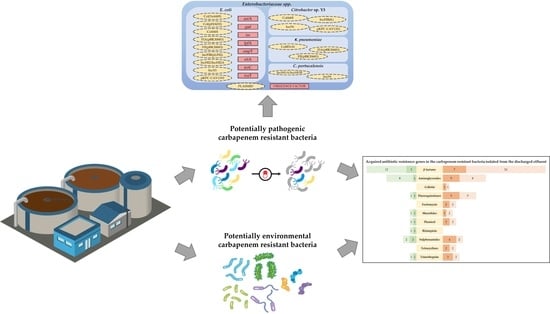
Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms
Abstract
:1. Introduction
2. Results
2.1. Concentrations of Carbapenem Resistant Bacteria
2.2. Species Identification by 16S rRNA Gene Sequencing and Screening of Carbapenem Resistance Genes
2.3. Taxonomic Confirmation Using the Whole Genome Sequencing Data
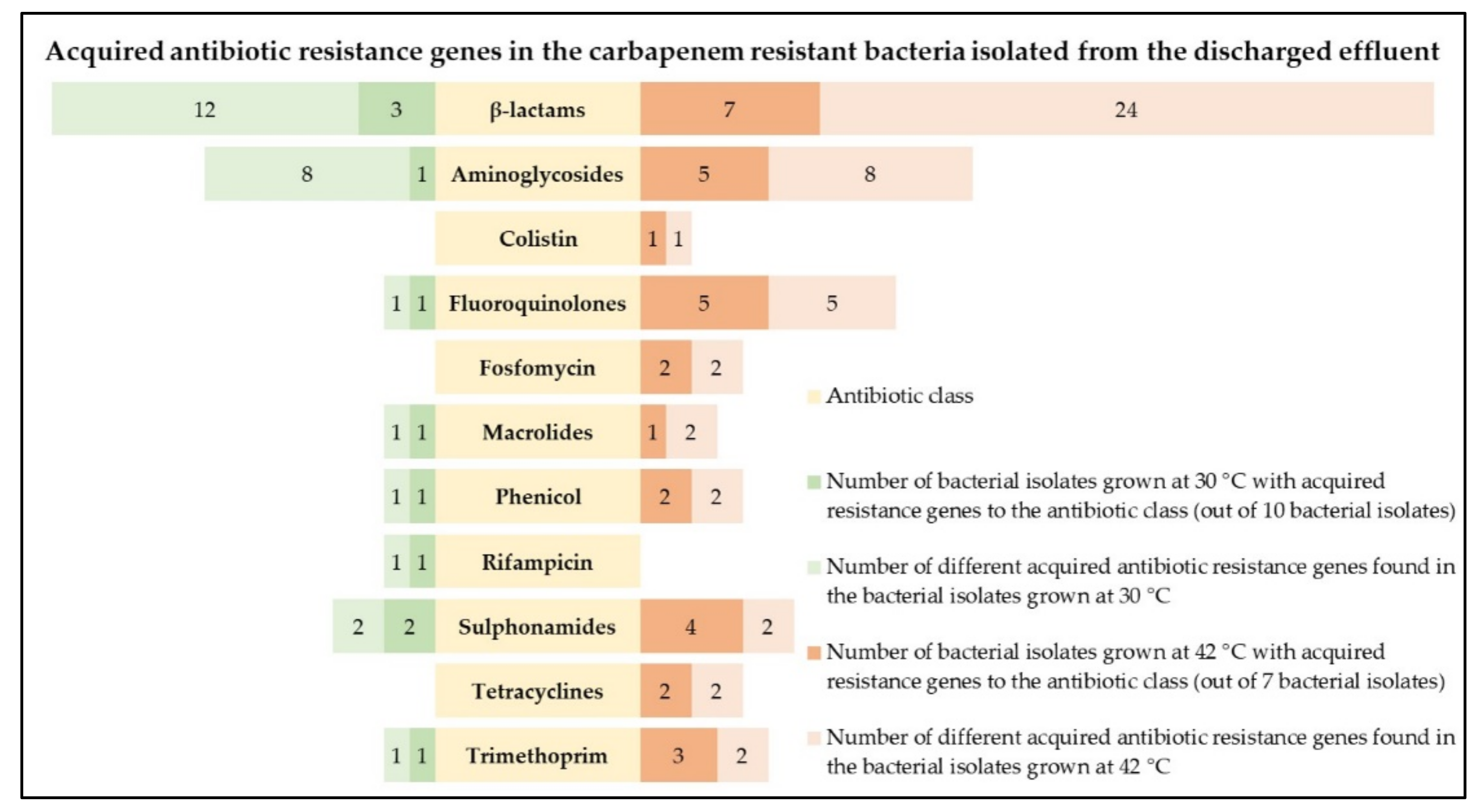
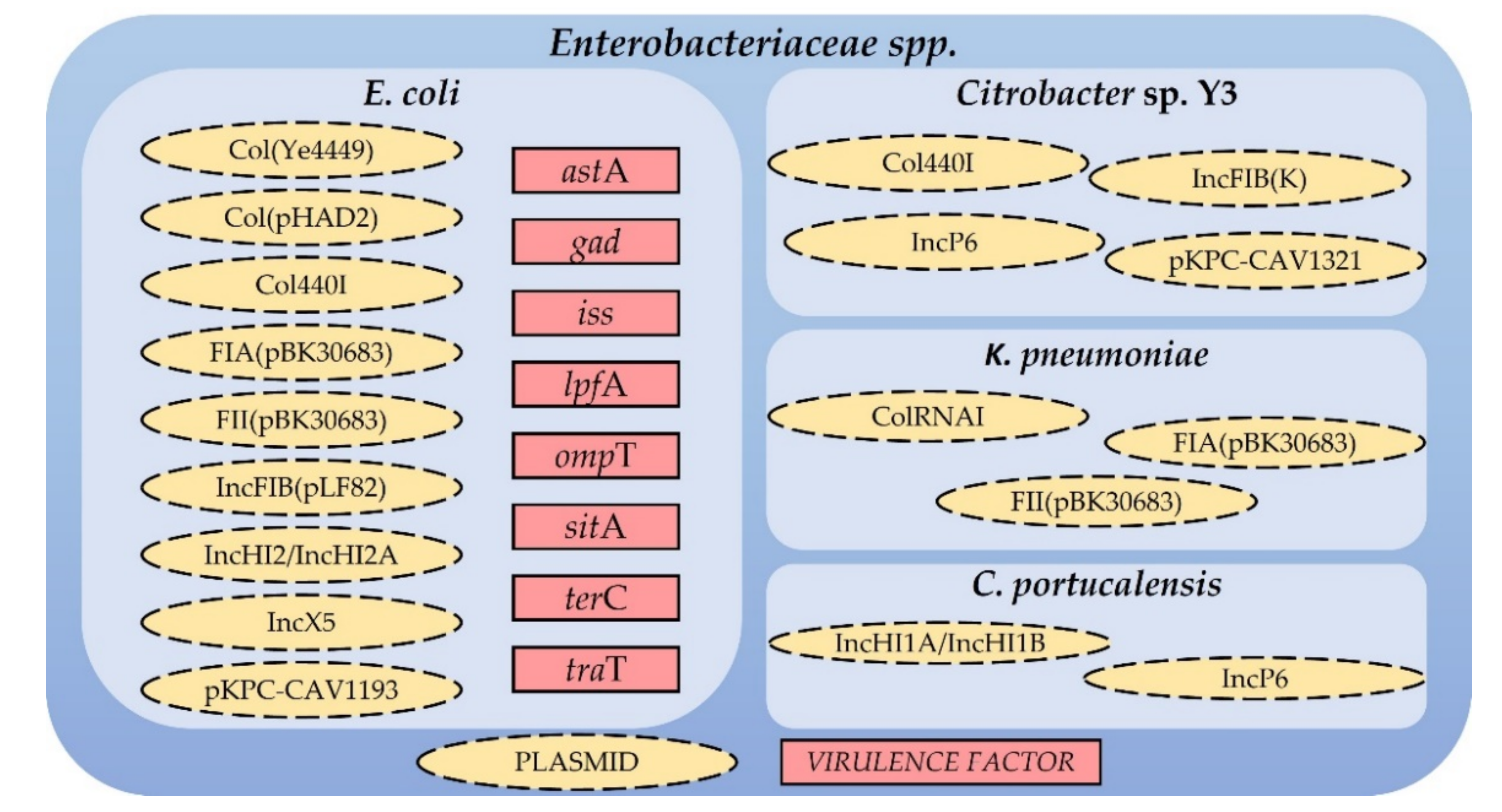
2.4. Identification of Acquired AB Resistance Genes, Conjugative Plasmids, and Virulence Factors
2.4.1. Acquired AB Resistance Genes
2.4.2. Conjugative Plasmids in Enterobacteriaceae spp. and Virulence Factors in E. coli
2.5. AB Susceptibility Testing
3. Discussion
4. Materials and Methods
4.1. WWTP Description and Sample Collection
4.2. Determination of Carbapenem Resistant Bacteria Concentrations
4.3. Isolation, DNA Extraction, and Species Identification of Carbapenem Resistant Bacteria
4.4. Screening of Carbapenem Resistance Genes
4.5. Whole Genome Sequencing and Assembly
4.6. Taxonomic Confirmation and Identification of Acquired AB Resistance Genes, Conjugative Plasmids, and Virulence Factors
4.7. AB Susceptibility Testing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Li, J.; Chen, H.; Bond, P.L.; Yuan, Z. Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Res. 2017, 123, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [Green Version]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Pitout, J.D.; Nordmann, P. Carbapenemases: Molecular diversity and clinical consequences. Future Microbiol. 2007, 2, 501–512. [Google Scholar] [CrossRef]
- Walsh, T.R. Emerging carbapenemases: A global perspective. Int. J. Antimicrob. Agents 2010, 36, S8–S14. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Schultsz, C.; Geerlings, S. Plasmid-Mediated Resistance in Enterobacteriaceae: Changing Landscape and Implications for Therapy. Drugs 2012, 72, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef]
- Lamba, M.; Ahammad, S.Z. Sewage treatment effluents in Delhi: A key contributor of Β-lactam resistant bacteria and genes to the environment. Chemosphere 2017, 188, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Mathys, D.A.; Mollenkopf, D.F.; Feicht, S.M.; Adams, R.J.; Albers, A.L.; Stuever, D.M.; Grooters, S.V.; Ballash, G.A.; Daniels, J.B.; Wittum, T.E. Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS ONE 2019, 14, e0218650. [Google Scholar] [CrossRef]
- Oliveira, M.; Nunes, M.; Barreto Crespo, M.T.; Silva, A.F. The environmental contribution to the dissemination of carbapenem and (fluoro)quinolone resistance genes by discharged and reused wastewater effluents: The role of cellular and extracellular DNA. Water Res. 2020, 182, 116011. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.M.; Narciso-Da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, aau9124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, F.; Mao, D.; Zhou, H.; Luo, Y. Prevalence and Fate of Carbapenemase Genes in a Wastewater Treatment Plant in Northern China. PLoS ONE 2016, 11, e0156383. [Google Scholar] [CrossRef]
- Hrenovic, J.; Ganjto, M.; Goic-Barisic, I. Carbapenem-resistant bacteria in a secondary wastewater treatment plant. Water SA 2017, 43, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Hrenovic, J.; Ivankovic, T.; Ivekovic, D.; Repec, S.; Stipanicev, D.; Ganjto, M. The fate of carbapenem-resistant bacteria in a wastewater treatment plant. Water Res. 2017, 126, 232–239. [Google Scholar] [CrossRef]
- Hrenovic, J.; Durn, G.; Kazazic, S.; Dekic, S.; Music, M.S. Untreated wastewater as a source of carbapenem-resistant bacteria to the riverine ecosystem. Water SA 2019, 45, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Hrenovic, J.; Ivankovic, T.; Durn, G.; Dekic, S.; Kazazic, S.; Kisic, I. Presence of carbapenem-resistant bacteria in soils affected by illegal waste dumps. Int. J. Environ. Health Res. 2019, 29, 154–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [Green Version]
- Tacão, M.; Correia, A.; Henriques, I.S. Low Prevalence of Carbapenem-Resistant Bacteria in River Water: Resistance is Mostly Related to Intrinsic Mechanisms. Microb. Drug Resist. 2015, 21, 497–506. [Google Scholar] [CrossRef] [PubMed]
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; WHO/EMP/IAU/2017.12; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Aires-de-sousa, M.; De la Rosa, J.M.O.; Gonçalves, M.L.; Pereira, A.L.; Nordmann, P.; Poirel, L. Epidemiology of Carbapenemase- Producing Klebsiella pneumoniae in a Hospital, Portugal. Emerg. Infect. Dis. 2019, 25, 1632–1638. [Google Scholar] [CrossRef] [Green Version]
- Manageiro, V.; Ferreira, E.; Almeida, J.; Barbosa, S.; Simões, C.; Antibiotic Resistance Surveillance Program in Portugal (ARSIP); Bonomo, R.A.; Caniça, M. Predominance of KPC-3 in a survey for carbapenemase-producing Enterobacteriaceae in Portugal. Antimicrob. Agents Chemother. 2015, 59, 3588–3592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manageiro, V.; Romão, R.; Moura, I.B.; Sampaio, D.A.; Vieira, L.; Ferreira, E.; The Network EuSCAPE-Portugal; Caniça, M. Molecular Epidemiology and Risk Factors of Carbapenemase-Producing Enterobacteriaceae Isolates in Portuguese Hospitals: Results from European Survey on Carbapenemase-Producing Enterobacteriaceae (EuSCAPE). Front. Microbiol. 2018, 9, 2834. [Google Scholar] [CrossRef] [Green Version]
- Vubil, D.; Figueiredo, R.; Reis, T.; Canha, C.; Boaventura, L.; Da Silva, G.J. Outbreak of KPC-3-producing ST15 and ST348 Klebsiella pneumoniae in a Portuguese hospital. Epidemiol. Infect. 2017, 145, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Y.; Lazaro-Perona, F.; Falgenhauer, L.; Valverde, A.; Imirzalioglu, C.; Dominguez, L.; Cantón, R.; Mingorance, J.; Chakraborty, T. Insights into a Novel bla KPC-2 -Encoding IncP-6 Plasmid Reveal Carbapenem-Resistance Circulation in Several Enterobacteriaceae Species from Wastewater and a Hospital Source in Spain. Front. Microbiol. 2017, 8, 1143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The Prevalence and Characterization of Extended-Spectrum β-lactamase-and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dai, X.; Zeng, J.; Gao, Y.; Zhang, Z.; Zhang, L. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 2020, 10, 8113. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Infect. 2017, 30, 557–596. [Google Scholar] [CrossRef] [Green Version]
- Amador, P.P.; Fernandes, R.M.; Prudêncio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health Part A 2015, 50, 26–39. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Zankari, E.; Aarestrup, F.M.; Lund, O. Benchmarking of methods for identification of antimicrobial resistance genes in bacterial whole genome data. J. Antimicrob. Chemother. 2016, 71, 2484–2488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- EUCAST. Disk Diffusion Method for Antimicrobial Susceptibility Testing. 2020. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2021_manuals/Manual_v_9.0_EUCAST_Disk_Test_2021.pdf (accessed on 5 February 2020).
- Kahlmeter, G. The EUCAST Steering Committee Redefining Susceptibility Testing Categories S, I and R. 2019. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_Presentations/2018/EUCAST_-_Intermediate_category_-_information_for_all.pdf (accessed on 5 February 2020).
| Bacteria | AMP10 | CTX5 | IPM10 | MEM10 | CIP5 | C30 | CN10 | TE30 | W5 | SXT25 |
|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter baumannii | R | R | R | R | R | R | R | R | R | S |
| Acinetobacter pittii | R | R | I | I | R | R | R | R | R | S |
| Acinetobacter oleivorans | R | R | I | I | R | R | S | R | R | R |
| Aeromonas caviae | * | * | * | * | S | * | * | * | * | R |
| Aeromonas veronii (1) | * | * | * | * | R | * | * | * | * | R |
| Aeromonas veronii (2) | * | * | * | * | R | * | * | * | * | S |
| Citrobacter portucalensis | R | R | R | R | R | R | R | R | R | R |
| Citrobacter sp. Y3 | R | R | R | R | R | R | R | R | S | S |
| Escherichia coli | R | R | R | R | R | S | R | R | R | R |
| Klebsiella pneumoniae | R | R | R | R | R | S | R | R | R | R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, M.; Leonardo, I.C.; Nunes, M.; Silva, A.F.; Barreto Crespo, M.T. Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms. Antibiotics 2021, 10, 1118. https://doi.org/10.3390/antibiotics10091118
Oliveira M, Leonardo IC, Nunes M, Silva AF, Barreto Crespo MT. Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms. Antibiotics. 2021; 10(9):1118. https://doi.org/10.3390/antibiotics10091118
Chicago/Turabian StyleOliveira, Micaela, Inês Carvalho Leonardo, Mónica Nunes, Ana Filipa Silva, and Maria Teresa Barreto Crespo. 2021. "Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms" Antibiotics 10, no. 9: 1118. https://doi.org/10.3390/antibiotics10091118
APA StyleOliveira, M., Leonardo, I. C., Nunes, M., Silva, A. F., & Barreto Crespo, M. T. (2021). Environmental and Pathogenic Carbapenem Resistant Bacteria Isolated from a Wastewater Treatment Plant Harbour Distinct Antibiotic Resistance Mechanisms. Antibiotics, 10(9), 1118. https://doi.org/10.3390/antibiotics10091118