Abstract
To analyse the epidemiology and population structure of third-generation cephalosporin-resistant (3GCR) and carbapenem-resistant (CR) Klebsiella pneumoniae complex isolates, patients were screened for rectal colonisation with 3GCR/CR K. pneumoniae complex on admission to six German university hospitals (2016–2019). Also collected were 3GCR/CR and susceptible K. pneumoniae isolates from patients with bloodstream infections (2016–2018). Whole-genome sequencing was performed followed by multilocus sequencing typing (MLST), core-genome MLST, and resistome and virulome analysis. The admission prevalence of 3GCR K. pneumoniae complex isolates during the 4-year study period was 0.8%, and 1.0 bloodstream infection per 1000 patient admissions was caused by K. pneumoniae complex (3GCR prevalence, 15.1%). A total of seven K. pneumoniae complex bloodstream isolates were CR (0.8%). The majority of colonising and bloodstream 3GCR isolates were identified as K. pneumoniae, 96.7% and 98.8%, respectively; the remainder were K. variicola and K. quasipneumoniae. cgMLST showed a polyclonal population of colonising and bloodstream isolates, which was also reflected by MLST and virulome analysis. CTX-M-15 was the most prevalent extended-spectrum beta-lactamase, and 29.7% of the colonising and 48.8% of the bloodstream isolates were high-risk clones. The present study provides an insight into the polyclonal 3GCR K. pneumoniae population in German hospitals.
1. Introduction
The Gram-negative bacterium Klebsiella pneumoniae, a member of the family Enterobacterales, is a natural inhabitant of the gastrointestinal tract of humans and animals. It is also encountered as a major cause of hospital- and community-acquired infections, such as urinary and respiratory tract infections, as well as bloodstream infections (BSI) [1,2]. The World Health Organisation listed third-generation cephalosporin-resistant (3GCR) and carbapenem-resistant (CR) K. pneumoniae in 2019 as a “Priority 1: Critical group” organism for which new antimicrobials are urgently needed [3]. Recently, the taxonomic classification of the K. pneumoniae complex, including seven phylogroups, has been revised. On the basis of population studies, the K. pneumoniae complex has been expanded, encompassing the bacterial species K. pneumoniae, K. quasipneumoniae subsp. quasipneumoniae, K. quasipneumoniae subsp. similipneumoniae, K. variicola subsp. variicola, K. variicola subsp. tropica, K. quasivariicola, and K. africana [4,5,6,7,8].
The rapid expansion of 3GCR and CR K. pneumoniae is an increasing public health threat in Europe. In 2020, 3GCR rates of invasive K. pneumoniae isolates were 50% or above in 20% of the countries reporting data to the European Antimicrobial Resistance Surveillance network (EARS-Net), mainly in Southern and Eastern Europe [9]. Furthermore, an admission prevalence study in Germany in 2014 reported a carriage rate of 9.5% for 3GCR Enterobacterales and 0.8% for K. pneumoniae [10]. The 3GCR phenotype is mainly attributed to the presence of extended-spectrum beta-lactamases (ESBLs), such as blaCTX-M-15, AmpC beta-lactamases, SHV hyperproduction, and decreased outer membrane permeability [11,12,13]. In addition, carbapenem resistance in K. pneumoniae mainly mediated by KPC, NDM, OXA-48, and VIM remains at relatively low levels for most European countries including Germany (0.5% in 2020) [9,14]. However, a North/South gradient is evident concerning CR K. pneumoniae, with higher resistance rates observed in Mediterranean countries (e.g., Greece and Italy) compared to Northern European countries [9]. K. pneumoniae is the major species within the complex; nevertheless, other members such as K. variicola or K. quasipneumoniae subsp. quasipneumoniae may also be involved in human infections and associated with carbapenemases (such as blaNDM-9 and blaOXA-181) and are gaining more recognition [15,16,17,18].
The burden of antimicrobial resistance in the hospital environment is tightly linked to the successful spread of certain bacterial clones, known as high-risk (HiR) clones, that are isolated in different geographic locations at different times and are more likely to cause outbreaks [19,20]. Numerous multidrug-resistant (MDR) or extensively drug-resistant (XDR) K. pneumoniae HiR clones have been identified as causing outbreaks around the world [1,21]. The presence of certain plasmids has contributed to the success of some HiR clones, e.g., sequence type (ST) 258 has spread worldwide after the acquisition of a blaKPC-encoding plasmid [1,22]. Furthermore, HiR clones have been reported harbouring hybrid plasmids containing both resistance and virulence genes [23]. Virulence determinants may enable K. pneumoniae to overcome physical and chemical barriers and evade host defence. Cases of hypervirulent strains, which are more virulent than classical K. pneumoniae strains causing severe infections in healthy individuals from the community, have been previously reported [24,25,26]. The capsule of K. pneumoniae is a well-known virulence factor, since it facilitates colonisation and infection by providing defence against the complement system and macrophages [27,28]. In addition, K. pneumoniae may gain an advantage over host cells in acquiring iron by expressing iron scavenging systems such as aerobactin and enterobactin [29,30].
A surveillance study was conducted as part of the multicentre Resistance-Network study (R-Net) of the German Centre for Infection Research (DZIF) at six German university hospitals. To understand the German-wide epidemiology and population structure of 3GCR and CR K. pneumoniae complex isolates, we conducted a genome-based analysis that included isolates from patients colonised on hospital admission, as well as invasive isolates from hospitalised patients with BSI, and focused on the molecular characterisation and the identification of HiR clones and resistance and virulence genes. Because of the large scale of the present multicentre study, the data gained about the colonising and bloodstream K. pneumoniae complex isolates will fill the knowledge gap about the genotypes circulating in the country, the molecular epidemiology and will provide data that could strengthen effective infection control measures.
2. Results
2.1. Antimicrobial Susceptibility and Species Distribution
Between 2016 and 2019, a total of 11,885 patients were screened for rectal colonisation with 3GCR/CR K. pneumoniae complex isolates within 3 days of hospital admission. Of these, 92 patients were found colonised with 3GCR K. pneumoniae complex isolates on hospital admission, accounting for a prevalence of 0.8% (Table 1). Overall, no increase was observed in the prevalence of 3GCR/CR K. pneumoniae complex isolates on hospital admission between 2016 and 2019 (2016, 0.7%; 2017, 0.5%; 2018, 1%; 2019, 0.8%) at the six study centres (Figure 1A and Supplementary Figure S1A). Among 3GCR isolates, 98.9% (n = 91) were resistant to cefotaxime and 69.6% (n = 64) to ceftazidime. A total of 91 K. pneumoniae complex isolates were available for molecular characterisation using WGS. Genotyping identified 88 colonising isolates as K. pneumoniae (96.7%), 2 isolates as K. quasipneumoniae subsp. quasipneumoniae (2.2%), and 1 isolate as K. variicola subsp. variicola (1.1%).

Table 1.
Number of colonising and bloodstream K. pneumoniae complex isolates recovered at six study centres in Germany.
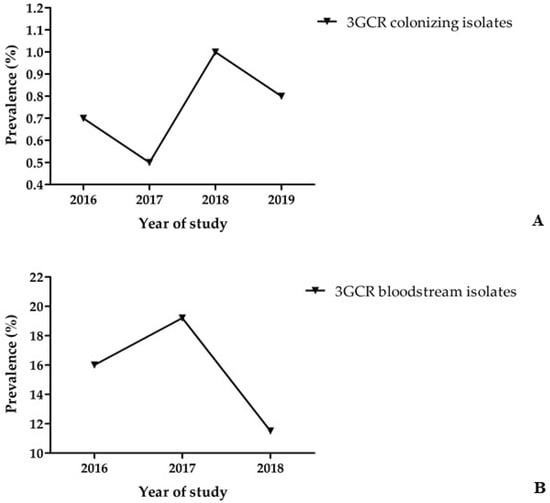
Figure 1.
(A) Prevalence of 3GCR K. pneumoniae complex carriage of patients on hospital admission. (B) Prevalence of 3GCR among 880 K. pneumoniae complex bloodstream isolates.
Within the surveillance study of nosocomial BSI, a total of 880 K. pneumoniae complex bloodstream isolates (Table 1) were recovered from patients at the six study centres between 2016 and 2018, accounting for an incidence of 1.0 per 1000 patient admissions (0.21 K. pneumoniae complex BSI per 1000 patient days). The incidence of 3GCR K. pneumoniae complex BSI was 0.15 per 1000 patient admissions (0.03 3GCR K. pneumoniae complex BSI per 1000 patient days). A total of 133 K. pneumoniae complex isolates were 3GCR, accounting for a prevalence of 15.1% (2016, 16%; 2017, 19.2%; 2018, 11.5%; Figure 1B and Supplementary Figure S1B), while only seven isolates were CR (prevalence, 0.8%). Among the 3GCR BSI isolates, 94% of isolates (n = 125) were resistant to cefotaxime, while 90.2% (n = 120) were resistant to ceftazidime, and 4.5% (n = 6) and 5.3% (n = 7) were resistant to imipenem and meropenem, respectively. Of 133 3GCR/CR isolates, 80 were available for molecular characterisation, of which, 79 isolates were identified as K. pneumoniae (98.8%) and 1 isolate as K. variicola subsp. variicola (1.2%). Finally, between 2016 and 2018, a total of 721 bloodstream infections (Table 1) were caused by 3GCS/CS K. pneumoniae complex, while another 26 bloodstream infections involved K. pneumoniae complex isolates that tested intermediate (cefotaxime MIC, 2 mg/L, and/or ceftazidime MIC, 2–4 mg/L; Table 1). Ninety-five 3GCS/CS isolates were subjected to WGS and molecular typing, and of these, 71 were identified as K. pneumoniae (74.7%), 20 as K. variicola subsp. variicola (21.1%), 3 isolates as K. quasipneumoniae subsp. similipneumoniae (3.2%), and 1 isolate as K. quasipneumoniae subsp. quasipneumoniae (1.1%).
2.2. MLST, HiR Clones, and cgMLST Analysis
Using 7-loci MLST, 91 3GCR K. pneumoniae complex isolates colonising patients on hospital admission were grouped into 58 STs (Table 2, Supplementary Tables S1 and S2). The most prevalent ST was ST307 (n = 10, 11%), followed by ST45 (n = 5, 5.5%) and ST219 (n = 5, 5.5%). Moreover, 29.7% of the identified STs were classified as HiR clones, i.e., ST307 (n = 10), ST14 (n = 4), ST17 (n = 4), ST15 (n = 3), ST20 (n = 2), ST37 (n = 2), ST48 (n = 1), and ST147 (n = 1). cgMLST analysis of the colonising isolates revealed only small clusters of closely related isolates including HiR clone ST307 (n = 6) recovered from study centres A and F in 2017, 2018, and 2019; a cluster of three ST1653 isolates from study centre A collected in 2019; and a cluster of three ST219 isolates from centres B, C, and D. Finally, two additional HiR clone clusters were detected, ST14 (n = 2) in study centre C and ST20 (n = 2) in study centre A (Figure 2 and Supplementary Figure S2). No clusters of closely related isolates were observed for the K. variicola and K. quasipneumoniae colonising isolates.

Table 2.
Overview of the most frequent sequence types of colonising and bloodstream K. pneumoniae complex isolates excluding most singletons.
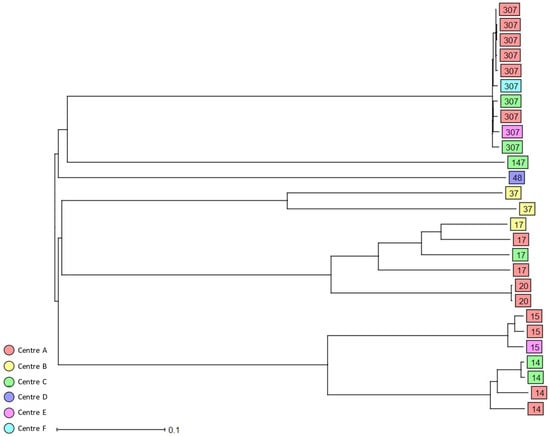
Figure 2.
Dendrogram generated using Ridom SeqSphere+ for the HiR 3GCR/CR colonising K. pneumoniae complex isolates (n = 27) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes.
Among the 3GCR/CR K. pneumoniae complex bloodstream isolates, 39 different STs were identified (Table 2, Supplementary Tables S1 and S2), the most common being HiR clone ST307 (n = 13, 16.2%), HiR clone ST15 (n = 7, 8.8%), HiR clone ST48 (n = 7, 8.8%), ST219 (n = 5, 6.3%), and HiR clone ST147 (n = 4, 5%) (Table 2). Other HiR clones identified were ST101 (n = 3), ST17 (n = 1), ST37 (n = 1), ST258 (n = 1), ST383 (n = 1), and ST395 (n = 1). Using cgMLST analysis, only a few clusters of closely related isolates could be identified, including a cluster of five K. pneumoniae bloodstream isolates assigned as ST219 that were recovered from three study centres, A, B, and C; a cluster of four HiR clone ST48 isolates recovered in 2017 from study centre D; and a cluster of four HiR clone ST307 isolates recovered from study centre A in 2017 and 2018. Finally, six small clusters of two K. pneumoniae isolates each, namely, ST13 (centre A and B), HiR clone ST15 (centre B), HiR clone ST101 (centre C), HiR clone ST147 (centre C), ST607 (centre F), and ST1825 (centres A and C), were also detected in the present study (Figure 3 and Supplementary Figure S3).
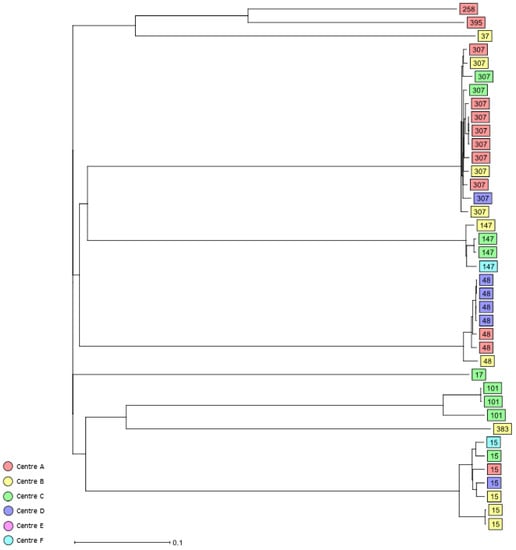
Figure 3.
Dendrogram generated using Ridom SeqSphere+ for the HiR 3GCR/CR K. pneumoniae complex bloodstream isolates (n = 39) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes.
A total of 77 STs were identified among the 3GCS and CS bloodstream isolates (Table 2, Supplementary Tables S1 and S2). HiR clone ST37 (n = 7, 7.4%) was the most common ST, followed by HiR clones ST14 and ST17 (each; n = 3, 3.2%). The HiR clones ST20 and ST101 were singletons. cgMLST analysis revealed the presence of only a few clusters of closely related 3GCS/CS K. pneumoniae isolates. Two ST160 isolates from study centre B and C and recovered in 2017 and 2018, respectively, clustered together, as well as two HiR clone ST14 isolates from centres C and F. Furthermore, two ST3640 isolates recovered from two different patients in centre C in 2018 were identical by cgMLST. Finally, two ST6069 K. pneumoniae isolates from centre D recovered in 2017 and 2018 were identical (Figure 4 and Supplementary Figure S4).
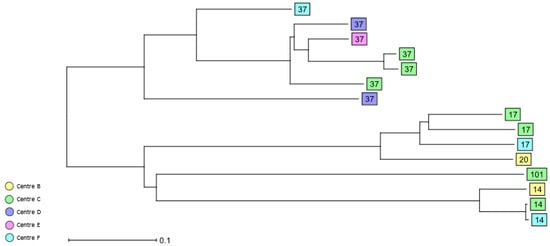
Figure 4.
Dendrogram generated using Ridom SeqSphere+ for the HiR 3GCS/CS K. pneumoniae complex bloodstream isolates (n = 15) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes.
2.3. Acquired Beta-Lactamases
Four families of beta-lactam resistance determinants were identified among the colonising 3GCR K. pneumoniae complex isolates, namely, blaCTX-M-like (n = 84), blaTEM-like (n = 47), blaOXA-1 (n = 24), and blaDHA-1 (n = 2) (Table 3). Among ESBL CTX-M beta-lactamases, the most predominant group was blaCTX-M-15 (n = 68), followed by blaCTX-M-14 (n = 11), blaCTX-M-1 (n = 2), blaCTX-M-27 (n = 1), blaCTX-M-55 (n = 1), and blaCTX-M-65 (n = 1). Furthermore, among the broad-spectrum TEM-1 beta-lactamases, blaTEM-1B (n = 46) was the most common variant identified, and blaTEM-1A was detected in one isolate. Finally, one ST1599 K. variicola isolate recovered from study centre A was CS and therefore excluded from the study but harboured blaOXA-181, an OXA-48-like carbapenemase.

Table 3.
Overview of acquired beta-lactamases of colonising and bloodstream K. pneumoniae complex isolates.
A total of seven families of acquired beta-lactamases (Table 3) were detected in the 3GCR K. pneumoniae complex bloodstream isolates, namely, blaCTX-M-like (n = 65), blaTEM-like (n = 38), blaOXA-like (n = 36), blaDHA-1 (5), blaCMY-4 (n = 1), blaKPC-2 (n = 1), and blaVIM-19 (n = 1). blaCTX-M-15 (n = 59) was the predominant variant among the blaCTX-M-like ESBLs, followed by blaCTX-M-14 (n = 3), blaCTX-M-1 (n = 1), blaCTX-M-3 (n = 1), and blaCTX-M-27 (n = 1). The ESBL blaOXA-1 was detected in 30 bloodstream isolates, while four isolates harboured the narrow-spectrum beta-lactamase blaOXA-9 and two isolates encoded the carbapenemase blaOXA-48. Furthermore, the broad-spectrum TEM beta-lactamase blaTEM-1B (n = 35) was the most common blaTEM-like variant identified, and blaTEM-1A was detected in three isolates. Finally, two families of acquired beta-lactamases were identified among the 3GCS/CS K. pneumoniae complex isolates: broad-spectrum beta-lactamase TEM, blaTEM-1A (n = 1), and blaTEM-1B (n = 2), and narrow-spectrum beta-lactamase OXA, blaOXA-9 (n = 1).
2.4. Virulence Genes
Since various virulence properties may contribute to infectivity and persistence of K. pneumoniae, several virulence-associated factors were analysed in the present study. Forty-nine 3GCR/CR colonising isolates were grouped into 28 different KL types, and 42 isolates were classified as unknowns (Table S2). KL102 (n = 9) was the most prevalent K locus and was identified in 6 of 10 HiR clone ST307 and three of three ST1653 colonising isolates. Furthermore, KL2 (n = 3) associated with biosynthesis of the K2 capsule serotype was identified in three of four HiR clone ST14. Finally, one of five ST45, one of one HiR clone ST48, and one of one ST791 were typed as KL62 (n = 3). The siderophore yersiniabactin ybt locus was identified in 41 3GCR/CR K. pneumoniae complex isolates. The most prevalent yersiniabactin locus was ybt 10 harboured by the integrative and conjugative element ICEKp4 (n = 10), followed by ICEKp3 harbouring ybt 9 (n = 6), ICEKp5 harbouring ybt 14 (n = 6), and plasmid-encoded ybt 4 (n = 4). Two colonising isolates encoded the yersiniabactin ybt 17 co-located in ICEKp10 with colibactin clb 3, while another isolate harboured the aerobactin locus icu 3.
The virulence gene content of the 3GCR/CR K. pneumoniae bloodstream isolates was also investigated. A total of 20 bloodstream isolates were grouped into 11 KL types, while 60 isolates were classified as unknown (Table S2). KL62 (n = 6) was the most common locus associated with the K62 capsular serotype and was identified only in HiR clone ST48 isolates. In addition, KL102 (n = 3) was carried by HiR clone ST307, KL24 (n = 2) by HiR clone ST15, and KL114 (n = 2) by ST219. A total of 34 3GCR/CR isolates carried a siderophore yersiniabactin ybt lineage. The most common locus was ybt 10 harboured by ICEKp4 (n = 18), while ICEKp3 encoding ybt 9 and ICEKp12 carrying ybt 16 were found in five and three bloodstream isolates, respectively. In addition, an ST25 isolate with unknown KL type carried the aerobactin icu 1 and salmochelin iro 1 loci. In the present study, a total of 14 KL loci were identified among the susceptible bloodstream isolates, while 77 were classified as unknown (Table S2). KL2 (n = 4) was the most common capsule type identified in two of four HiR clone ST14, one of two ST39, and one of one ST380 isolates followed by KL14 (n = 2) in one of seven HiR clone ST37 and one of one ST76 isolates. A total of 32 3GCS/CS K. pneumoniae isolates harboured the virulence locus yersiniabactin, and the most common was ybt 9 harboured by ICEKp3 (n = 6), followed by ybt 10 encoded by ICEKp4 (n = 5) and ybt 14 carried by ICEKp5 (n = 4), while five 3GCS/CS bloodstream isolates harboured a colibactin locus. Furthermore, five K. pneumoniae isolates harboured aerobactin loci and six isolates salmochelin, while a total of three isolates harboured the hypermucoidy locus rmpADC.
3. Discussion
Increasing rates of 3GCR K. pneumoniae causing bloodstream infections have been reported by several countries according to EARS-Net. In detail, in 2020, 15% of the reporting countries 3GCR rates were <10%, while in 44% of the countries reporting data to EARS-Net, resistance rates were ≥50%, including mainly countries in the southern and eastern parts of Europe, e.g., Bulgaria (79.1%), Greece (74.5%), and Romania (67.9%) [9]. In the present multicentre surveillance study addressing the prevalence of 3GCR K. pneumoniae complex colonising patients on hospital admission, the carriage rate over the four-year study period remained stable, ranging between 0.5 and 1%. This result is in agreement with a previous survey conducted in Germany in 2014 reporting an admission prevalence of 0.8% for 3GCR K. pneumoniae [10]. In the present three-year surveillance study of patients with bloodstream infections, the prevalence of 3GCR isolates of K. pneumoniae complex was found to be 15.1% (2016, 16%; 2017, 19.2%; 2018, 11.5%), showing a slight decrease over the study period. Resistance to third-generation cephalosporins was mainly mediated by blaCTX-M-like betalactamases, with no difference between colonising and bloodstream isolates. EARS-Net reported for Germany slightly lower 3GCR rates in K. pneumoniae isolates causing invasive infections, i.e., 13.6% in 2016, 14.6% in 2017, and 12.9% in 2018 [9]. Finally, no colonising CR K. pneumoniae complex isolates were detected in the present study, while the prevalence of carbapenem resistance among bloodstream isolates was rather low (0.8%). Similar results were also reported by EARS-Net and other studies, suggesting that carbapenem resistance in K. pneumoniae complex is currently not a threat in Germany [9,10].
In the present study, the vast majority of 3GCR/CR bloodstream isolates (98.8%) were K. pneumoniae, confirming that this species is the major cause of invasive K. pneumoniae complex infections in healthcare settings in Germany [4,31]. Similarly, 96.7% of the colonising isolates were identified as K. pneumoniae. However, 25.4% 3GCS/CS bloodstream isolates presumptively identified as K. pneumoniae by the participating study centres were re-identified as non-K. pneumoniae by sequencing, highlighting that susceptible K. variicola and K. quasipneumoniae isolates are often causing bloodstream infections in Germany and are underreported. Similar frequencies of non-K. pneumoniae bloodstream infections, i.e., 24.4% K. variicola and 6.5% K. quasipneumoniae, were reported in the Stockholm area [16].
Overall, both colonising and bloodstream K. pneumoniae complex isolates were classified into a large number of STs (3GCR/CR colonising, n = 58; 3GCR/CR, n = 39; and 3GCS/CS, n = 77 bloodstream isolates), indicating that the population structure of K. pneumoniae in German hospitals is heterogenous and consists of multiple genotypes with even more diversity among 3GCS/CS vs. 3GCR/CR bloodstream isolates. K. pneumoniae HiR ST307 was the most frequent clone among the 3GCR/CR colonising and bloodstream isolates in the present study, accounting for 10 (11%) and 13 (16.2%) of isolates, respectively. HiR ST307 has a global distribution and has caused numerous outbreaks in healthcare settings worldwide [32]. Furthermore, an increasing prevalence has been observed for ST307 carbapenemase-producing K. pneumoniae isolates, even replacing other successful HiR clones such as ST258 in Italy and Colombia [21,33]. Outbreaks of XDR ST307 carrying resistance plasmids encoding either NDM-1, CTX-M-15, or OXA-48, have been reported previously in four medical facilities in north-eastern Germany [34,35]. Among the 3GCR colonising K. pneumoniae isolates, 29.7% were identified as HiR, as were 48.8% of the 3GCR/CR bloodstream isolates. When only the ST is used as inclusion criterion and the susceptible phenotype is not taken into account, HiR clones were also identified among 3GCS/CS bloodstream isolates, including ST14, ST17, ST20, ST37, and ST101, accounting for 15.8% of these isolates. These data suggest that although various K. pneumoniae HiR clones are circulating in the six German university hospitals participating in the study, no particular clone was predominant.
The cgMLST analysis of both colonising and bloodstream K. pneumoniae complex isolates revealed only a few small clusters of closely related K. pneumoniae isolates, indicating potential transmission events, whereas the non-K. pneumoniae isolates were all singletons. Representatives of HiR clones such as ST13, ST15, ST48, ST101, ST147, and ST307 also formed small clusters comprising between two and six isolates that were mainly centre-specific and likely reflect the local epidemiology of each study centre, while isolates representing other well-known HiR clones such as ST258 were singletons. Numerous previous studies have reported the presence of HiR clones such as ST101, ST147, or ST258 in different hospitals in Germany, highlighting that these strains were already circulating in the country [36,37,38].
Resistance to 3GC was caused mainly by ESBLs of the CTX-M-1 group (CTX-M-1, CTX-M-3, CTX-M-15), found in 76.9% of 3GCR colonising and 76.2% of bloodstream isolates, and of the CTX-M-9 group (CTX-M-14, CTX-M-27, CTX-M-55, CTX-M-65), found in 15.4% of colonising and 5% of bloodstream isolates. Consistent with our results, the CTX-M-1 group was the predominant ESBL group in K. pneumoniae in a survey of admission prevalence in Germany in 2014 and 2015 as also the case in other regions of Europe [10,39,40]. The high prevalence of ESBLs, such as CTX-M-15, in the polyclonal population of colonising and bloodstream isolates of the present study may suggest that horizontal plasmid transmission or dissemination of other mobile genetic elements may be responsible for the spread of acquired beta-lactamases, which needs to be further investigated. Of note, three of four carbapenemases detected in bloodstream isolates at study sites B and C were found in HiR clones, i.e., OXA-48, ST101; KPC-2, ST258; and VIM-19, ST383, which also co-harboured ESBLs and other antimicrobial resistance genes. In Enterobacterales, OXA-48, VIM-1, and KPC-2 are the most frequently detected carbapenemases in Germany, as reported by the German National Center for Multidrug-Resistant Gram-Negative Bacteria and have been frequently involved in healthcare-associated infections and outbreaks [37,38,41,42].
Numerous studies have reported that hypervirulent K. pneumoniae, a pathotype that is more virulent than classical K. pneumoniae, carries large virulence plasmids and causes infections in healthy individuals in the community. Furthermore, increased virulence has been associated with certain capsular serotypes, e.g., K1 and K2 [4,24,26]. In the present study, KL102 was the most frequent capsule locus among the colonising isolates, mainly detected in HiR ST307 (67%), concurring with previous findings from a K. pneumoniae ST307 outbreak in north-eastern Germany [35]. Conversely, KL62 was the most frequent capsular serotype among 3GCR/CR bloodstream isolates and was only detected in HiR ST48 isolates as previously reported in China [43]. Different acquired virulence factors such as capsule and aerobactin production have been linked to hypervirulent strains [24,26]. Siderophores such as yersiniabactin were identified in the present study in 45%, 42.5%, and 33.7% of the 3GCR/CR colonising, 3GCR/CR, and 3GCS/CS bloodstream isolates, respectively. Furthermore, aerobactin, colibactin, salmochelin, and the hypermucoidy determinant rmpA were also detected in a small number of isolates. In the present study, the diverse population structure of Klebsiella spp. isolates was also confirmed by the heterogeneity of acquired virulence factors.
To the best of our knowledge, the present work represents the largest study on the molecular epidemiology of 3GCR/CR colonising and bloodstream K. pneumoniae complex isolates in Germany; however, our study has several limitations. The data obtained from the admission prevalence study are not representative of the general population because many patients admitted to tertiary care hospitals represent a patient population with frequent hospital contacts and colonisation may have occurred during previous healthcare contacts. In addition, the present study did not investigate whether 3GCR/CR bloodstream infections were caused by the same isolates found at hospital admission.
In summary, the results of the present study describe the polyclonal population of 3GCR K. pneumoniae complex isolates that colonise patients on hospital admission and cause bloodstream infections. Although certain K. pneumoniae genotypes were more frequent, no extensive clonality was detected among the isolates, suggesting that a diverse 3GCR K. pneumoniae population circulates in German hospitals. MLST and virulome analysis supported these findings. Finally, our data demonstrate the presence of HiR clones circulating in Germany and provide epidemiological data on 3GCR and CR K. pneumoniae complex isolates that may contribute to more effective infection control.
4. Materials and Methods
4.1. Study Participants and Design
Two independent epidemiologic surveys involving patients (i) at hospital admission and (ii) with nosocomial BSI were conducted at six German tertiary care university hospitals in eastern (centre A), western (centre B), south-western (centres C and F), central (centre D), and northern (centre E) Germany.
In the prospective epidemiological study on the prevalence of 3GCR and/or CR (3GCR/CR) K. pneumoniae complex on hospital admission, patients aged ≥18 years were included between June 2016 and December 2019. Centre F did not participate in the survey in 2016. Excluded were patients from the departments of ophthalmology, paediatrics, psychiatry, and from intensive care units (ICUs). 3GCR/CR K. pneumoniae complex bloodstream isolates were collected from patients aged ≥18 years between October 2016 and December 2018. Patients from the departments of ophthalmology, paediatrics, and psychiatry were excluded. Finally, a random sample of third-generation cephalosporin-susceptible (3GCS) and carbapenem-susceptible (CS) K. pneumoniae complex bloodstream isolates collected between 2016 and 2018 at the same study centres were used as controls.
4.2. Species Identification and Antimicrobial Susceptibility Testing
Species identification was performed using MALDI-TOF MS (Bruker Daltonics GmbH, Bremen, Germany). Antimicrobial susceptibility testing (AST) for cefotaxime, ceftazidime, imipenem, and meropenem was carried out using VITEK®2 (bioMérieux, Nürtingen, Germany). VITEK®2 was routinely validated using characterised quality control strains. Isolates that were non-susceptible to cefotaxime and/or ceftazidime (MIC ≥ 2 mL/L) as well as isolates non-susceptible to imipenem and/or meropenem (MIC ≥ 4 mL/L) were included in the study and further characterised on the basis of the EUCAST resistance breakpoints for Enterobacterales (Version 6.0, January 2016, http://www.eucast.org/, accessed on 1 April 2022).
4.3. Whole-Genome Sequencing (WGS)
Total DNA was extracted using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Sequencing libraries were prepared using the Nextera XT library prep kit (Illumina GmbH, Munich, Germany) for a 250 bp paired-end sequencing run on a MiSeq (Illumina GmbH) platform. The genomes were assembled de novo using Velvet [44].
4.4. Molecular Species Identification, Molecular Epidemiology, and High-Risk Clones
The species-intrinsic beta-lactamase gene (blaSHV in K. pneumoniae, blaLEN in K. variicola, blaOKP in K. quasipneumoniae) were used as indicators for species identification. Furthermore, the genotyping tool Kleborate (https://github.com/katholt/Kleborate, accessed on 1 March 2022) was used to identify the species using genome assemblies [31]. Finally, JSpeciesWS was used to further confirm the species of the isolates identified as non-K. pneumoniae [45]. The Klebsiella spp. Multi-locus sequence typing (MLST) scheme was used to assign the ST (https://bigsdb.web.pasteur.fr/, accessed on 1 March 2022). The molecular epidemiology was investigated using a K. pneumoniae sensu lato core genome MLST (cgMLST) scheme (https://www.ridom.de/seqsphere/u/Task_Template_Sphere.html, accessed on 1 April 2022), including 2358 target alleles, using the Ridom SeqSphere+ v.8.3.0 software (Ridom GmbH, Münster, Germany) [46]. Genotypes differing in ≤15 alleles were defined as closely related. Isolates with the following STs: ST11, ST14, ST15, ST16, ST17, ST20, ST37, ST48, ST101, ST147, ST258, ST307, ST336, ST340, ST383, ST395, ST512 were classified as HiR clones, i.e., clones that caused at least four recognised outbreaks and were reported in ≥10 countries [1], but also including well described emerging K. pneumoniae lineages previously designated as HiR by various researchers [21,23,47,48,49].
4.5. Antimicrobial Resistance and Virulence Genes
ABRicate v1.0.1 (https://github.com/tseemann/ABRicate (accessed on 1 February 2022)) (with options--minid 90--mincov 90) was used to screen for acquired antimicrobial resistance genes with ResFinder (database date 1 February 2022) [50]. Kaptive, integrated in Kleborate, was used with the default options to assign the K. pneumoniae isolates to capsular locus types (KL). In detail, results with a K locus confidence “perfect”, “very high”, “high”, and “good” were counted, while genomes with “low” or “none” confidence Kaptive calls were classified as unknowns. Finally, the same platform was used to screen the isolates for five important acquired virulence loci, i.e., the siderophores yersiniabactin (ybt), aerobactin (iuc), and salmochelin (iro); the genotoxin colibactin (clb); and the hypermucoidy locus rmpADC [31,51].
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics11101286/s1. Table S1: Overview of the STs of the K. pneumoniae complex colonising and bloodstream isolates including singletons. Table S2: Overview of the species, STs, and virulence factors of the K. pneumoniae complex 3GCR/CR colonising (a), 3GCR/CR (b), and 3GCS/CS (c) bloodstream isolates. Figure S1. (A) Prevalence of 3GCR K. pneumoniae complex carriage of patients on hospital admission per year and centre. (B) Prevalence of 3GCR among 880 K. pneumoniae complex bloodstream isolates per year and centre. Figure S2. Dendrogram generated using Ridom SeqSphere+ for the 3GCR/CR colonising K. pneumoniae complex isolates including HiR clones (n = 91) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes. Figure S3. Dendrogram generated using Ridom SeqSphere+ for the 3GCR/CR K. pneumoniae complex bloodstream isolates including HiR clones (n = 80) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes. Figure S4. Dendrogram generated using Ridom SeqSphere+ for the 3GCS/CS K. pneumoniae complex bloodstream isolates including HiR clones (n = 95) coloured by study centre and identified by ST type, ignoring missing values. Each box represents one isolate from an individual patient according to sequence analysis of 2358 cgMLST target genes.
Author Contributions
Conceptualisation, P.G. and H.S.; data curation, K.X., C.I., S.V.W., M.B., A.G.D., S.E., P.G., H.G., N.K., W.V.K., A.K., E.K., K.L., A.M., S.P., A.M.R., J.R., E.T., D.T., M.J.G.T.V., J.W. and P.G.H., DZIF R-Net Study Group; investigation, K.X., C.I., S.V.W., M.B., A.G.D., S.E., P.G., H.G., N.K., W.V.K., A.K., E.K., K.L., A.M., S.P., A.M.R., J.R., E.T., D.T., M.J.G.T.V., J.W. and P.G.H., DZIF R-Net Study Group; supervision, H.S.; writing—original draft, K.X. and P.G.H.; writing—review and editing, C.I., S.V.W. and H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the German Centre for Infection Research (DZIF), project number TTU 08.811.
Institutional Review Board Statement
The study addressing the prevalence of 3GCR and/or CR K. pneumoniae complex on hospital admission was approved by an institutional review board (approval number 14–170).
Informed Consent Statement
Surveillance was performed with patients’ informed consent.
Data Availability Statement
The raw sequencing reads generated in this project were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena/) under the study accession number PRJEB39867.
Acknowledgments
We would like to thank Ahmad Saleh, Vivien Persy, Carina Müller, and Michael Sonnabend for excellent technical assistance. The results have been partially presented as poster presentation at the Joint Annual Meeting German Society of Infectious Diseases and German Centre for Infection Research (2019, Bad Nauheim, Germany and 2022, Stuttgart, Germany) and at the 31st and 32nd European Congress of Clinical Microbiology and Infectious Diseases (2021, online conference and 2022, Lisbon, Portugal). We thank the Institute Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at http://bigsdb.pasteur.fr/ (accessed on 17 March 2022). In addition to the authors, the following members of the DZIF R-Net Study Group (in alphabetical order) contributed to the study: Lena Biehl, Cologne; Trinad Chakraborty, Giessen; Nadine Hoffmann, Tübingen; Florian Hölzl, Tübingen; Baris Bader, Tübingen; Michael Buhl, Tübingen; Frieder Fuchs, Cologne; Georg Häcker, Freiburg; Nathalie Jazmati, Cologne; Dana Lenke, Lübeck; Luis Alberto Peña Diaz, Berlin; Gabriele Peyerl-Hoffmann, Freiburg; Georg Pilarski, Berlin; Susanna Proske, Cologne; Sabine Schuster, Freiburg; Norbert Thoma, Berlin; Martina Vavra, Freiburg; Anna Weber, Berlin. We acknowledge support for the Article Processing Charge from the DFG (German Research Foundation, 491454339).
Conflicts of Interest
C.I. has served as a consultant to MSD Sharp and Dohme and Shionogi and has received lecture fees from MSD Sharp and Dohme. S.P. consulted for IDbyDNA and received speaker’s honoraria from bioMérieux. MJGTV received research grants from 3M, Astellas Pharma, Biontech, DaVolterra, Evonik, Gilead Sciences, Glycom, Immunic, MaaT Pharma, Merck/MSD, Organobalance, Seres Therapeutics, and Takeda Pharmaceutical and speaker fees or consultation fees from Alb Fils Kliniken GmbH, Arderypharm, Astellas Pharma, Basilea, bioMérieux, DaVolterra, Farmak International Holding GmbH, Ferring, Gilead Sciences, Immunic AG, MaaT Pharma, Merck/MSD, Pfizer, Roche, Organobalance, and SocraTec R&D GmbH. The remaining authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Holt, K.E. Klebsiella pneumoniae Population Genomics and Antimicrobial-Resistant Clones. Trends Microbiol. 2016, 24, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Holt, K.E.; Wertheim, H.; Zadoks, R.N.; Baker, S.; Whitehouse, C.A.; Dance, D.; Jenney, A.; Connor, T.R.; Hsu, L.Y.; Severin, J.; et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 2015, 112, E3574–E3581. [Google Scholar] [CrossRef]
- Blin, C.; Passet, V.; Touchon, M.; Rocha, E.P.C.; Brisse, S. Metabolic diversity of the emerging pathogenic lineages of Klebsiella pneumoniae. Environ. Microbiol. 2017, 19, 1881–1898. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez, L.; Silva, J.; Martinez-Romero, E. Klebsiella variicola, A Novel Species with Clinical and Plant-Associated Isolates. Syst. Appl. Microbiol. 2004, 27, 27–35. [Google Scholar] [CrossRef]
- Brisse, S.; Passet, V.; Grimont, P.A.D. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 2014, 64, 3146–3152. [Google Scholar] [CrossRef]
- Long, S.W.; Linson, S.E.; Saavedra, M.O.; Cantu, C.; Davis, J.J.; Brettin, T.; Olsen, R.J. Whole-Genome Sequencing of a Human Clinical Isolate of the Novel Species Klebsiella quasivariicola sp. nov. Genome Announc. 2017, 5, e01057-17. [Google Scholar] [CrossRef]
- WHO Regional Office for Europe and European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe, 2020 data. In Executive Summary; WHO Regional Office for Europe: Copenhagen, Denmark, 2021. [Google Scholar]
- Hamprecht, A.; Rohde, A.M.; Behnke, M.; Feihl, S.; Gastmeier, P.; Gebhardt, F.; Kern, W.V.; Knobloch, J.K.; Mischnik, A.; Obermann, B.; et al. Colonization with third-generation cephalosporin-resistant Enterobacteriaceae on hospital admission: Prevalence and risk factors. J. Antimicrob. Chemother. 2019, 71, 2957–2963. [Google Scholar] [CrossRef][Green Version]
- Peirano, G.; Pitout, J.D.D. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Petit, A.; Ben Yaghlane-Bouslama, H.; Sofer, L.; Labia, R. Does high level production of SHV-type penicillinase confer resistance to ceftazidime in enterobacteriaceae? FEMS Microbiol. Lett. 1992, 92, 89–94. [Google Scholar] [CrossRef][Green Version]
- Paterson, D.L. Resistance in Gram-Negative Bacteria: Enterobacteriaceae. Am. J. Med. 2006, 119, S20–S28. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69 (Suppl. 7), S521–S528. [Google Scholar] [CrossRef] [PubMed]
- Breurec, S.; Melot, B.; Hoen, B.; Passet, V.; Schepers, K.; Bastian, S.; Brisse, S. Liver Abscess Caused by Infection with Community-Acquired Klebsiella quasipneumoniae subsp. quasipneumoniae. Emerg. Infect. Dis. 2016, 22, 529–531. [Google Scholar] [CrossRef] [PubMed]
- Maatallah, M.; Vading, M.; Kabir, M.H.; Bakhrouf, A.; Kalin, M.; Nauclér, P.; Brisse, S.; Giske, C.G. Klebsiella variicola Is a Frequent Cause of Bloodstream Infection in the Stockholm Area, and Associated with Higher Mortality Compared to K. pneumoniae. PLoS ONE 2014, 9, e113539. [Google Scholar] [CrossRef]
- Zurfluh, K.; Poirel, L.; Nordmann, P.; Klumpp, J.; Stephan, R. First detection of Klebsiella variicola producing OXA-181 carbapenemase in fresh vegetable imported from Asia to Switzerland. Antimicrob. Resist. Infect. Control 2015, 4, 3. [Google Scholar] [CrossRef]
- Di, D.Y.W.; Jang, J.; Unno, T.; Hur, H.-G. Emergence of Klebsiella variicola positive for NDM-9, a variant of New Delhi metallo-β-lactamase, in an urban river in South Korea. J. Antimicrob. Chemother. 2017, 72, 1063–1067. [Google Scholar] [CrossRef][Green Version]
- Woodford, N.; Turton, J.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef]
- Chmelnitsky, I.; Shklyar, M.; Hermesh, O.; Navon-Venezia, S.; Edgar, R.; Carmeli, Y. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J. Antimicrob. Chemother. 2013, 68, 2178. [Google Scholar] [CrossRef][Green Version]
- Peirano, G.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D.D. Emerging Antimicrobial-Resistant High-Risk Klebsiella pneumoniae Clones ST307 and ST147. Antimicrob. Agents Chemother. 2020, 64, e01148-20. [Google Scholar] [CrossRef]
- Bowers, J.R.; Kitchel, B.; Driebe, E.M.; MacCannell, D.R.; Roe, C.; Lemmer, D.; De Man, T.; Rasheed, J.K.; Engelthaler, D.M.; Keim, P.; et al. Genomic Analysis of the Emergence and Rapid Global Dissemination of the Clonal Group 258 Klebsiella pneumoniae Pandemic. PLoS ONE 2015, 10, e0133727. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.; Davies, F.; Turton, J.; Perry, C.; Payne, Z.; Pike, R. Hybrid Resistance and Virulence Plasmids in “High-Risk” Clones of Klebsiella pneumoniae, Including Those Carrying blaNDM-5. Microorganisms 2019, 7, 326. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2021, 12, 642484. [Google Scholar] [CrossRef]
- Shon, A.S.; Bajwa, R.P.S.; Russo, T.A. Hypervirulent (hypermucoviscous)Klebsiella pneumoniae: A new and dangerous breed. Virulence 2013, 4, 107–118. [Google Scholar] [CrossRef]
- Cortés, G.; Borrell, N.; de Astorza, B.; Gómez, C.; Sauleda, J.; Albertí, S. Molecular Analysis of the Contribution of the Capsular Polysaccharide and the Lipopolysaccharide O Side Chain to the Virulence of Klebsiella pneumoniae in a Murine Model of Pneumonia. Infect. Immun. 2002, 70, 2583–2590. [Google Scholar] [CrossRef]
- Favre-Bonté, S.; Licht, T.R.; Forestier, C.; Krogfelt, K.A. Klebsiella pneumoniae Capsule Expression Is Necessary for Colonization of Large Intestines of Streptomycin-Treated Mice. Infect. Immun. 1999, 67, 6152–6156. [Google Scholar] [CrossRef]
- Bachman, M.A.; Miller, V.L.; Weiser, J.N. Mucosal Lipocalin 2 Has Pro-Inflammatory and Iron-Sequestering Effects in Response to Bacterial Enterobactin. PLOS Pathog. 2009, 5, e1000622. [Google Scholar] [CrossRef]
- Russo, T.A.; Olson, R.; MacDonald, U.; Metzger, D.; Maltese, L.M.; Drake, E.J.; Gulick, A. Aerobactin Mediates Virulence and Accounts for Increased Siderophore Production under Iron-Limiting Conditions by Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 2014, 82, 2356–2367. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Esteban-Cantos, A.; Aracil, B.; Bautista, V.; Ortega, A.; Lara, N.; Saez, D.; Fernández-Romero, S.; Perez-Vazquez, M.; Navarro, F.; Grundmann, H.; et al. The Carbapenemase-Producing Klebsiella pneumoniae Population Is Distinct and More Clonal than the Carbapenem-Susceptible Population. Antimicrob. Agents Chemother. 2017, 61, e02520-16. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; Feudi, C.; Fortini, D.; Brisse, S.; Passet, V.; Bonura, C.; Endimiani, A.; Mammina, C.; Ocampo, A.M.; Jiménez, J.N.; et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 2017, 3, e000110. [Google Scholar] [CrossRef]
- Haller, S.; Kramer, R.; Becker, K.; A Bohnert, J.; Eckmanns, T.; Hans, J.B.; Hecht, J.; Heidecke, C.-D.; Hübner, N.-O.; Kramer, A.; et al. Extensively drug-resistant Klebsiella pneumoniae ST307 outbreak, north-eastern Germany, June to October 2019. Eurosurveillance 2019, 24, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Heiden, S.E.; Hübner, N.-O.; Bohnert, J.A.; Heidecke, C.-D.; Kramer, A.; Balau, V.; Gierer, W.; Schaefer, S.; Eckmanns, T.; Gatermann, S.; et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020, 12, 113. [Google Scholar] [CrossRef]
- Becker, L.; Kaase, M.; Pfeifer, Y.; Fuchs, S.; Reuss, A.; Von Laer, A.; Abu Sin, M.; Korte-Berwanger, M.; Gatermann, S.; Werner, G. Genome-based analysis of Carbapenemase-producing Klebsiella pneumoniae isolates from German hospital patients, 2008-2014. Antimicrob. Resist. Infect. Control 2018, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulou, K.; Carattoli, A.; Wille, J.; Biehl, L.M.; Rohde, H.; Farowski, F.; Krut, O.; Villa, L.; Feudi, C.; Seifert, H.; et al. Antibiotic Resistance and Mobile Genetic Elements in Extensively Drug-Resistant Klebsiella pneumoniae Sequence Type 147 Recovered from Germany. Antibiotics 2020, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- E Zautner, A.; Bunk, B.; Pfeifer, Y.; Spröer, C.; Reichard, U.; Eiffert, H.; Scheithauer, S.; Groß, U.; Overmann, J.; Bohne, W. Monitoring microevolution of OXA-48-producing Klebsiella pneumoniae ST147 in a hospital setting by SMRT sequencing. J. Antimicrob. Chemother. 2017, 72, 2737–2744. [Google Scholar] [CrossRef]
- Mugnaioli, C.; Luzzaro, F.; De Luca, F.; Brigante, G.; Perilli, M.; Amicosante, G.; Stefani, S.; Toniolo, A.; Rossolini, G.M. CTX-M-Type Extended-Spectrum β-Lactamases in Italy: Molecular Epidemiology of an Emerging Countrywide Problem. Antimicrob. Agents Chemother. 2006, 50, 2700–2706. [Google Scholar] [CrossRef]
- Eckert, C.; Gautier, V.; Saladin-Allard, M.; Hidri, N.; Verdet, C.; Ould-Hocine, Z.; Barnaud, G.; Delisle, F.; Rossier, A.; Lambert, T.; et al. Dissemination of CTX-M-Type β-Lactamases among Clinical Isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 2004, 48, 1249–1255. [Google Scholar] [CrossRef]
- Pfennigwerth, N.; Schauer, J. Bericht des Nationalen Referenzzentrums für gramnegative Krankenhauserreger–Zeitraum 1. Januar 2020 bis 31. Dezember 2020. Epid Bull. 2021, 36, 4–11. [Google Scholar]
- Koppe, U.; Von Laer, A.; Kroll, L.E.; Noll, I.; Feig, M.; Schneider, M.; Claus, H.; Eckmanns, T.; Abu Sin, M. Carbapenem non-susceptibility of Klebsiella pneumoniae isolates in hospitals from 2011 to 2016, data from the German Antimicrobial Resistance Surveillance (ARS). Antimicrob. Resist. Infect. Control 2018, 7, 1–12. [Google Scholar] [CrossRef]
- Wang, B.; Pan, F.; Wang, C.; Zhao, W.; Sun, Y.; Zhang, T.; Shi, Y.; Zhang, H. Molecular epidemiology of Carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int. J. Infect. Dis. 2020, 93, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haeseler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Weber, R.E.; Pietsch, M.; Frühauf, A.; Pfeifer, Y.; Martin, M.; Luft, D.; Gatermann, S.; Pfennigwerth, N.; Kaase, M.; Werner, G.; et al. IS26-Mediated Transfer of blaNDM–1 as the Main Route of Resistance Transmission During a Polyclonal, Multispecies Outbreak in a German Hospital. Front. Microbiol. 2019, 10, 2817. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Jousset, A.B.; Chiarelli, A.; Emeraud, C.; Glaser, P.; Naas, T.; Dortet, L. Emergence of New Non–Clonal Group 258 High-Risk Clones among Klebsiella pneumoniae Carbapenemase–Producing K. pneumoniae Isolates, France. Emerg. Infect. Dis. 2020, 26, 1212–1220. [Google Scholar] [CrossRef]
- Palmieri, M.; Wyres, K.L.; Mirande, C.; Qiang, Z.; Liyan, Y.; Gang, C.; Goossens, H.; van Belkum, A.; Ping, L.Y. Genomic evolution and local epidemiology of Klebsiella pneumoniae from a major hospital in Beijing, China, over a 15 year period: Dissemination of known and novel high-risk clones. Microb. Genom. 2021, 7, 14. [Google Scholar] [CrossRef]
- Di Pilato, V.; Errico, G.; Monaco, M.; Giani, T.; Del Grosso, M.; Antonelli, A.; David, S.; Lindh, E.; Camilli, R.; Aanensen, D.M.; et al. The changing epidemiology of carbapenemase-producingKlebsiella pneumoniaein Italy: Toward polyclonal evolution with emergence of high-risk lineages. J. Antimicrob. Chemother. 2021, 76, 355–361. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Wick, R.R.; Heinz, E.; Holt, K.E.; Wyres, K.L. Kaptive web: User-Friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018, 56, e00197-18. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).