Synergy between Mecillinam and Ceftazidime/Avibactam or Avibactam against Multi-Drug-Resistant Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae
Abstract
1. Introduction
2. Results
2.1. A Synergistic Effect between Mecillinam and Ceftazidime/Avibactam or Avibactam on Carbapenemase-Producing Strains
2.2. Mecillinam and Avibactam Combination Treatment in Time–Kill Assays of K. pneumoniae Indicates Synergism
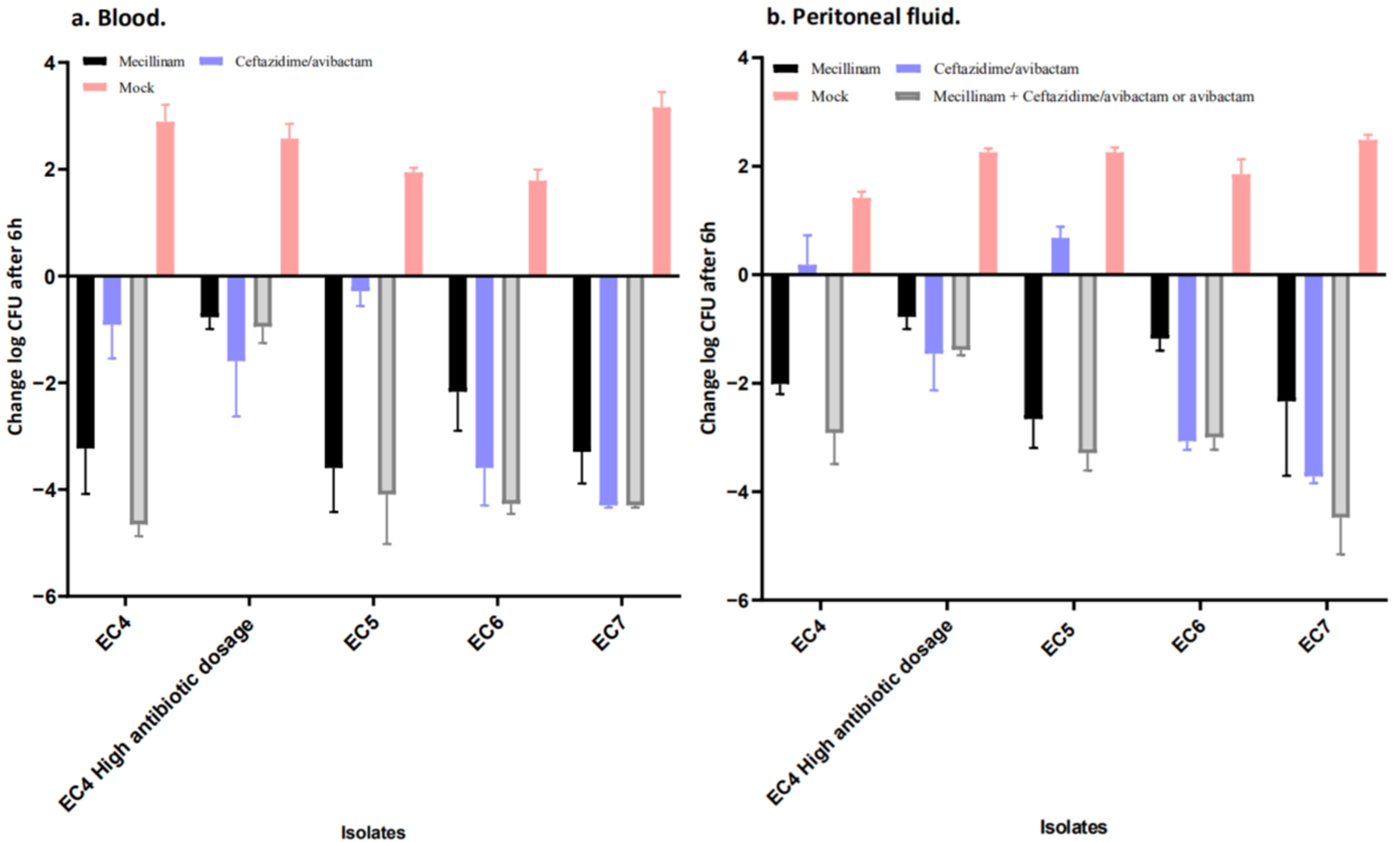
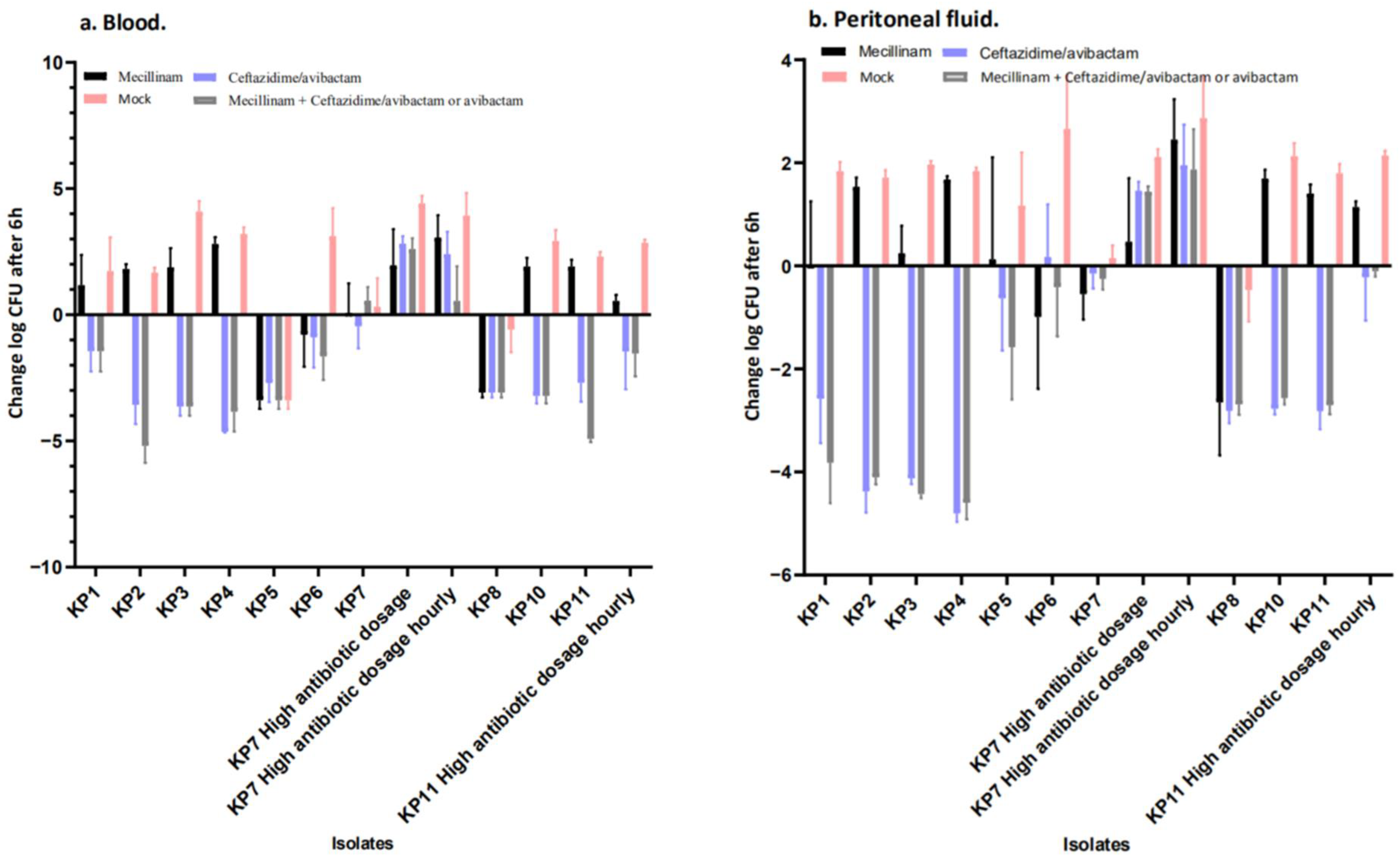
2.3. Mecillinam Combination Treatment with Avibactam or Ceftazidime/Avibactam In Vivo
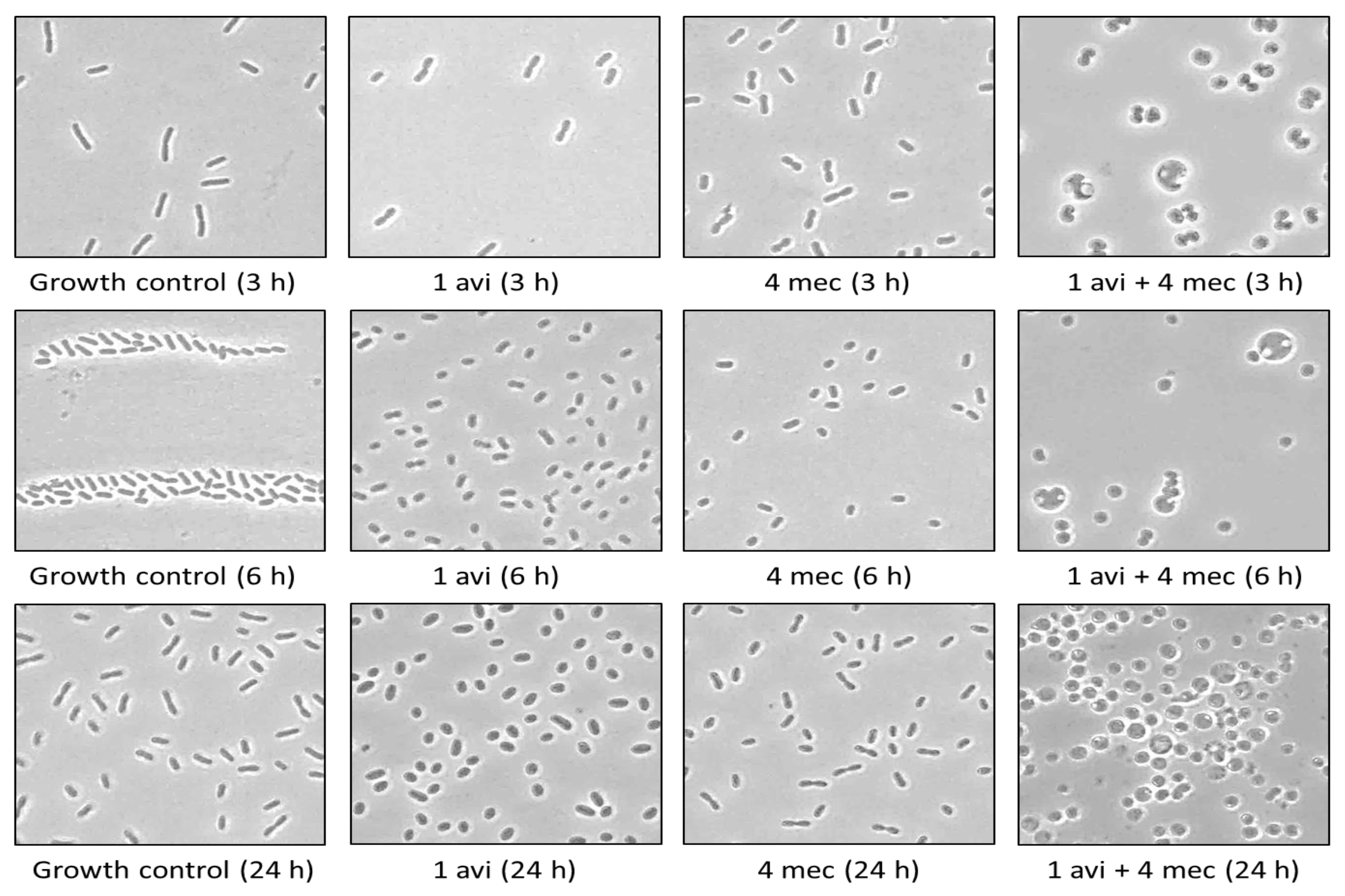
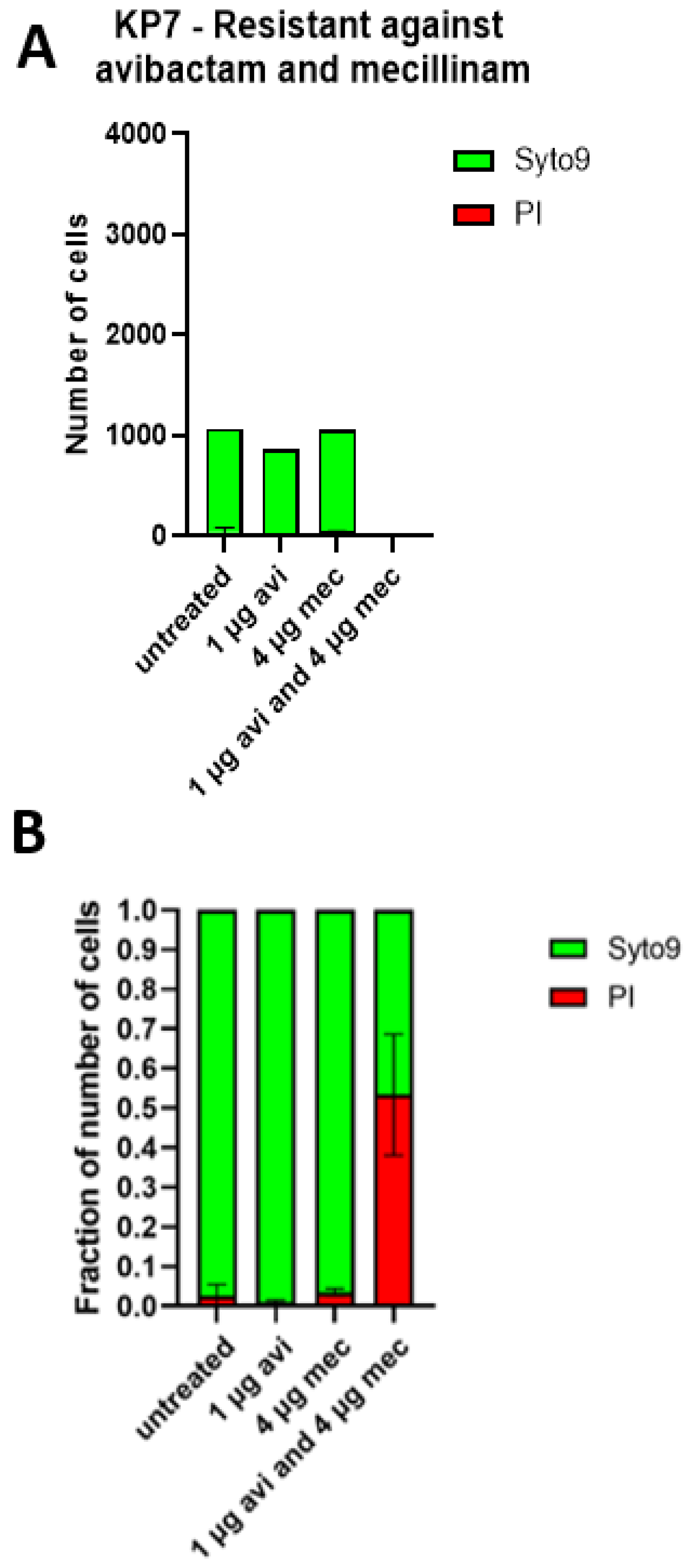
2.4. The Cell Shapes of KP7 and KP11 Are Strongly Affected by the Mecillinam and Avibactam Combination In Vitro and Ex Vivo
2.5. Combination of Mecillinam and Avibactam Does Not Alter Their Specific Interaction with Penicillin-Binding Protein In Vitro
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Antibiotics
4.3. Broth and Growth Conditions
4.4. Whole-Genome Sequencing
4.5. MIC Determination
4.6. Binding of Mecillinam and Avibactam to Penicillin-Binding Proteins (PBPs) Measured with Bocillin FL Assay
4.7. Phase-Contrast Microscopy
4.8. Confocal Laser Scanning Microscopy
4.9. Single and Combination Antibiotic Treatment In Vivo in the Mouse Peritonitis/Sepsis Model
- Setup 3: 200 mg kg−1 avibactam alone, 100 mg kg−1 mecillinam alone, 200 mg kg−1 avibactam and 100 mg kg−1 mecillinam together or saline.
4.10. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Transparency Declaration
Previously Presented
References
- Kaur, J.; Dhama, A.S.; Buttolia, H.; Kaur, J.; Walia, K.; Ohri, V.; Kumar, V.; Lynn, A.M.; Srivastava, A.; Singh, H. ICMR’s Antimicrobial Resistance Surveillance system (i -AMRSS): A promising tool for global antimicrobial resistance surveillance. JAC-Antimicrob. Resist. 2021, 3, dlab023. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Marrs, E.C.L.; Day, K.M.; Perry, J.D. In vitro activity of mecillinam against Enterobacteriaceae with NDM-1 carbapenemase. J. Antimicrob. Chemother. 2014, 69, 2873–2875. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, F.; Ahmadzada, A.; Plambeck, L.; Wille, T.; Hamprecht, A. Susceptibility of Clinical Enterobacterales Isolates with Common and Rare Carbapenemases to Mecillinam. Front. Microbiol. 2021, 11, 627267. [Google Scholar] [CrossRef] [PubMed]
- Zykov, I.N.; Frimodt-Møller, N.; Småbrekke, L.; Sundsfjord, A.; Samuelsen, Ø. Efficacy of mecillinam against clinical multidrug-resistant Escherichia coli in a murine urinary tract infection model. Int. J. Antimicrob. Agents 2020, 55, 105851. [Google Scholar] [CrossRef] [PubMed]
- Tsakris, A.; Koumaki, V.; Baka, S.; Balakrishnan, I. Activity of mecillinam against OXA-48-like carbapenemase-producing Enterobacterales. J. Antimicrob. Chemother. 2021, 77, 537–538. [Google Scholar] [CrossRef] [PubMed]
- Lagacé-wiens, P.; Walkty, A.; Karlowsky, J.A. Ceftazidime—Avibactam: An evidence-based review of its pharmacology and potential use in the treatment of Gram-negative bacterial infections. Core Evid. 2014, 9, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Hansen, K.H.; Andreasen, M.R.; Pedersen, M.S.; Westh, H.; Jelsbak, L.; Schønning, K. Resistance to piperacillin/tazobactam in Escherichia coli resulting from extensive IS 26-associated gene amplification of bla TEM-1. J. Antimicrob. Chemother. 2019, 74, 3179–3183. [Google Scholar] [CrossRef] [PubMed]
- Lampri, N.; Galani, I.; Poulakou, G.; Katsarolis, I.; Petrikkos, G.; Giamarellou, H.; Souli, M. Mecillinam/clavulanate combination: A possible option for the treatment of community-acquired uncomplicated urinary tract infections caused by extended-spectrum β-lactamase-producing Escherichia coli. J. Antimicrob. Chemother. 2012, 67, 2424–2428. [Google Scholar] [CrossRef] [PubMed]
- Asli, A.; Brouillette, E.; Krause, K.M.; Nichols, W.W.; Malouin, F. Distinctive Binding of Avibactam to Penicillin-Binding Proteins of Gram-Negative and Gram-Positive Bacteria. Antimicrob. Agents Chemother. 2016, 60, 752–756. [Google Scholar] [CrossRef] [PubMed]
- Sutaria, D.S.; Moya, B.; Green, K.B.; Kim, T.H.; Tao, X.; Jiao, Y.; Louie, A.; Drusano, G.L.; Bulitta, J.B. First Penicillin-Binding Protein Occupancy Patterns of β- Lactams and β-Lactamase Inhibitors in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2018, 62, e00282-18. [Google Scholar] [CrossRef] [PubMed]
- Kocaoglu, O.; Carlson, E.E. Profiling of β-lactam selectivity for penicillin-binding proteins in Escherichia coli strain DC2. Antimicrob. Agents Chemother. 2015, 59, 2785–2790. [Google Scholar] [CrossRef] [PubMed]
- Ulloa, E.R.; Dillon, N.; Tsunemoto, H.; Pogliano, J.; Sakoulas, G.; Nizet, V. Avibactam Sensitizes Carbapenem-Resistant NDM-1–Producing Klebsiella pneumoniae to Innate Immune Clearance. J. Infect. Dis. 2019, 220, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Kragh, K.N.; Gijón, D.; Maruri, A.; Antonelli, A.; Coppi, M.; Kolpen, M.; Crone, S.; Tellapragada, C.; Hasan, B.; Radmer, S.; et al. Effective antimicrobial combination in vivo treatment predicted with microcalorimetry screening. J. Antimicrob. Chemother. 2021, 76, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Brouwers, R.; Vass, H.; Dawson, A.; Squires, T.; Tavaddod, S.; Allen, R.J. Stability of β-lactam antibiotics in bacterial growth media. PLoS ONE 2020, 15, e0236198. [Google Scholar] [CrossRef] [PubMed]
- Souvorov, A.; Agarwala, R.; Lipman, D.J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018, 19, 153. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, J.; Melchers, M.J.; van Mil, A.C.; Seyedmousavi, S.; Lagarde, C.M.; Schuck, V.J.; Nichols, W.W.; Mouton, J.W. Pharmacodynamics of ceftazidime and avibactam in neutropenic mice with thigh or lung infection. Antimicrob. Agents Chemother. 2016, 60, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Kerrn, M.B.; Espersen, F. Effects of Sulfamethizole and Amdinocillin against Escherichia coli Strains (with Various Susceptibilities) in an Ascending Urinary Tract Infection Mouse Model. Antimicrob. Agents Chemother. 2003, 47, 1002–1009. [Google Scholar] [CrossRef] [PubMed]
| Strains | Carbapenemase | MIC Mecillinam (mg/L) | MIC Ceftazidime/Avibactam (mg/L) | MIC Ceftazidime/Avibactam (mg/L) Performed on Agar Containing Mecillinam | MIC Ceftazidime (mg/L) | MIC Ceftazidime (mg/L) Performed on Agar Containing Mecillinam | ||
|---|---|---|---|---|---|---|---|---|
| Mecillinam: 0.5 mg/L | Mecillinam: 1 mg/L | Mecillinam: 2 mg/L | Mecillinam: 2 mg/L | |||||
| KP1 | KPC-2 | 16 | 1 | 0.032 | <0.016 | <0.016 | 8 | 8 |
| KP2 | KPC-3 | >256 | 4 | 0.064 | <0.016 | <0.016 | >256 | >256 |
| KP3 | KPC-3 | 32 | 1 | <0.016 | <0.016 | <0.016 | >256 | >256 |
| KP4 | KPC-3 | >256 | 2 | 2 | 0.064 | <0.016 | >256 | >256 |
| KP5 | KPC-3 | >256 | 2 | 0.016 | <0.016 | <0.016 | >256 | >256 |
| KP6 | NDM-1 | 2 | >256 | >256 | <0.016 | <0.016 | >256 | >256 |
| KP7 | NDM-1, OXA-48 | >256 | >256 | >256 | <0.016 | <0.016 | >256 | >256 |
| KP8 | NDM-5 | 2 | >256 | >256 | >256 | <0.016 | >256 | >256 |
| KP9 | NDM-7, OXA-181 | 8 | >256 | >256 | >256 | >256 | >256 | >256 |
| KP10 | OXA-232 | 16 | 1 | 0.25 | <0.016 | <0.016 | 32 | 32 |
| KP11 | OXA-436 | 4 | 0.5 | 0.032 | <0.016 | <0.016 | 64 | 32 |
| KP12 | IMP-1 | >256 | 32 | 32 | 32 | 32 | >256 | >256 |
| ATCC 13883 | >256 | 0.25 | 0.032 | 0.016 | 0.016 | 0.25 | 0.016 | |
| EC2 | NDM-1 | 1 | >256 | NG | NG | NG | >256 | NG |
| EC3 | NDM-5 | 8 | >256 | >256 | >256 | >256 | >256 | >256 |
| EC4 | NDM-5, OXA-181 | 4 | >256 | >256 | >256 | <0.016 | >256 | >256 |
| EC5 | NDM-7 | 4 | >256 | >256 | <0.016 | <0.016 | >256 | >256 |
| EC6 | OXA-244 | 2 | 0.5 | <0.016 | <0.016 | <0.016 | 2 | 2 |
| EC7 | OXA-48 | 2 | 0.25 | <0.016 | <0.016 | <0.016 | 0.25 | 0.25 |
| ATCC 25922 | 0.064 | 0.125 | NG | NG | NG | 0.125 | NG | |
| Strain | Carbapenemase | MIC Mecillinam (mg/L) | MIC Ceftazidime/Avibactam (mg/L) | Mecillinam MIC (mg/L) Performed on Agar Containing Avibactam | Mecillinam (mg/L) Performed on Agar Containing Ceftazidime/Avibactam | ||
|---|---|---|---|---|---|---|---|
| Avibactam: 1 mg/L | Avibactam: 4 mg/L | Ceftazidime/Avibactam: 4 mg/L | Ceftazidime/Avibactam: 16 mg/L | ||||
| KP2 | KPC-3 | >256 | 4 | 1 | 0.25 | 1 | NG |
| KP5 | KPC-3 | >256 | 2 | 1 | 0.25 | 2 | NG |
| KP6 | NDM-1 | 2 | >256 | 1 | 0.5 | 1 | 1 |
| KP7 | NDM-1, OXA-48 | >256 | >256 | 2 | 1 | 2 | 2 |
| KP8 | NDM-5 | 2 | >256 | 1 | 0.5 | 1 | 1 |
| ATCC 13883 | >256 | 0.25 | 8 | 8 | NG | NG | |
| EC4 | NDM-5, OXA-181 | 4 | >256 | 1 | 0.25 | 2 | 1 |
| EC5 | NDM-7 | 4 | >256 | 1 | 0.5 | 1 | 1 |
| ATCC 25922 | 0.064 | 0.125 | 0.064 | 0.016 | NG | NG | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
List, K.K.; Kolpen, M.; Kragh, K.N.; Charbon, G.; Radmer, S.; Hansen, F.; Løbner-Olesen, A.; Frimodt-Møller, N.; Hertz, F.B. Synergy between Mecillinam and Ceftazidime/Avibactam or Avibactam against Multi-Drug-Resistant Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae. Antibiotics 2022, 11, 1280. https://doi.org/10.3390/antibiotics11101280
List KK, Kolpen M, Kragh KN, Charbon G, Radmer S, Hansen F, Løbner-Olesen A, Frimodt-Møller N, Hertz FB. Synergy between Mecillinam and Ceftazidime/Avibactam or Avibactam against Multi-Drug-Resistant Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae. Antibiotics. 2022; 11(10):1280. https://doi.org/10.3390/antibiotics11101280
Chicago/Turabian StyleList, Karoline Knudsen, Mette Kolpen, Kasper Nørskov Kragh, Godefroid Charbon, Stine Radmer, Frank Hansen, Anders Løbner-Olesen, Niels Frimodt-Møller, and Frederik Boetius Hertz. 2022. "Synergy between Mecillinam and Ceftazidime/Avibactam or Avibactam against Multi-Drug-Resistant Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae" Antibiotics 11, no. 10: 1280. https://doi.org/10.3390/antibiotics11101280
APA StyleList, K. K., Kolpen, M., Kragh, K. N., Charbon, G., Radmer, S., Hansen, F., Løbner-Olesen, A., Frimodt-Møller, N., & Hertz, F. B. (2022). Synergy between Mecillinam and Ceftazidime/Avibactam or Avibactam against Multi-Drug-Resistant Carbapenemase-Producing Escherichia coli and Klebsiella pneumoniae. Antibiotics, 11(10), 1280. https://doi.org/10.3390/antibiotics11101280







