Multi-Drug and β-Lactam Resistance in Escherichia coli and Food-Borne Pathogens from Animals and Food in Portugal, 2014–2019
Abstract
:1. Introduction
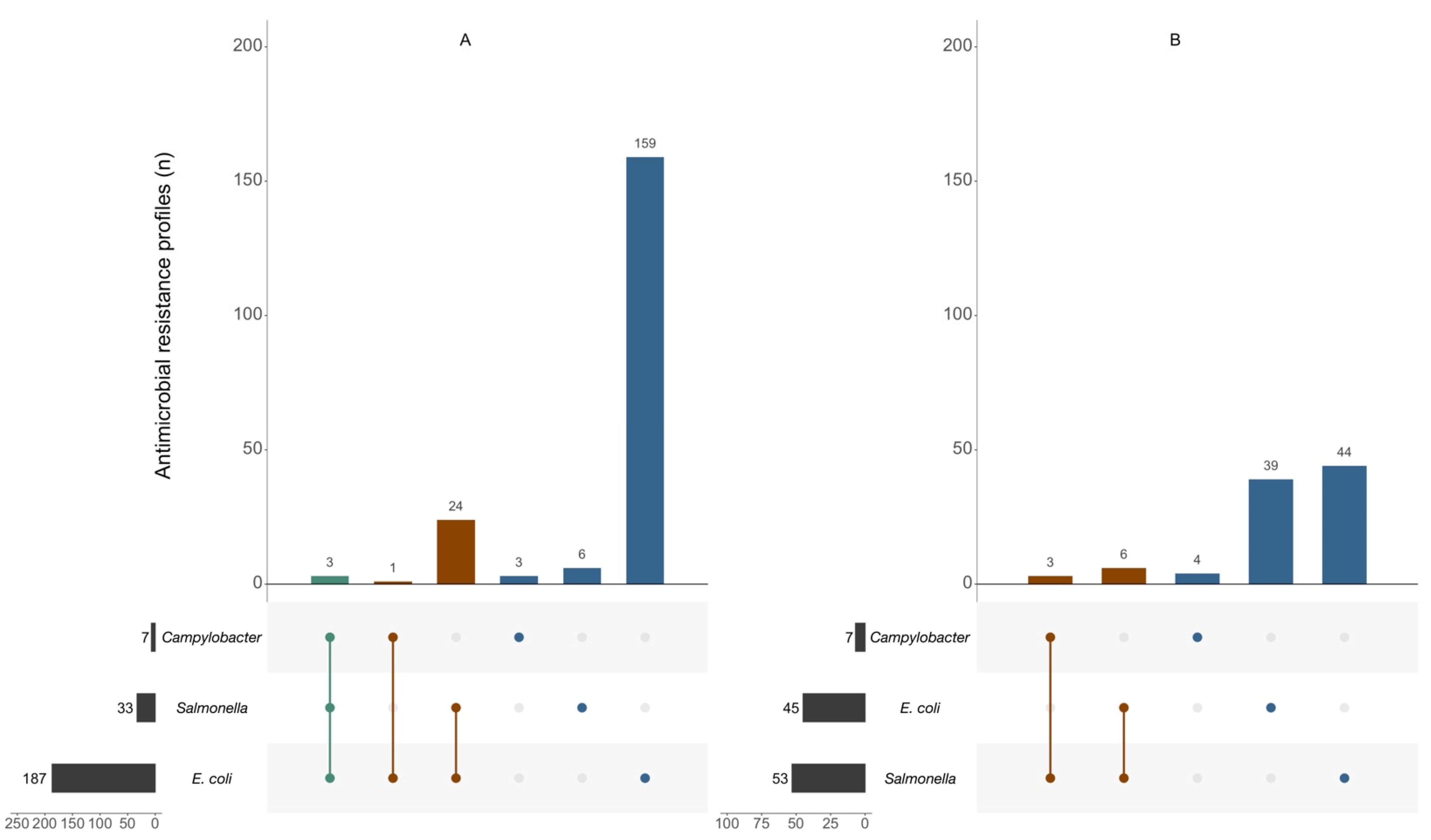
2. Results
3. Discussion
4. Material and Methods
4.1. Study Design, Setting, and Data Collection
4.2. Microbiology Surveillance Data
4.3. Study Variables
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brinkac, L.; Voorhies, A.; Gomez, A.; Nelson, K.E. The threat of antimicrobial resistance on the human microbiome. Microb. Ecol. 2017, 74, 1001–1008. [Google Scholar] [CrossRef]
- van den Bogaard, A.; London, N.; Driessen, C.; Stobberingh, E. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 2001, 47, 763–771. [Google Scholar] [CrossRef]
- Penders, J.; Stobberingh, E.; Savelkoul, P.; Wolffs, P. The human microbiome as a reservoir of antimicrobial resistance. Front. Microb. 2013, 4, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adator, E.; Narvaez-Bravo, C.; Zaheer, R.; Cook, S.; Tymensen, L.; Hannon, S.; Booker, C.; Deirdre, C.; Read, R.; McAllister, T. A One Health comparative assessment of antimicrobial resistance in generic and extended-spectrum cephalosporin-resistant Escherichia coli from beef production, sewage and clinical settings. Microorganisms 2020, 8, 885. [Google Scholar] [CrossRef]
- Iriti, M.; Vitalini, S.; Varoni, E.M. Humans, animals, and environment: One Health approach against global antimicrobial resistance. Antibiotics 2020, 9, 346. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J. Antimicrobial resistance, from bench-to-publicside. Microbes Infect. Chemother. 2021, 1, e1182. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R. Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J. Infect. Dis. 2017, 215 (Suppl. S1), S9–S17. [Google Scholar] [CrossRef] [Green Version]
- Tagliabue, A.; Rappuoli, R. Changing priorities in vaccinology: Antibiotic resistance moving to the top. Front. Immunol. 2018, 9, 1068. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the pig production chain: A comprehensive analysis of its impact on human health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Parsonage, B.; Hagglund, P.; Keogh, L.; Wheelhouse, N.; Brown, R.; Dancer, S. Control of antimicrobial resistance requires an ethical approach. Front. Microbiol. 2017, 8, 2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourély, C.; Chauvin, C.; Jouy, É.; Cazeau, G.; Jarrige, N.; Leblond, A.; Gay, É. Comparative epidemiology of E. coli resistance to third-generation cephalosporins in diseased food-producing animals. Vet. Microb. 2018, 223, 72–78. [Google Scholar] [CrossRef]
- EFSA; ECDC. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, 179. [Google Scholar] [CrossRef]
- Clemente, L.; Menageiro, V.; Ferreira, E.; Jones-Dias, D.; Correia, I.; Themudo, P.; Alburquerque, T.; Caniça, M. Occurrence of extended-spectrum β-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 2013, 167, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, N.; Figueiredo, R.; Mendes, C.; Card, R.; Anjum, M.; Silva, G. Microarray evaluation of antimicrobial resistance and virulence of Escherichia coli isolates from Portuguese Poultry. Antibiotics 2016, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of antibiotic resistance genes in multidrug-resistant Enterobacteriaceae on Portuguese livestock manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Abdalla, S.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From farm-to-fork: E. Coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef]
- Haulisah, N.; Hassan, L.; Bejo, S.; Jajere, S.; Ahmad, N. High levels of antibiotic resistance in isolates from diseased livestock. Front. Vet. Sci. 2021, 8, 652351. [Google Scholar] [CrossRef]
- Rahman, M.; Husna, A.; Elshabrawy, H.; Alam, J.; Runa, N.; Badruzzaman, A.; Banu, N.; Mamun, M.; Paul, B.; Das, S.; et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci. Rep. 2020, 10, 21999. [Google Scholar] [CrossRef] [PubMed]
- Aworh, M.; Kwaga, J.; Hendriksen, R.; Okolocha, E.; Thakur, S. Genetic relatedness of multidrug resistant Escherichia coli isolated from humans, chickens and poultry environments. Antimicrob. Resist. Infect. Control 2021, 10, 58. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Leverstein-van Hall, M.A.; Dierikx, C.M.; Stuart, C.; Voets, G.M.; van den Munckhof, M.P.; van Essen-Zandbergen, A.; Plattel, T.; Fluit, A.C.; van de Sande-Bruisma, N.; Scharinga, J.; et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin. Microbiol. Infect. 2011, 17, 873–880. [Google Scholar] [CrossRef] [Green Version]
- Dorado-García, A.; Smid, J.H.; van Pelt, W.; Bonten, M.; Fluit, A.; van den Bunt, G.; Wagenaar, J.; Hordijk, J.; Dierikx, C.; Veldman, K.; et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis. J. Antimicrob. Chemother. 2018, 73, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Egevarn, M.; Borjesson, S.; Byfors, S.; Finn, M.; Kaipe, C.; Englund, S.; Lindbald, M. Escherichia coli with extended-spectrum beta-lactamases or transferable AmpC beta-lactamases and Salmonella on meat imported intro Sweden. Int. J. Food Microbiol. 2014, 171, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum b-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [Green Version]
- Ramos, S.; Silva, V.; Dapkevicius, M.; Caniça, M.; Tejedor-Junco, M.; Igrejas, G.; Poeta, P. Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β -lactamase (ESBL) production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [Green Version]
- Lalak, A.; Wasyl, D.; Zajac, M.; Skarzynska, M.; Hoszowski, A.; Samcik, I.; Wozniakowski, G.; Szulowski, K. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet. Microbiol. 2016, 194, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, W.; Bonten, M.; Bos, M.; van Marm, S.; Scharringa, J.; Wagenaar, J.; Heederik, D. Carriage of extended-spectrum β-lactamases in pig farmers is associated with occurrence in pigs. Clin. Microbiol. Infect. 2015, 21, 917–923. [Google Scholar] [CrossRef] [Green Version]
- Bridier, A.; Le Grandois, P.; Moreau, M.-H.; Prénom, C.; Le Roux, A.; Feurer, C.; Soumet, C. Impact of cleaning and disinfection procedures on microbial ecology and Salmonella antimicrobial resistance in a pig slaughterhouse. Sci. Rep. 2019, 9, 12947. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly prevalent multidrug-resistant Salmonella from chicken and pork meat at retail markets in Guangdong, China. Front. Microbiol. 2018, 9, 2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ibrahim, S.; WeiHoong, L.; Lai, S.Y.; Mustapha, Z.; Zalati, C.; Aklilu, E.; Mohamad, M.; Kamaruzzaman, N. Prevalence of antimicrobial resistance (AMR) Salmonella spp. and Escherichia coli isolated from broilers in the East Coast of Peninsular Malaysia. Antibiotics 2021, 10, 579. [Google Scholar] [CrossRef]
- Woźniak-Biel, A.; Bugla-Płoskońska, G.; Kielsznia, A.; Korzekwa, K.; Tobiasz, A.; Korzeniowska-Kowal, A.; Wieliczko, A. High prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb. Drug Resist. 2018, 24, 314–322. [Google Scholar] [CrossRef]
- Yang, Y.; Feye, K.; Shi, Z.; Pavlidis, H.; Kogut, M.; Ashworth, A.; Ricke, S. A historical review on antibiotic resistance of foodborne Campylobacter. Front. Microbiol. 2019, 10, 1509. [Google Scholar] [CrossRef] [Green Version]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Zhang, Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oniciuc, E.-A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; López, M.; Alvarez-Ordóñez, A. Food processing as a risk factor for antimicrobial resistance spread along the food chain. Curr. Opi. Food Sci. 2019, 30, 21–26. [Google Scholar] [CrossRef]
- Huth, M.; Weich, K.; Grimm, H. Veterinarians between the frontlines?! The concept of One Health and three frames of health in Veterinary Medicine. Food Ethics 2019, 3, 91–108. [Google Scholar] [CrossRef] [Green Version]
- Bonilla-Aldana, D.K.; Dhama, K.; Rodriguez-Morales, A.J. Revisiting the One Health approach in the context of COVID-19: A look into the ecology of this emerging disease. Adv. Anim. Vet. Sci. 2020, 8, 234–237. [Google Scholar] [CrossRef] [Green Version]
- Destoumieux-Garzón, D.; Mavingui, P.; Boetsch, G.; Boissier, J.; Darriet, F.; Duboz, P.; Fritsch, C.; Giraudoux, P.; Le Roux, F.; Morand, S.; et al. The One Health concept: 10 years old and a long road ahead. Front. Vet. Sci. 2018, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- European Commission. Commission implementing decision of 12 November 2013 on the monitoring and reporting of antimicrobial resistance in zoonotic and commensal bacteria (2013/652/EU). Off. J. Eur. Union 2013, 303, 26–39. [Google Scholar]
- European Parliament and Council of the European Union. Directive 2003/99/EC of the European Parliament and of the Council of 17 November 2003 on the monitoring of zoonoses and zoonotic agents, amending Council Decision 90/424/EEC and repealing Council Directive 92/117/EEC. Off. J. Eur. Union 2003, 325, 31–40. [Google Scholar]
- Commission of the European communities. Commission regulation No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off. J. Eur. Union 2005, 338, 1–26. [Google Scholar]
- EFSA. Scientific report of EFSA: Technical specifications on randomised sampling for harmonised monitoring of antimicrobial resistance in zoonotic and commensal bacteria. EFSA J. 2014, 12, 3686. [Google Scholar] [CrossRef] [Green Version]
- ECDC. Surveillance of Antimicrobial Resistance in Europe 2017: Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2017; ECDC: Stockholm, Sweden, 2018. [Google Scholar]
- EUCAST. Consultation on Breakpoint Changes Necessary in Conjunction with Introducing New Definitions of S, I and R in the EUCAST Breakpoint Table v 9.0. Consultation 4 October–4 November 2018. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/2018/Breakpoint_changes_with_introducing_new_definitions_of_SIR_4OCT2018_v2.pdf (accessed on 20 June 2020).
- Aerts, M.; Battisti, A.; Hendriksen, R.; Kempf, I.; Teale, C.; Tenhagen, B.-A.; Veldman, K.; Wasyl, D.; Guerra, B.; Liébana, E.; et al. Technical specifications on harmonised monitoring of antimicrobial resistance in zoonotic and indicator bacteria from food-producing animals and food. EFSA J. 2019, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z. Extensions to the k-Means algorithm for clustering large data sets with categorical values. Data Min. Knowl. Discov. 1998, 2, 283–304. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. Data Analysis and Decision Support. klaR Analyzing German Business Cycles; Baier, D., Decker, R., Schmidt-Thieme, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar] [CrossRef]
- Conway, J.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [Green Version]

| Unique Profiles | Shared Profiles | ||||||
|---|---|---|---|---|---|---|---|
| E. coli n = 1462 | Salmonella n = 10 | Campylobacter n = 67 | * E. coli Salmonella n = 947 | * E. coli Campylobacter n = 3 | * E. coli Salmonella Campylobacter n = 727 | ||
| Food-producing animals n = 3216 | 1 | TET-FQ-C3G-CHL-SLP-PEN (124) | CHL (3) | MAC-TET-FQ (57) | TET-FQ-CHL-SLP-PEN-TMP (189) | MAC-FQ (3) | TET-FQ (354) |
| 2 | TET-FQ-C3G-SLP-PEN-TMP (84) | FQ-AMN (2) | TET-FQ-AMN (9) | TET-FQ-C3G-CHL-SLP-PEN-TMP (186) | - | FQ (301) | |
| 3 | TET-FQ-PEN (76) | MAC (2) | MAC-FQ-AMN (1) | TET-FQ-SLP-PEN-TMP (133) | - | TET (72) | |
| E. coli n = 181 | Salmonella n = 232 | Campylobacter n = 57 | * E. coli Salmonella n = 92 | * Campylobacter Salmonella n = 128 | * All three bacteria n = 0 | ||
| Food products n = 690 | 1 | TET-FQ-C3G-SLP-PEN-TMP (35) | TET-SLP-PEN (69) | MAC-TET-FQ (26) | TET-FQ-C3G-CHL-SLP-PEN-TMP (49) | FQ (59) | - |
| 2 | TET-FQ-C3G-CHL-SLP-PEN (27) | TET-CHL-SLP-PEN-TMP (13) | MAC-TET-FQ-AMN (15) | TET-C3G-SLP-PEN-TMP (11) | TET-FQ (43) | - | |
| 3 | FQ-C3G-PEN (21) | MAC-TET-SLP-TMP (12) | TET-FQ-AMN (14) | TET-FQ-C3G-SLP-PEN (11) | TET (26) | - | |
| Clusters | AMN (%) | MAC (%) | F(Q) (%) | TET (%) | CHL (%) | PLM (%) | PEN (%) | SLP (%) | TMP (%) | C3G (%) | CARB (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | 1 n = 1292 | 9.0 | 14.6 | 73.4 | 91.3 | 73.3 | 15.0 | 92.0 | 95.7 | 88.7 | 28.9 | 1.2 |
| n = 2777 | 2 n = 535 | 3.9 | 3.4 | 39.1 | 58.5 | 2.6 | 4.1 | 57.2 | 8.0 | 5.0 | 16.6 | 1.1 |
| 3 n = 799 | 3.9 | 12.9 | 81.1 | 79.2 | 26.4 | 7.8 | 97.4 | 82.0 | 32.2 | 91.9 | 0.3 | |
| 4 n = 151 | 100.0 | 45.7 | 88.7 | 94.7 | 82.8 | 32.5 | 100.0 | 96.0 | 92.1 | 100.0 | 0.0 | |
| Salmonella | 1 n = 141 | 6.4 | 2.1 | 12.8 | 97.2 | 14.9 | 5.7 | 92.2 | 84.4 | 0.0 | 2.1 | |
| n = 919 | 2 n = 619 | 2.3 | 1.3 | 43.8 | 0.0 | 0.6 | 5.0 | 6.8 | 1.3 | 1.5 | 0.3 | |
| 3 n = 30 | 0.0 | 0.0 | 6.7 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ||
| 4 n = 129 | 5.4 | 24.8 | 41.9 | 77.5 | 38.8 | 3.9 | 77.5 | 99.2 | 100.0 | 14.0 | ||
| Campylobacter | 1 n = 479 | 96.5 | 18.2 | 97.5 | 100.0 | |||||||
| n = 541 | 2 n = 11 | 0.0 | 0.0 | 0.0 | 0.0 | |||||||
| 3 n = 51 | 98.0 | 5.9 | 100.0 | 0.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, M.M.; Cardo, M.; Soares, P.; Cara d’Anjo, M.; Leite, A. Multi-Drug and β-Lactam Resistance in Escherichia coli and Food-Borne Pathogens from Animals and Food in Portugal, 2014–2019. Antibiotics 2022, 11, 90. https://doi.org/10.3390/antibiotics11010090
Costa MM, Cardo M, Soares P, Cara d’Anjo M, Leite A. Multi-Drug and β-Lactam Resistance in Escherichia coli and Food-Borne Pathogens from Animals and Food in Portugal, 2014–2019. Antibiotics. 2022; 11(1):90. https://doi.org/10.3390/antibiotics11010090
Chicago/Turabian StyleCosta, Miguel Mendes, Miguel Cardo, Patricia Soares, Maria Cara d’Anjo, and Andreia Leite. 2022. "Multi-Drug and β-Lactam Resistance in Escherichia coli and Food-Borne Pathogens from Animals and Food in Portugal, 2014–2019" Antibiotics 11, no. 1: 90. https://doi.org/10.3390/antibiotics11010090
APA StyleCosta, M. M., Cardo, M., Soares, P., Cara d’Anjo, M., & Leite, A. (2022). Multi-Drug and β-Lactam Resistance in Escherichia coli and Food-Borne Pathogens from Animals and Food in Portugal, 2014–2019. Antibiotics, 11(1), 90. https://doi.org/10.3390/antibiotics11010090







