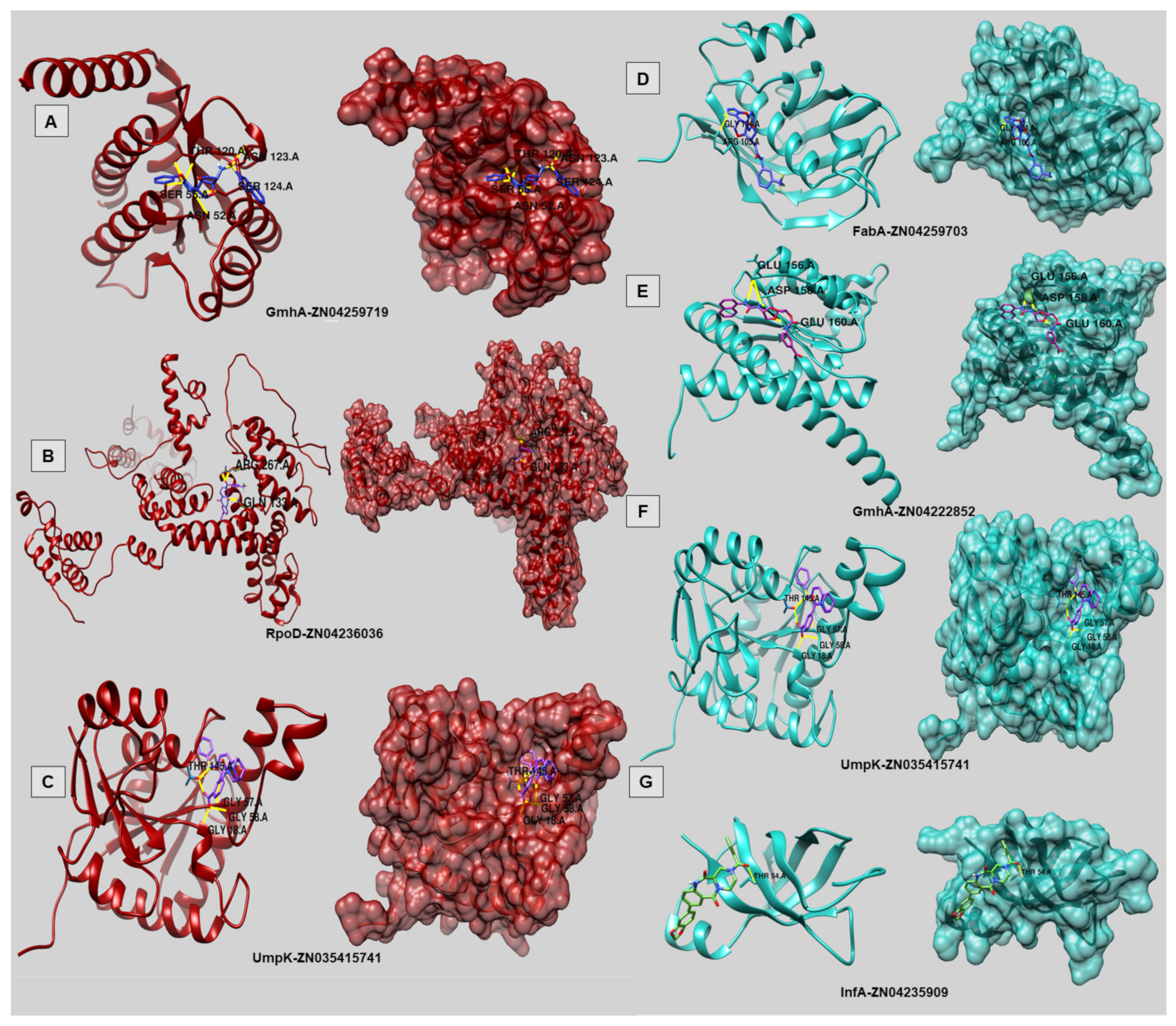
| Docking Results | Targets | ZINC Compound ID | AutoDock Vina Binding Affinity (kcal/mol) | Num. of HBonds | Residues |
|---|---|---|---|---|---|
| FP | InfA (WP_001040192.1) | ZN04235909 | −7.5 | 1 | THR54 |
| FabA (WP_005390081.1) | ZN04259703 | −9.2 | 2 | GLY 104, ARG 105 | |
| GmhA (WP_005372901.1) | ZN04222852 | −8.2 | 4 | GLU156 (2×); ASP158; GLU160 | |
| UmpK(WP_005380329.1) | ZN03541574 | −10.6 | 6 | THR145 (3×); GLY57; GLY58; GLY18 | |
| HP | UmpK (WP_005380329.1) | ZN03541574 | −10.6 | 6 | THR145 (3×); GLY57; GLY58; GLY18 |
| GmhA (WP_005380392.1) | ZN04259719 | −8.9 | 7 | THR120 (3x); SER55; ASN52; SER124; ASN123 | |
| RpoD (WP_005395986.1) | ZN04236036 | −10.7 | 3 | ARG267 (2×); GLN133 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques, P.H.; Prado, L.C.d.S.; Felice, A.G.; Rodrigues, T.C.V.; Pereira, U.d.P.; Jaiswal, A.K.; Azevedo, V.; Oliveira, C.J.F.; Soares, S. Insights into the Vibrio Genus: A One Health Perspective from Host Adaptability and Antibiotic Resistance to In Silico Identification of Drug Targets. Antibiotics 2022, 11, 1399. https://doi.org/10.3390/antibiotics11101399
Marques PH, Prado LCdS, Felice AG, Rodrigues TCV, Pereira UdP, Jaiswal AK, Azevedo V, Oliveira CJF, Soares S. Insights into the Vibrio Genus: A One Health Perspective from Host Adaptability and Antibiotic Resistance to In Silico Identification of Drug Targets. Antibiotics. 2022; 11(10):1399. https://doi.org/10.3390/antibiotics11101399
Chicago/Turabian StyleMarques, Pedro Henrique, Lígia Carolina da Silva Prado, Andrei Giacchetto Felice, Thaís Cristina Vilela Rodrigues, Ulisses de Padua Pereira, Arun Kumar Jaiswal, Vasco Azevedo, Carlo José Freire Oliveira, and Siomar Soares. 2022. "Insights into the Vibrio Genus: A One Health Perspective from Host Adaptability and Antibiotic Resistance to In Silico Identification of Drug Targets" Antibiotics 11, no. 10: 1399. https://doi.org/10.3390/antibiotics11101399
APA StyleMarques, P. H., Prado, L. C. d. S., Felice, A. G., Rodrigues, T. C. V., Pereira, U. d. P., Jaiswal, A. K., Azevedo, V., Oliveira, C. J. F., & Soares, S. (2022). Insights into the Vibrio Genus: A One Health Perspective from Host Adaptability and Antibiotic Resistance to In Silico Identification of Drug Targets. Antibiotics, 11(10), 1399. https://doi.org/10.3390/antibiotics11101399











