Effects of Broad-Spectrum Antibiotic (Florfenicol) on Resistance Genes and Bacterial Community Structure of Water and Sediments in an Aquatic Microcosm Model
Abstract
1. Introduction
2. Results
2.1. Abundance of Florfenicol ARGs
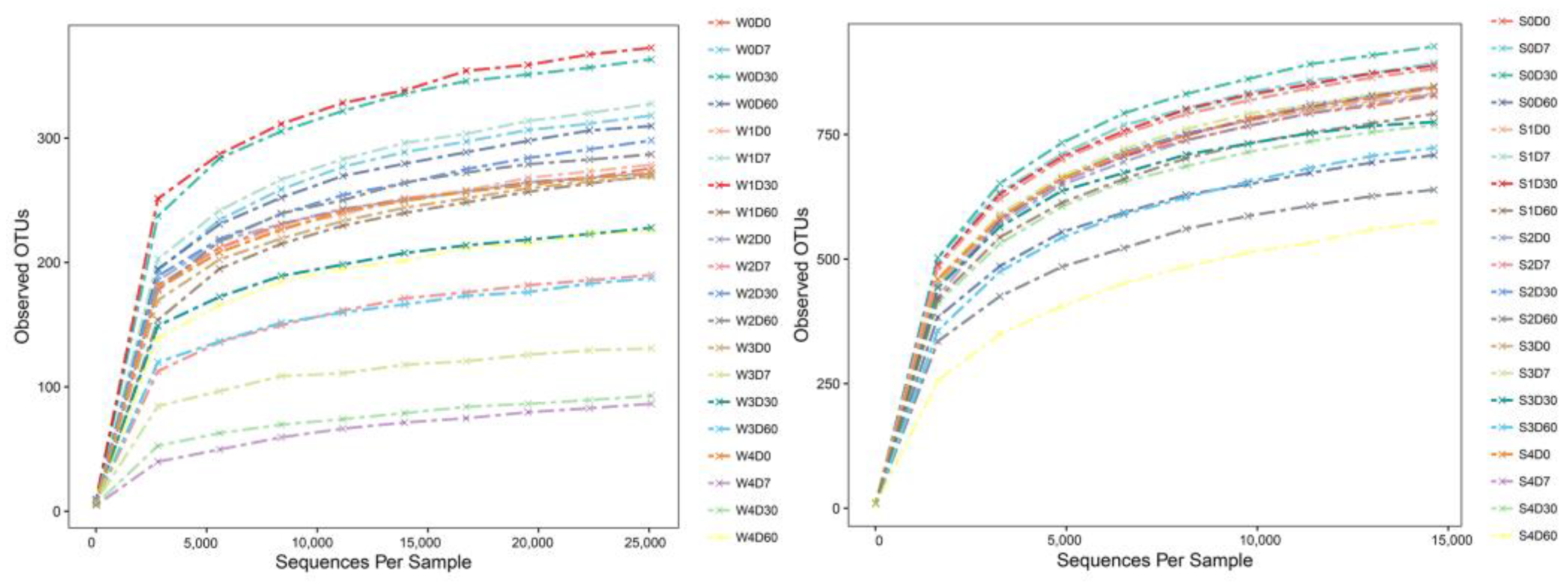
2.2. Analysis of OTUs
2.3. Alpha Diversity Index Analysis
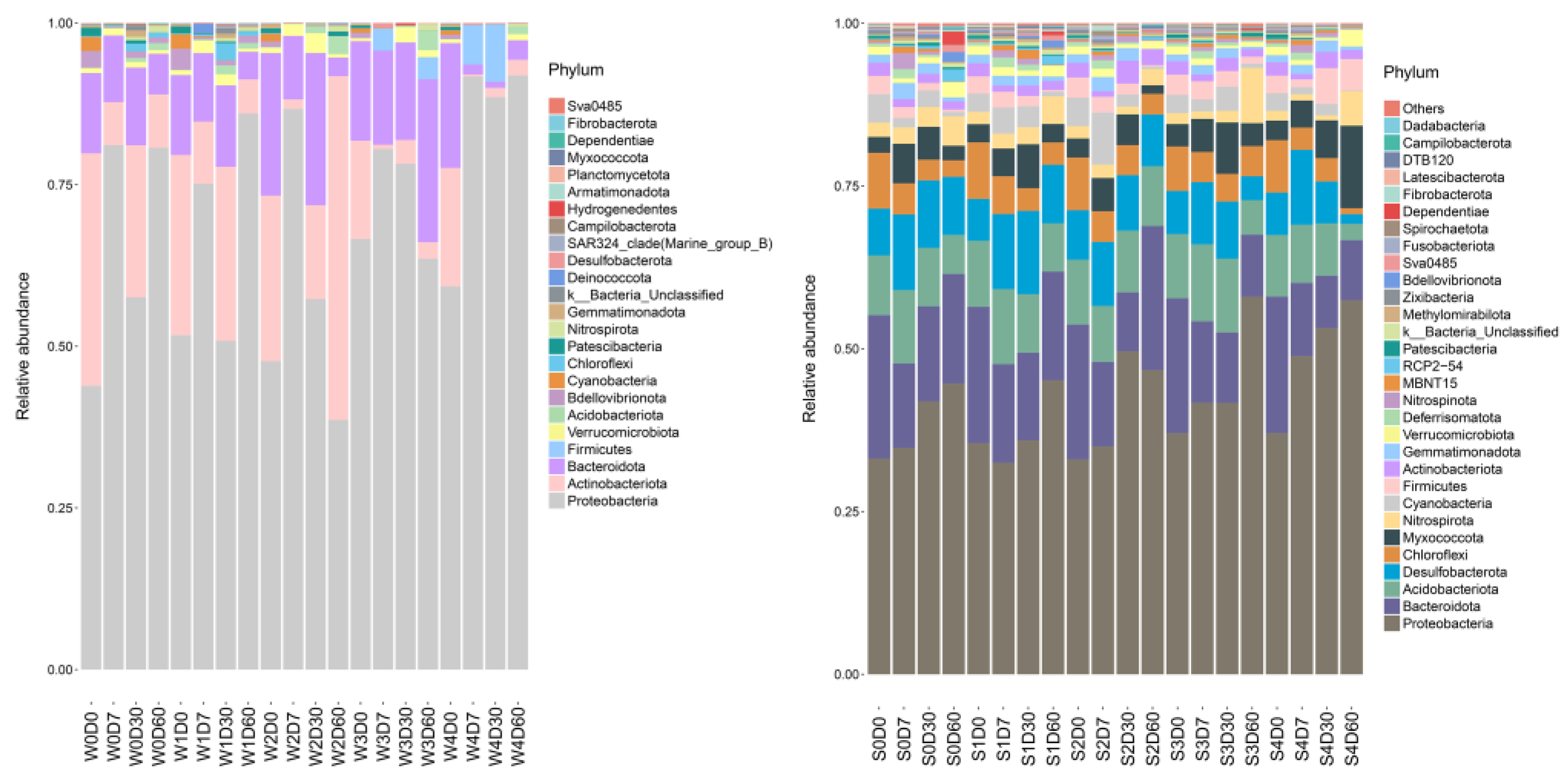
2.4. Bacterial Community Structure Analysis
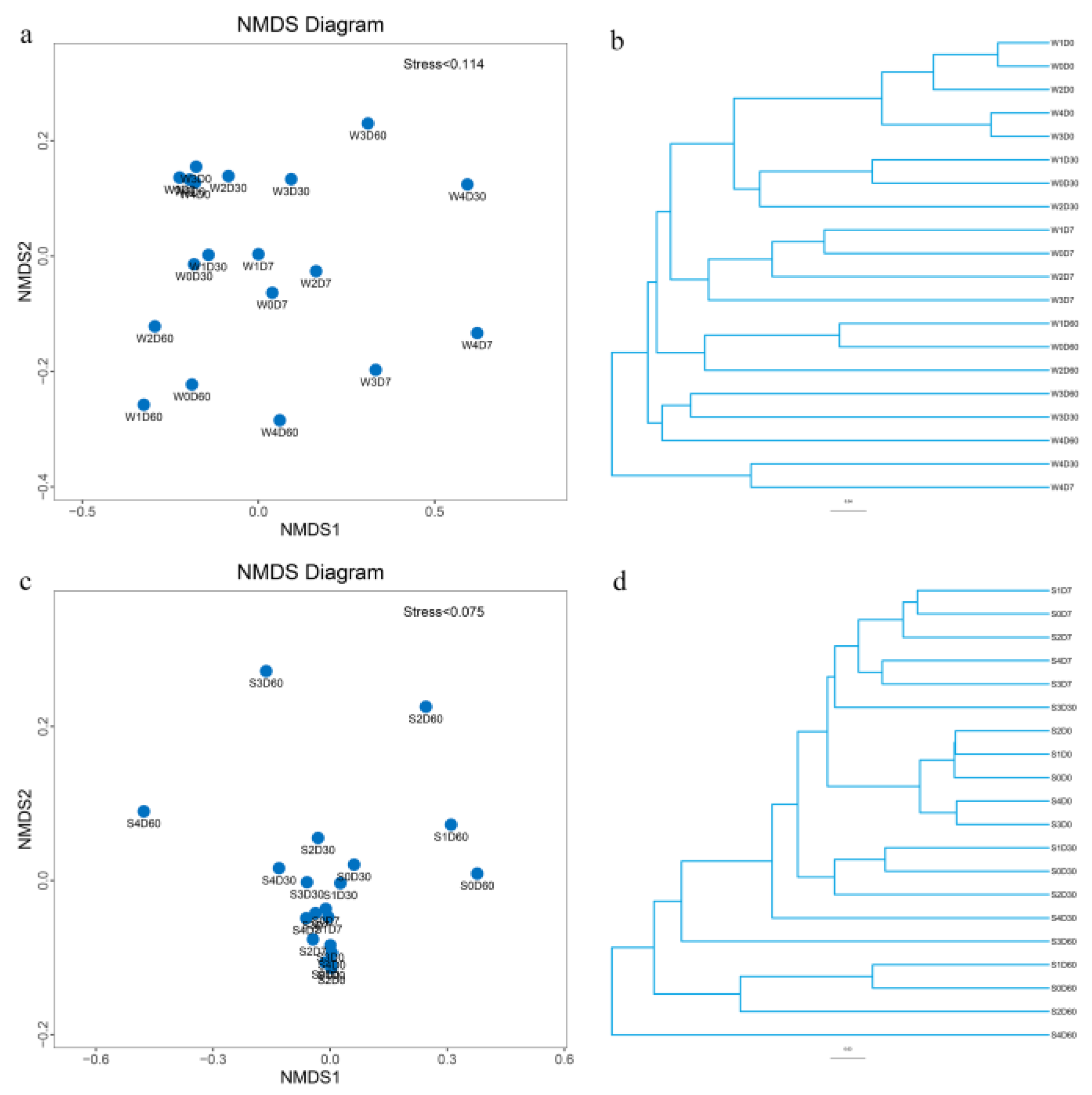
2.5. Multisample Comparative Analysis
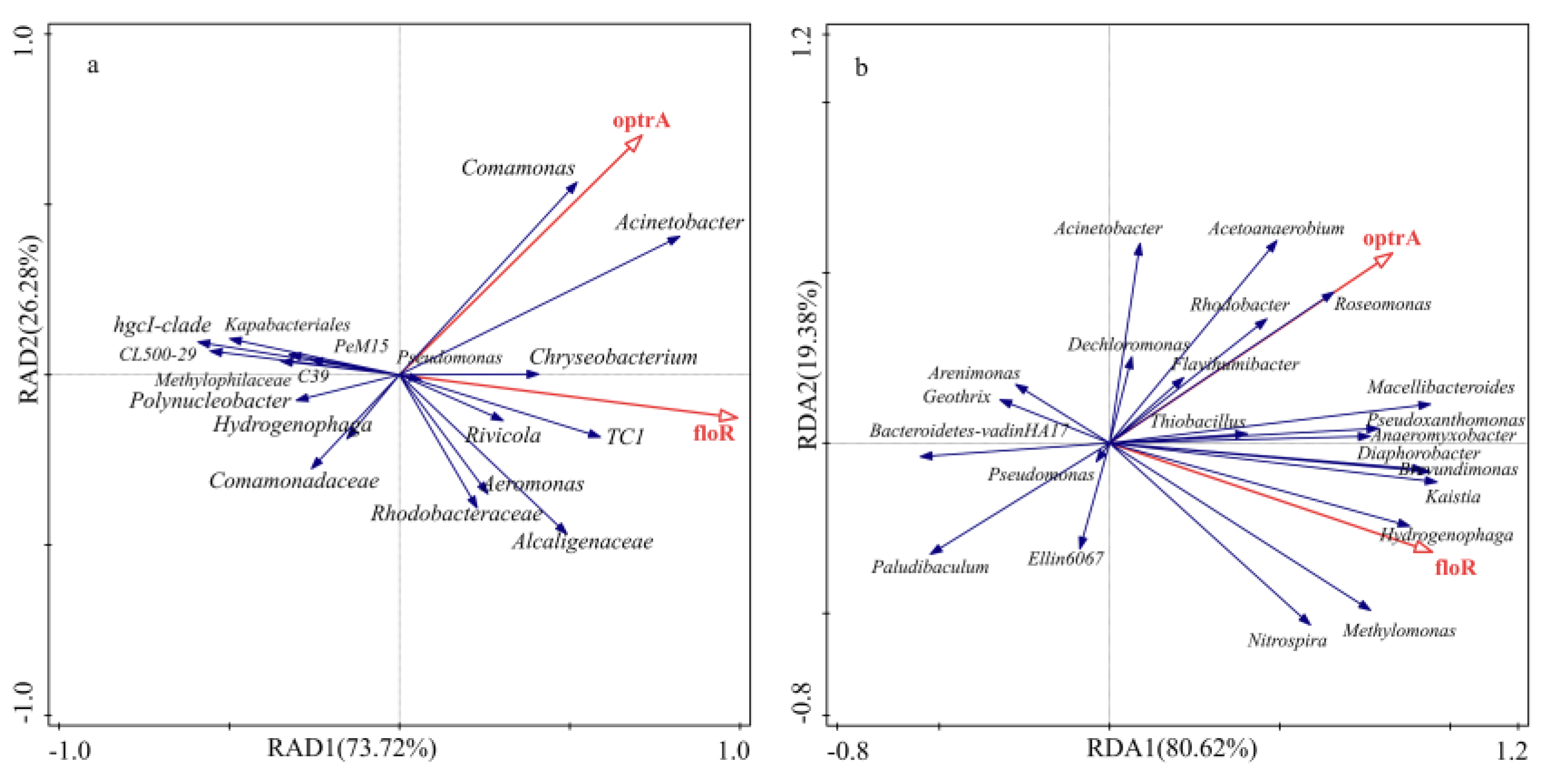
2.6. Correlation Analysis between Bacterial Communities and ARGs
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Experimental Design and Sampling
4.3. Detection of ARGs
4.4. 16S rDNA Sequencing
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, Y.; Zhao, Z.; Pan, Z.; An, L.; Balasubramanian, B.; Liu, W. New insights into the role of dietary marine-derived polysaccharides on productive performance, egg quality, antioxidant capacity, and jejunal morphology in late-phase laying hens. Poult. Sci. 2020, 99, 2100–2107. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhu, Y.; Zhao, Z.; Jiang, P.; Yin, F. Effects of dietary supplementation of algae-derived polysaccharides on morphology, tight junctions, antioxidant capacity and immune response of duodenum in broilers under heat stress. Animals 2021, 11, 2279. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Shen, Q.; Zhao, F. Antibiotics and antibiotic resistance from animal manures to soil: A review. Eur. J. Soil Sci. 2018, 69, 181–195. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, S.; Balasubramanian, B.; Zeng, F.; Sun, C.; Pang, H. Dietary seaweed (Enteromorpha) polysaccharides improves growth performance involved in regulation of immune responses, intestinal morphology and microbial community in banana shrimp Fenneropenaeus merguiensis. Fish Shellfish Immun. 2020, 104, 202–212. [Google Scholar] [CrossRef] [PubMed]
- FAO. The State of World Fisheries and Aquaculture; Sustainability in Action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in fish: A review. J. Fish Dis. 2014, 37, 163–188. [Google Scholar] [CrossRef]
- Giguère, S.; Prescott, J.F.; Dowling, P.M. Antimicrobial Therapy in Veterinary Medicine; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Huang, X.; Feng, Y.; Hu, C.; Xiao, X.; Yu, D.; Zou, X. Mechanistic model for interpreting the toxic effects of sulfonamides on nitrification. J. Hazard. Mater. 2016, 305, 123–129. [Google Scholar] [CrossRef]
- Zong, H.; Ma, D.; Wang, J.; Hu, J. Research on florfenicol residue in coastal area of Dalian (northern China) and analysis of functional diversity of the microbial community in marine sediment. Bull. Environ. Contam. Toxicol. 2010, 84, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Jara, B.; Tucca, F.; Srain, B.M.; Méjanelle, L.; Aranda, M.; Fernández, C.; Pantoja-Gutiérrez, S. Antibiotics florfenicol and flumequine in the water column and sediments of Puyuhuapi Fjord, Chilean Patagonia. Chemosphere 2021, 275, 130029. [Google Scholar] [CrossRef]
- Yan, C.; Yang, Y.; Zhou, J.; Liu, M.; Nie, M.; Shi, M.; Gu, L. Antibiotics in the surface water of the Yangtze Estuary: Occurrence, distribution and risk assessment. Environ. Pollut. 2013, 175, 22–29. [Google Scholar] [CrossRef]
- Zhang, Q.; Ying, G.; Pan, C.; Liu, Y.; Zhao, J. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef]
- Na, G.; Fang, X.; Cai, Y.; Ge, L.; Zong, H.; Yuan, X.; Yao, Z.; Zhang, Z. Occurrence, distribution, and bioaccumulation of antibiotics in coastal environment of Dalian, China. Mar. Pollut. Bull. 2013, 69, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhou, J. Occurrence and behavior of antibiotics in water and sediments from the Huangpu River, Shanghai, China. Chemosphere 2014, 95, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Pei, M.; Wang, D.; Cao, S.; Xiao, X.; Sun, B. Improvement of soil ecosystem multifunctionality by dissipating manure-induced antibiotics and resistance genes. Environ. Sci. Technol. 2017, 51, 4988–4998. [Google Scholar] [CrossRef] [PubMed]
- Crecchio, C.; Ruggiero, P.; Curci, M.; Colombo, M.; Palumbo, G.; Stotzky, G. Binding of DNA from Bacillus subtilis on montmorillonite–humic acids–aluminum or iron hydroxypolymers: Effects on transformation and protection against DNase. Soil Sci. Soc. Am. J. 2005, 69, 834–841. [Google Scholar] [CrossRef]
- Ma, W.; Wang, L.; Xu, X.; Huo, M.; Zhou, K.; Mi, K.; Tian, X.; Cheng, G.; Huang, L. Fate and exposure risk of florfenicol, thiamphenicol, and antibiotic resistance genes during composting of swine manure. Sci. Total Environ. 2022, 839, 156243. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Xie, J.; Wang, G.; Cheng, X.; Zhang, J.; Sang, C.; Liu, Z. Impact of microecological agents on water environment restoration and microbial community structures of trench system in a Baiyangdian wetland ecosystem. J. Appl. Microbiol. 2022, 132, 2450–2463. [Google Scholar] [CrossRef]
- Kim, J.; Hong, Y.; Yang, J.; Kwon, O.; Kim, S. Bioaccumulation and mass balance analysis of veterinary antibiotics in an agricultural environment. Toxics 2022, 10, 213. [Google Scholar] [CrossRef]
- Zeng, S.; Sun, J.; Chen, Z.; Xu, Q.; Wei, W.; Wang, D.; Ni, B. The impact and fate of clarithromycin in anaerobic digestion of waste activated sludge for biogas production. Environ. Res. 2021, 195, 110792. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.; Singer, A.C.; Zhu, Y. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Zeng, Q.; Liao, C.; Terhune, J.; Wang, L. Impacts of florfenicol on the microbiota landscape and resistome as revealed by metagenomic analysis. Microbiome 2019, 7, 155. [Google Scholar] [CrossRef]
- Xie, H.; Du, J.; Chen, J. Concerted efforts are needed to control and mitigate antibiotic pollution in coastal waters of China. Antibiotics 2020, 9, 88. [Google Scholar] [CrossRef]
- Jampani, M.; Gothwal, R.; Mateo-Sagasta, J.; Langan, S. Water quality modelling framework for evaluating antibiotic resistance in aquatic environments. J. Hazard. Mater. 2022, 3, 100056. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, J.; Xu, L.; Liu, Y.; Li, P.; Zhu, T.; Cheng, C.; Lu, S.; Xu, T.; Yi, H.; et al. Spread of the florfenicol resistance floR gene among clinical Klebsiella pneumoniae isolates in China. Antimicrob. Resist. Infect. Control 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.D.; Rojas, R.; Geisse, J.; Romero, J.; González-Rocha, G. Scallop larvae hatcheries as source of bacteria carrying genes encoding for non-enzymatic phenicol resistance. Mar. Pollut. Bull. 2015, 95, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.C.; Rossi, C.C.; Santana, M.F.; Langford, P.R.; Bossé, J.T.; Bazzolli, D.M.S. p518, a small floR plasmid from a South American isolate of Actinobacillus pleuropneumoniae. Vet. Microbiol. 2017, 204, 129–132. [Google Scholar] [CrossRef]
- Fan, R.; Li, D.; Wang, Y.; He, T.; Feßler, A.T.; Schwarz, S.; Wu, C. Presence of the optrA gene in methicillin-resistant Staphylococcus sciuri of porcine origin. Antimicrob. Agents Chemother. 2016, 60, 7200–7205. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Campo, R.D.; Coque, T.M.; Aarestrup, F.M.; Schwarz, S.; Shen, J.; Cavaco, L. Antimicrobial resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, T.; Liang, Y.; Li, G.; Sun, Y. Response characteristics of nirS-type denitrifier Paracoccus denitrificans under florfenicol stress. Ecotox. Environ. Safe. 2021, 219, 112355. [Google Scholar] [CrossRef]
- Suzuki, S.; Nakanishi, S.; Tamminen, M.; Yokokawa, T.; Sato-Takabe, Y.; Ohta, K.; Chou, H.; Muziasari, W.I.; Virta, M. Occurrence of sul and tet (M) genes in bacterial community in Japanese marine aquaculture environment throughout the year: Profile comparison with Taiwanese and Finnish aquaculture waters. Sci. Total Environ. 2019, 669, 649–656. [Google Scholar] [CrossRef]
- Wang, M.; Xie, X.; Wang, M.; Wu, J.; Zhou, Q.; Sun, Y. The bacterial microbiota in florfenicol contaminated soils: The antibiotic resistome and the nitrogen cycle. Environ. Pollut. 2020, 259, 113901. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Bi, Y.; Deng, Y.; He, Z.; Wu, L.; Van, N.J.D.; Shi, Z.; Li, J.; Wang, X.; Hu, Z.; et al. Impacts of the Three Gorges Dam on microbial structure and potential function. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, C.; Lian, K.; Liu, C. Effects of chronic exposure of antibiotics on microbial community structure and functions in hyporheic zone sediments. J. Hazard. Mater. 2021, 416, 126141. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shu, L.; Mitchell, S.M.; Yu, L.; Zhao, Q.; Frear, C. Effects of different antibiotic operation modes on anaerobic digestion of dairy manure: Focus on microbial population dynamics. J. Environ. Chem. Eng. 2021, 9, 105521. [Google Scholar] [CrossRef]
- Al Hag, W.A.; Elbadawi, H.; Abdel, H.M.M.; A’Court, C.; Garrard, C.S.; AbdulWahab, A.; Taj Aldeen, S.J.; Ibrahim, E.B.; Akbar, M.; Zahid, M.; et al. Use of 16s rRNA to identify non-lactose-fermenting bacilli and molecular detection of ESBL resistance genes associated with hospital-acquired infection in Soba University Hospital, Sudan. F1000Research 2020, 9, 1311. [Google Scholar] [CrossRef]
- Endo, S.; Yano, H.; Kanamori, H.; Inomata, S.; Aoyagi, T.; Hatta, M.; Gu, Y.; Tokuda, K.; Kitagawa, M.; Kaku, M. High frequency of Acinetobacter soli among Acinetobacter isolates causing bacteremia at a tertiary hospital in Japan. J. Clin. Microbiol. 2014, 52, 911–915. [Google Scholar] [CrossRef]
- Luo, J.; McIntyre, E.A.; Bedore, S.R.; Santala, V.; Neidle, E.L.; Santala, S. Characterization of highly ferulate-tolerant Acinetobacter baylyi ADP1 isolates by a rapid reverse engineering method. Appl. Environ. Microb. 2022, 88, e0178021. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Z.; Pang, Z.; Sun, Y.; Zhao, Y.; Hu, M.; Lin, L.; Yan, Z.; Huang, K.; Si, H. Study on the effects of stephania delavayi diels water extracts combined with antibiotics on NDM-1-producing Acinetobacter in Vitro. China Anim. Husb. Vet. Med. 2016, 43, 1092–1097. [Google Scholar]
- Liu, C.; Meng, Y.; Zhu, X.; Li, B.; Xia, L.; Xu, Z.; Shen, M.; Chen, X.; Bian, W. Elimination and migration of florfenicol and its metabolic residues in Letaurus punetaus and aquaculture environment under pond conditions. Chin. Fish. Qual. Stand. 2019, 9, 30–40. [Google Scholar]
- Li, S.; Huang, Z.; Wang, Y.; Liu, Y.; Luo, R.; Shang, J.; Liao, Q. Migration of two antibiotics during resuspension under simulated wind-wave disturbances in a water-sediment system. Chemosphere 2018, 192, 234–243. [Google Scholar] [CrossRef]
- Wang, F.; Stedtfeld, R.D.; Kim, O.; Chai, B.; Yang, L.; Stedtfeld, T.M.; Hong, S.G.; Kim, D.; Lim, H.S.; Hashsham, S.A.; et al. Influence of soil characteristics and proximity to Antarctic research stations on abundance of antibiotic resistance genes in soils. Environ. Sci. Technol. 2016, 50, 12621–12629. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, H. First report of carbapenem-resistant Acinetobacter soli isolates coharboring blaNDM-1 and blaOXA-58 genes from China. Diagn. Microbiol. Infec. Dis. 2015, 83, 407–408. [Google Scholar] [CrossRef] [PubMed]
- Hantsis-Zacharov, E.; Senderovich, Y.; Halpern, M. Chryseobacterium bovis sp. nov., isolated from raw cow’s milk. Int. J. Syst. Evol. Microbiol. 2008, 58, 1024–1028. [Google Scholar] [CrossRef] [PubMed]
- Mwanza, E.P.; Hugo, A.; Charimba, G.; Hugo, C.J. Pathogenic potential and control of Chryseobacterium species from clinical, fish, food and environmental sources. Microorganisms 2022, 10, 895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Zhang, Y.; Wu, X.; Zhou, Z.; Huang, Y.; Zhao, Y.; Mishra, S.; Bhatt, P.; Chen, S. Characterization of a novel glyphosate-degrading bacterial species, Chryseobacterium sp. Y16C, and evaluation of its effects on microbial communities in glyphosate-contaminated soil. J. Hazard. Mater. 2022, 432, 128689. [Google Scholar] [CrossRef]
- Fu, J.; Zhong, C.; Zhou, Y.; Lu, M.; Zong, G.; Zhang, P.; Cheng, M.; Cao, G. The integrative and conjugative element ICECspPOL2 contributes to the outbreak of multi-antibiotic-resistant bacteria for Chryseobacterium Spp. and Elizabethkingia Spp. Microbiol. Spectr. 2021, 9, e02005–e02021. [Google Scholar] [CrossRef]
- Nadimpalli, M.L.; Marks, S.J.; Montealegre, M.C.; Gilman, R.H.; Pajuelo, M.J.; Saito, M.; Tsukayama, P.; Njenga, S.M.; Kiiru, J.; Swarthout, J.; et al. Urban informal settlements as hotspots of antimicrobial resistance and the need to curb environmental transmission. Nat. Microbiol. 2020, 5, 787–795. [Google Scholar] [CrossRef]
- Cabassi, C.S.; Sala, A.; Santospirito, D.; Alborali, G.L.; Carretto, E.; Ghibaudo, G.; Taddei, S. Activity of AMP2041 against human and animal multidrug resistant Pseudomonas aeruginosa clinical isolates. Ann. Clin. Microb. Anti. 2017, 16, 1–9. [Google Scholar] [CrossRef]
- Cazares, A.; Moore, M.P.; Hall, J.P.J.; Wright, L.L.; Grimes, M.; Emond-Rhéault, J.; Pongchaikul, P.; Santanirand, P.; Levesque, R.C.; Fothergill, J.L.; et al. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Haenni, M.; Bour, M.; Châtre, P.; Madec, J.; Plésiat, P.; Jeannot, K. Resistance of animal strains of Pseudomonas aeruginosa to carbapenems. Front. Microbiol. 2017, 8, 1847. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.; Hu, Y.; Coates, Y. The efficacy of using combination therapy against multi-drug and extensively drug-resistant Pseudomonas aeruginosa in clinical settings. Antibiotics 2022, 11, 323. [Google Scholar] [CrossRef] [PubMed]
- Kracalik, I.; Ham, D.C.; McAllister, G.; Smith, A.R.; Vowles, M.; Kauber, K.; Zambrano, M.; Rodriguez, G.; Garner, K.; Chorbi, K.; et al. Extensively drug-resistant carbapenemase-producing Pseudomonas aeruginosa and medical tourism from the United States to Mexico, 2018–2019. Emerg. Infect. Dis. 2022, 28, 51. [Google Scholar] [CrossRef] [PubMed]
- Faldynova, M.; Pravcova, M.; Sisak, F.; Havlickova, H.; Kolackova, I.; Cizek, A.; Karpiskova, R.; Rychlik, I. Evolution of antibiotic resistance in Salmonella enterica serovar typhimurium strains isolated in the Czech Republic between 1984 and 2002. Antimicrob. Agents Chemother. 2003, 47, 2002–2005. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ying, Y.; Wu, F.; Wu, C.; Jiang, Y.; Yin, M.; Zhou, W.; Zhu, X.; Cheng, C.; Zhu, L.; Li, K.; et al. Florfenicol resistance in Enterobacteriaceae and whole-genome sequence analysis of florfenicol-resistant Leclercia adecarboxylata Strain R25. Int. J. Genom. 2019, 2019, 9828504. [Google Scholar] [CrossRef]
- Jin, Y.; Guo, Y.; Zheng, C.; Liu, W. Effect of heat stress on ileal microbial community of indigenous yellow-feather broilers based on 16S rRNA gene sequencing. Vet. Med. Sci. 2022, 8, 642–653. [Google Scholar] [CrossRef]



| Sample | OTUs | ACE | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|
| W0D0 | 279 | 320.58 | 328.79 | 5.99 | 0.97 |
| W1D0 | 283 | 328.62 | 334.04 | 5.86 | 0.96 |
| W2D0 | 275 | 293.72 | 294.33 | 5.96 | 0.97 |
| W3D0 | 273 | 300.98 | 302.64 | 5.05 | 0.90 |
| W4D0 | 275 | 296.88 | 304.53 | 5.36 | 0.92 |
| W0D7 | 323 | 350.91 | 349.28 | 5.48 | 0.95 |
| W1D7 | 333 | 376.69 | 396.58 | 5.85 | 0.96 |
| W2D7 | 194 | 219.66 | 217.62 | 4.90 | 0.94 |
| W3D7 | 135 | 164.48 | 164.25 | 4.62 | 0.93 |
| W4D7 | 91 | 152.33 | 137.75 | 2.59 | 0.73 |
| W0D30 | 368 | 396.82 | 397.13 | 6.50 | 0.98 |
| W1D30 | 379 | 424.92 | 438.11 | 6.95 | 0.99 |
| W2D30 | 303 | 349.49 | 340.50 | 6.12 | 0.97 |
| W3D30 | 231 | 261.72 | 260.75 | 5.73 | 0.96 |
| W4D30 | 95 | 127.48 | 131.11 | 2.96 | 0.76 |
| W0D60 | 314 | 351.89 | 351.60 | 5.31 | 0.91 |
| W1D60 | 274 | 308.32 | 308.14 | 4.05 | 0.85 |
| W2D60 | 291 | 308.85 | 307.53 | 4.93 | 0.89 |
| W3D60 | 191 | 230.08 | 217.25 | 5.24 | 0.95 |
| W4D60 | 230 | 261.66 | 267.05 | 4.13 | 0.84 |
| Sample | OTUs | ACE | Chao1 | Shannon | Simpson |
|---|---|---|---|---|---|
| S0D0 | 841 | 922.03 | 955.24 | 8.37 | 0.99 |
| S1D0 | 847 | 918.58 | 939.26 | 8.41 | 1.00 |
| S2D0 | 844 | 928.59 | 941.04 | 8.36 | 0.99 |
| S3D0 | 857 | 953.93 | 977.28 | 8.37 | 0.99 |
| S4D0 | 858 | 944.58 | 952.10 | 8.39 | 0.99 |
| S0D7 | 904 | 982.03 | 996.36 | 8.67 | 1.00 |
| S1D7 | 908 | 1002.84 | 1017.90 | 8.61 | 1.00 |
| S2D7 | 902 | 1004.74 | 1049.35 | 8.58 | 1.00 |
| S3D7 | 862 | 941.05 | 954.55 | 8.28 | 0.99 |
| S4D7 | 860 | 944.85 | 947.53 | 7.83 | 0.98 |
| S0D30 | 940 | 1016.88 | 1027.78 | 8.67 | 1.00 |
| S1D30 | 900 | 970.75 | 967.22 | 8.58 | 1.00 |
| S2D30 | 859 | 942.94 | 965.21 | 8.31 | 0.99 |
| S3D30 | 785 | 825.71 | 829.69 | 8.31 | 0.99 |
| S4D30 | 782 | 848.19 | 860.47 | 7.87 | 0.99 |
| S0D60 | 721 | 810.66 | 817.40 | 7.95 | 0.99 |
| S1D60 | 806 | 896.28 | 908.75 | 8.16 | 0.99 |
| S2D60 | 653 | 748.21 | 733.68 | 7.53 | 0.99 |
| S3D60 | 741 | 865.00 | 877.07 | 7.57 | 0.99 |
| S4D60 | 590 | 727.29 | 731.81 | 6.72 | 0.98 |
| Genes | Sequences (5′-3′) | Product size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| floR | F: GCGATATTCATTACTTTGGC | 425 | 54 |
| R: TAGGATGAAGGTGAGGAATG | |||
| optrA | F: CTTATGGATGGTGTGGCAGC | 310 | 59 |
| R: CCATGTGGTTTGTCGGTTCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Ding, Y.; Peng, J.; Dai, Y.; Luo, S.; Liu, W.; Ma, Y. Effects of Broad-Spectrum Antibiotic (Florfenicol) on Resistance Genes and Bacterial Community Structure of Water and Sediments in an Aquatic Microcosm Model. Antibiotics 2022, 11, 1299. https://doi.org/10.3390/antibiotics11101299
Zhang T, Ding Y, Peng J, Dai Y, Luo S, Liu W, Ma Y. Effects of Broad-Spectrum Antibiotic (Florfenicol) on Resistance Genes and Bacterial Community Structure of Water and Sediments in an Aquatic Microcosm Model. Antibiotics. 2022; 11(10):1299. https://doi.org/10.3390/antibiotics11101299
Chicago/Turabian StyleZhang, Tengyue, Yuexia Ding, Jinju Peng, Yue Dai, Shuaishuai Luo, Wenchao Liu, and Yi Ma. 2022. "Effects of Broad-Spectrum Antibiotic (Florfenicol) on Resistance Genes and Bacterial Community Structure of Water and Sediments in an Aquatic Microcosm Model" Antibiotics 11, no. 10: 1299. https://doi.org/10.3390/antibiotics11101299
APA StyleZhang, T., Ding, Y., Peng, J., Dai, Y., Luo, S., Liu, W., & Ma, Y. (2022). Effects of Broad-Spectrum Antibiotic (Florfenicol) on Resistance Genes and Bacterial Community Structure of Water and Sediments in an Aquatic Microcosm Model. Antibiotics, 11(10), 1299. https://doi.org/10.3390/antibiotics11101299






