Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections
Abstract
:1. Staphylococcus aureus Biofilms: Slow Growing Organisms Highly Resistant to Drugs
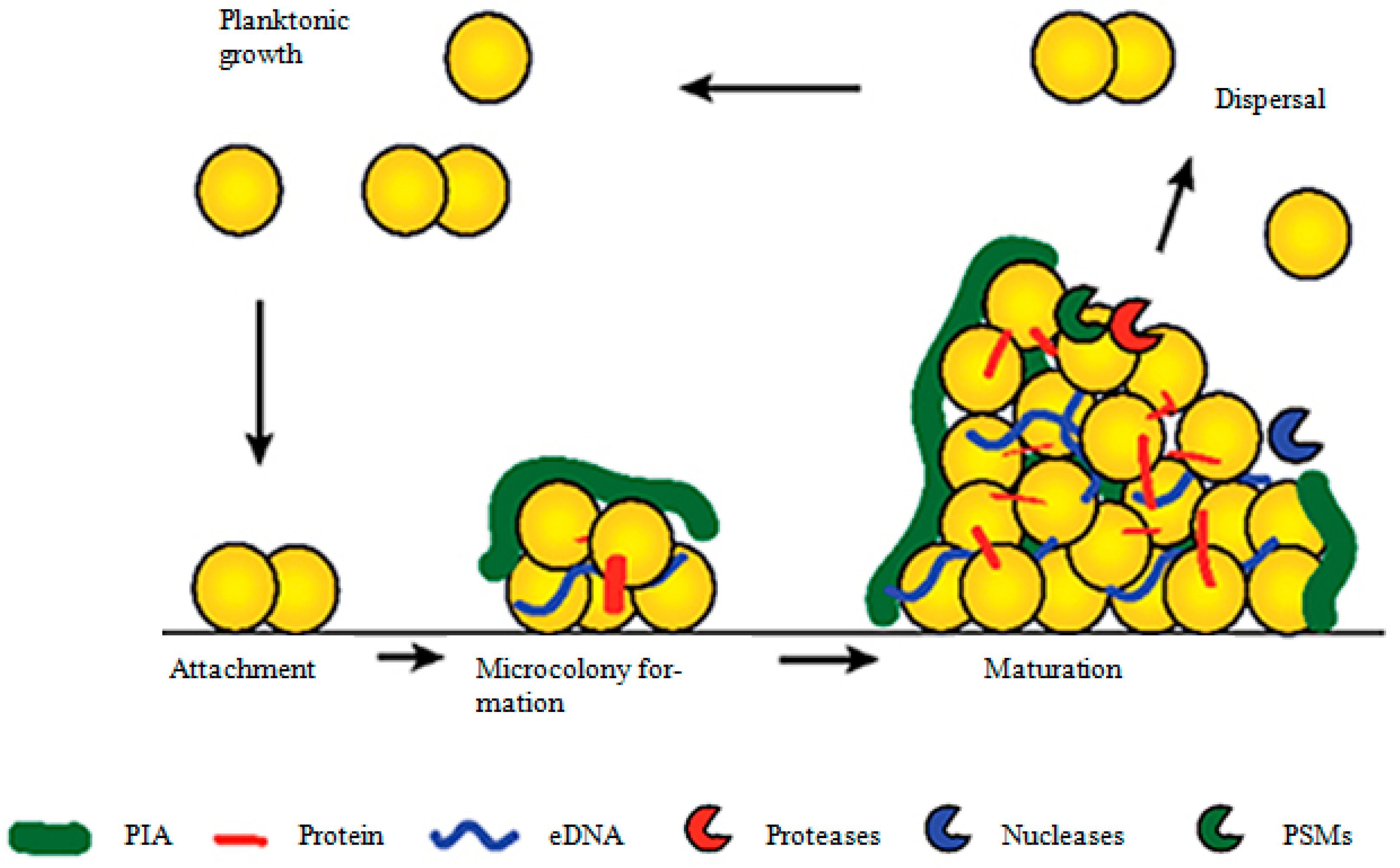
2. Stages of Biofilm Formation in S. aureus
2.1. The First Step: Attachment of S. aureus to Surfaces
2.2. Maturation of the S. aureus Biofilm
2.3. Triggering of the Biofilm Dispersal Response
| Biofilm/Cell Components | Biofilm Stages | Functions | References |
|---|---|---|---|
| eDNA | Attachment | Development of electrostatic interaction for initial attachment | [1] |
| Maturation | Biofilm matrix formation and biofilm stabilization | [28] | |
| Cell-wall-anchored proteins | Attachment | Initial attachment | [9,10,11,12,13] |
| Maturation | Intercellular binding and bacterial cell accumulation | [13] | |
| Sortase A | Attachment | Cleavage of cell-wall-anchored proteins to catalyze initial attachment | [14] |
| Teichoic acid | Attachment | Initial attachment | [17] |
| Maturation | Biofilm matrix formation | [22] | |
| Cytoplasmic proteins | Maturation | Biofilm matrix formation and biofilm stabilization by binding with eDNA | [26] |
| PSMs | Maturation | Biofilm stabilization by forming insoluble amyloid fibers and binding with eDNA | [30,45] |
| Dispersal | Biofilm dispersal by interacting with biofilm matrix | [43,44] | |
| Nucleoid-associated proteins | Maturation | Biofilm stabilization by binding with eDNA | [31] |
| Nucleases | Dispersal | Biofilm dispersal through degradation of eDNA | [34] |
| Proteases | Dispersal | Biofilm dispersal through degradation of protein component of biofilm | [42] |
| AIPs | Dispersal | Biofilm dispersal through activation of agr quorum sensing system | [37,39] |
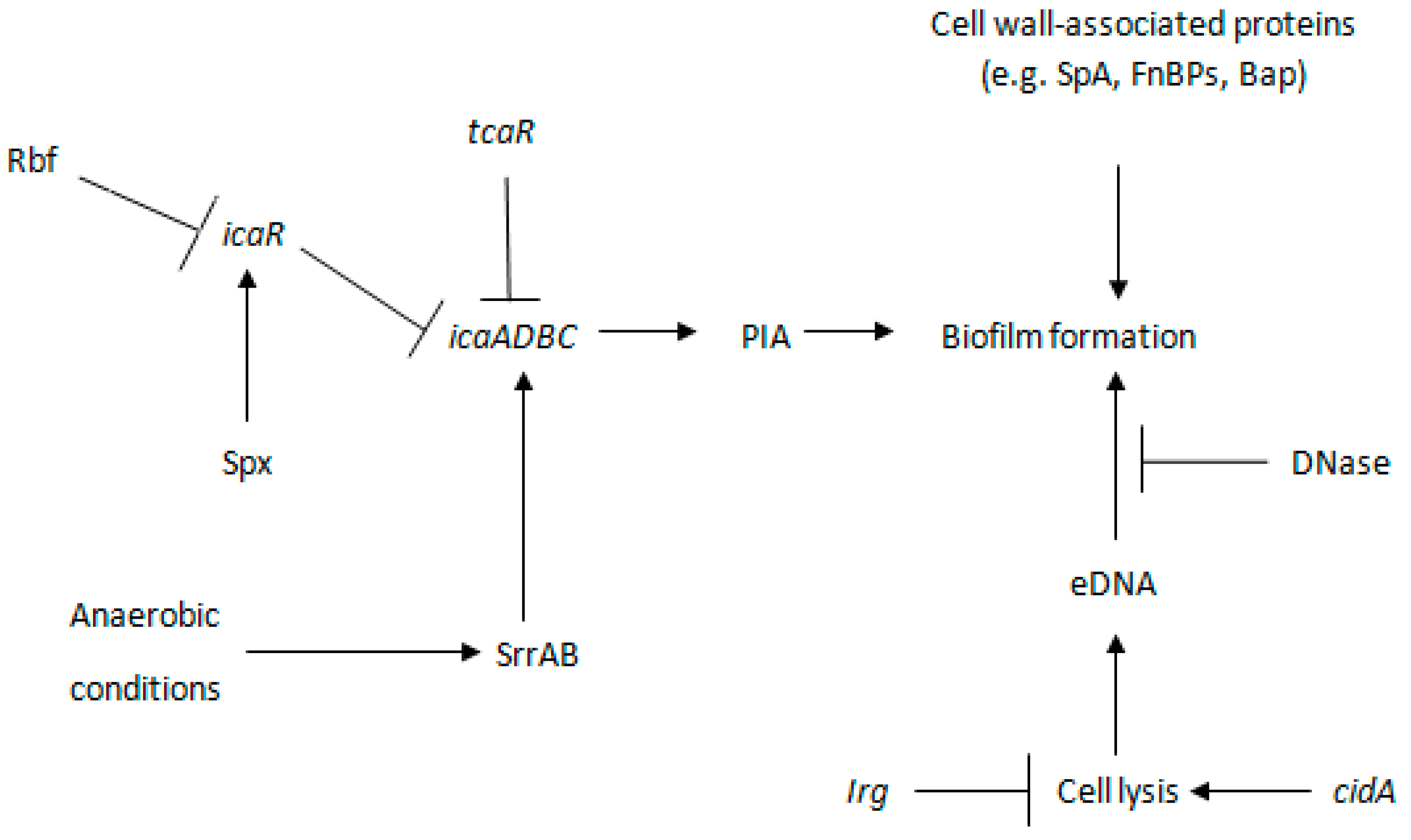
3. Biofilm Formation through PIA/PNAG-Dependent Mechanism
4. Biofilm Formation through PIA-Independent Mechanisms
5. Regulation of S. aureus Biofilm Formation: The Master Controllers and Their Targets
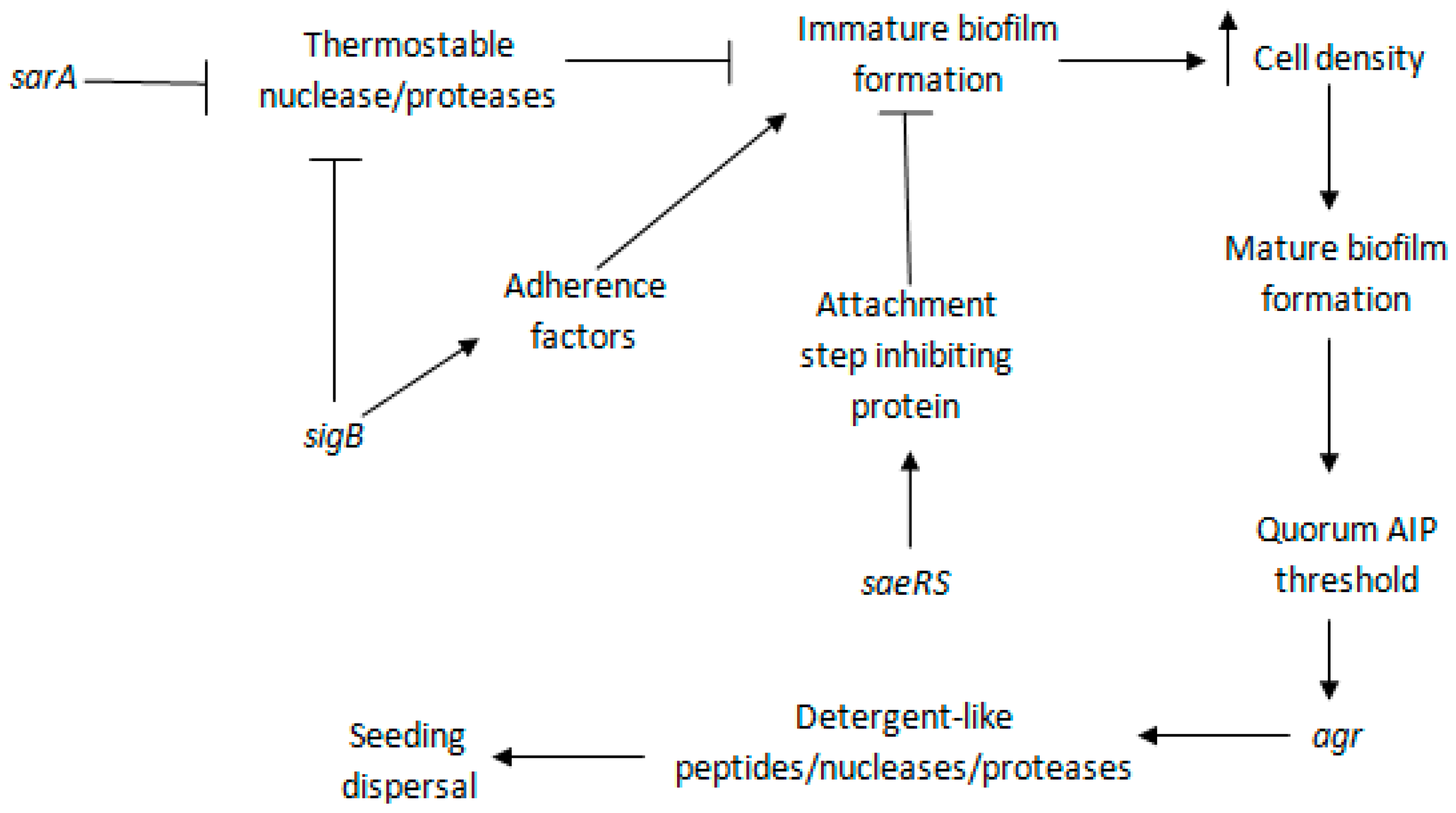
5.1. sarA
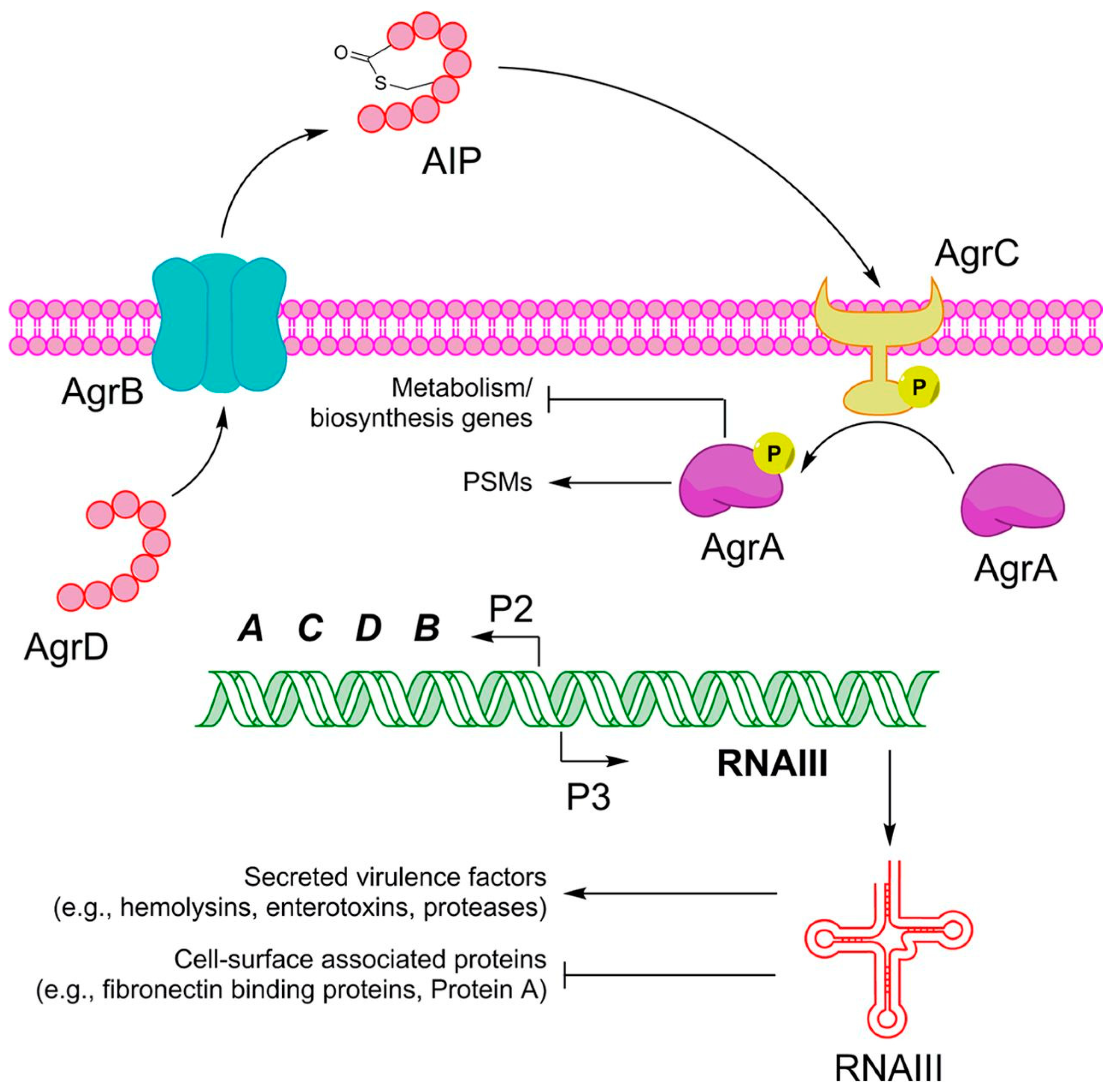
5.2. agr
5.3. sigB
5.4. saeRS
6. Clinical Context of Biofilm-Related S. aureus Prosthetic Joint Infections: Failure of Life-Enhancing Prosthetic Joints
7. Possible Adjuvant Treatments for Biofilm-Related S. aureus Prosthetic Joint Infections—The Search for a Novel Approach to an Intractable Problem
7.1. Quorum Sensing and Quorum Sensing Inhibitors: Stopping the Bacterial Communication
7.1.1. RNAIII-Inhibiting Peptide
7.1.2. Hamamelitannin
7.1.3. Auto-Inducing Peptides
7.1.4. Savirin
7.2. Drug Repurposing: Can Old Become New Again?
7.2.1. Auranofin
7.2.2. Aspirin
7.2.3. Ticagrelor
7.2.4. Simvastatin
7.2.5. Thioridazine
8. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wertheim, H.F.; Vos, M.C.; Ott, A.; van Belkum, A.; Voss, A.; Kluytmans, J.A.; van Keulen, P.H.; Vandenbroucke-Grauls, C.M.; Meester, M.H.; Verbrugh, H.A. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004, 364, 703–705. [Google Scholar] [CrossRef]
- Minasyan, H. Sepsis: Mechanisms of bacterial injury to the patient. Scand. J. Trauma Resusc. Emerg. Med. 2019, 27, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, a010306. [Google Scholar] [CrossRef] [Green Version]
- Parastan, R.; Kargar, M.; Solhjoo, K.; Kafilzadeh, F. Staphylococcus aureus biofilms: Structures, antibiotic resistance, inhibition, and vaccines. Gene Rep. 2020, 20, 100739. [Google Scholar] [CrossRef]
- Van Gestel, J.; Vlamakis, H.; Kolter, R. Division of labor in biofilms: The ecology of cell differentiation. Microbiol. Spectr. 2015, 3, 3.2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Dunne, W.M., Jr. Bacterial adhesion: Seen any good biofilms lately? Clin. Microbiol. Rev. 2002, 15, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Pollitt, E.J.G.; Crusz, S.A.; Diggle, S.P. Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci. Rep. 2015, 5, 17698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef] [Green Version]
- McDevitt, D.; Francois, P.; Vaudaux, P.; Foster, T.J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 1994, 11, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Ghebrehiwet, B.; Peerschke, E.I. Staphylococcus aureus protein A recognizes platelet gC1qR/p33: A novel mechanism for staphylococcal interactions with platelets. Infect. Immun. 2000, 68, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Speziale, P.; Pietrocola, G.; Foster, T.J.; Geoghegan, J.A. Protein-based biofilm matrices in Staphylococci. Front. Cell. Infect. Microbiol. 2014, 4, 171. [Google Scholar] [CrossRef] [Green Version]
- Mazmanian, S.K.; Liu, G.; Ton-That, H.; Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 1999, 285, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, C.A.; O’Gara, J.P. Contribution of culture media and chemical properties of polystyrene tissue culture plates to biofilm development by Staphylococcus aureus. J. Med. Microbiol. 2004, 53, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Gross, M.; Cramton, S.E.; Gotz, F.; Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 2001, 69, 3423–3426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Tseng, B.S.; Beckerman, B.; Jin, F.; Gibiansky, M.L.; Harrison, J.J.; Luijten, E.; Parsek, M.R.; Wong, G.C.L. Psl trails guide exploration and microcolony formation in Pseudomonas aeruginosa biofilms. Nature 2013, 497, 388–391. [Google Scholar] [CrossRef] [Green Version]
- Otto, K.; Silhavy, T.J. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 2287–2292. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar]
- Cramton, S.E.; Gerke, C.; Schnell, N.F.; Nichols, W.W.; Gotz, F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999, 67, 5427–5433. [Google Scholar] [CrossRef] [Green Version]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penades, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penades, J.R.; Lasa, I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulston, L.; Elsholz, A.K.; DeFrancesco, A.S.; Losick, R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. mBio 2014, 5, e01667-14. [Google Scholar] [CrossRef] [Green Version]
- Sadykov, M.R.; Bayles, K.W. The control of death and lysis in staphylococcal biofilms: A coordination of physiological signals. Curr. Opin. Microbiol. 2012, 15, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Dengler, V.; Foulston, L.; DeFrancesco, A.S.; Losick, R. An Electrostatic Net Model for the Role of Extracellular DNA in Biofilm Formation by Staphylococcus aureus. J. Bacteriol. 2015, 197, 3779–3787. [Google Scholar] [CrossRef] [Green Version]
- DeFrancesco, A.S.; Masloboeva, N.; Syed, A.K.; DeLoughery, A.; Bradshaw, N.; Li, G.-W.; Gilmore, M.S.; Walker, S.; Losick, R. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, E5969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, K.; Ganesan, M.; Payne, D.E.; Solomon, M.J.; Boles, B.R. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol. Microbiol. 2016, 99, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Goodman, S.D.; Obergfell, K.P.; Jurcisek, J.A.; Novotny, L.A.; Downey, J.S.; Ayala, E.A.; Tjokro, N.; Li, B.; Justice, S.S.; Bakaletz, L.O. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011, 4, 625–637. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Mann, E.E.; Endres, J.L.; Weiss, E.C.; Cassat, J.E.; Smeltzer, M.S.; Bayles, K.W. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2007, 104, 8113–8118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moormeier, D.E.; Bose, J.L.; Horswill, A.R.; Bayles, K.W. Temporal and stochastic control of Staphylococcus aureus biofilm development. mBio 2014, 5, e01341-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vuong, C.; Saenz, H.L.; Gotz, F.; Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef] [Green Version]
- Nealson, K.H.; Platt, T.; Hastings, J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970, 104, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Ji, G.; Beavis, R.C.; Novick, R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 1995, 92, 12055–12059. [Google Scholar] [CrossRef] [Green Version]
- Novick, R.P.; Projan, S.J.; Kornblum, J.; Ross, H.F.; Ji, G.; Kreiswirth, B.; Vandenesch, F.; Moghazeh, S. The agr P2 operon: An autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 1995, 248, 446–458. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975. [Google Scholar] [CrossRef]
- Abdelnour, A.; Arvidson, S.; Bremell, T.; Rydén, C.; Tarkowski, A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 1993, 61, 3879–3885. [Google Scholar] [CrossRef] [Green Version]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Khan, B.A.; Cheung, G.Y.; Bach, T.-H.L.; Jameson-Lee, M.; Kong, K.-F.; Queck, S.Y.; Otto, M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Investig. 2011, 121, 238–248. [Google Scholar] [CrossRef]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012, 8, e1002744. [Google Scholar] [CrossRef]
- Salam, A.M.; Quave, C.L. Targeting virulence in Staphylococcus aureus by chemical inhibition of the accessory gene regulator system in vivo. mSphere 2018, 3, e00500-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramton, S.E.; Ulrich, M.; Gotz, F.; Doring, G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69, 4079–4085. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, M.; Bastian, M.; Cramton, S.E.; Ziegler, K.; Pragman, A.A.; Bragonzi, A.; Memmi, G.; Wolz, C.; Schlievert, P.M.; Cheung, A.; et al. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 2007, 65, 1276–1287. [Google Scholar] [CrossRef]
- Cue, D.; Lei, M.G.; Lee, C.Y. Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell. Infect. Microbiol. 2012, 2, 38. [Google Scholar] [CrossRef] [Green Version]
- Pamp, S.J.; Frees, D.; Engelmann, S.; Hecker, M.; Ingmer, H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2006, 188, 4861–4870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cue, D.; Lei, M.G.; Luong, T.T.; Kuechenmeister, L.; Dunman, P.M.; O’Donnell, S.; Rowe, S.; O’Gara, J.P.; Lee, C.Y. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. 2009, 191, 6363–6373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferson, K.K.; Pier, D.B.; Goldmann, D.A.; Pier, G.B. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 2004, 186, 2449–2456. [Google Scholar] [CrossRef] [Green Version]
- Kiem, S.; Oh, W.S.; Peck, K.R.; Lee, N.Y.; Lee, J.Y.; Song, J.H.; Hwang, E.S.; Kim, E.C.; Cha, C.Y.; Choe, K.W. Phase variation of biofilm formation in Staphylococcus aureus by IS 256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 2004, 19, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Debarbouille, M.; Penades, J.R.; Lasa, I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef] [Green Version]
- Fitzpatrick, F.; Humphreys, H.; O’Gara, J.P. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 2005, 43, 1973–1976. [Google Scholar] [CrossRef] [Green Version]
- Boles, B.R.; Thoendel, M.; Roth, A.J.; Horswill, A.R. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5, e10146. [Google Scholar] [CrossRef] [Green Version]
- Houston, P.; Rowe, S.E.; Pozzi, C.; Waters, E.M.; O’Gara, J.P. Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 2011, 79, 1153–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huseby, M.J.; Kruse, A.C.; Digre, J.; Kohler, P.L.; Vocke, J.A.; Mann, E.E.; Bayles, K.W.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 14407–14412. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.; Cockayne, A.; Morrissey, J.A. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires icaand the secreted protein Emp. Infect. Immun. 2008, 76, 1756. [Google Scholar] [CrossRef] [Green Version]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadurugamuwa, J.L.; Beveridge, T.J. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: A novel mechanism of enzyme secretion. J. Bacteriol. 1995, 177, 3998–4008. [Google Scholar] [CrossRef] [Green Version]
- Rice, K.C.; Firek, B.A.; Nelson, J.B.; Yang, S.-J.; Patton, T.G.; Bayles, K.W. The Staphylococcus aureus cidAB operon: Evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 2003, 185, 2635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moormeier, D.E.; Endres, J.L.; Mann, E.E.; Sadykov, M.R.; Horswill, A.R.; Rice, K.C.; Fey, P.D.; Bayles, K.W. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 2013, 79, 3413–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma-Kuinkel, B.K.; Mann, E.E.; Ahn, J.S.; Kuechenmeister, L.J.; Dunman, P.M.; Bayles, K.W. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 2009, 191, 4767–4775. [Google Scholar] [CrossRef] [Green Version]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Fehrenbacher, B.; Eisele, K.; Schaller, M.; Gotz, F. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol. Lett. 2005, 252, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapotoczna, M.; O’Neill, E.; O’Gara, J.P. Untangling the diverse and redundant mechanisms of Staphylococcus aureus biofilm formation. PLoS Pathog. 2016, 12, e1005671. [Google Scholar] [CrossRef] [PubMed]
- Thomer, L.; Emolo, C.; Thammavongsa, V.; Kim, H.K.; McAdow, M.E.; Yu, W.; Kieffer, M.; Schneewind, O.; Missiakas, D. Antibodies against a secreted product of Staphylococcus aureus trigger phagocytic killing. J. Exp. Med. 2016, 213, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Zapotoczna, M.; McCarthy, H.; Rudkin, J.K.; O’Gara, J.P.; O’Neill, E. An essential role for coagulase in Staphylococcus aureus biofilm development reveals new therapeutic possibilities for device-related infections. J. Infect. Dis. 2015, 212, 1883–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation, and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resch, A.; Leicht, S.; Saric, M.; Pasztor, L.; Jakob, A.; Gotz, F.; Nordheim, A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 2006, 6, 1867–1877. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Dunman, P.; Kormanec, J.; Macapagal, D.; Murphy, E.; Mounts, W.; Berger-Bachi, B.; Projan, S. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 2004, 186, 4085–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cue, D.; Junecko, J.M.; Lei, M.G.; Blevins, J.S.; Smeltzer, M.S.; Lee, C.Y. SaeRS-dependent inhibition of biofilm formation in Staphylococcus aureus Newman. PLoS ONE 2015, 10, e0123027. [Google Scholar] [CrossRef] [PubMed]
- Fournier, B.; Klier, A.; Rapoport, G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 2001, 41, 247–261. [Google Scholar] [CrossRef]
- Valle, J.; Toledo-Arana, A.; Berasain, C.; Ghigo, J.M.; Amorena, B.; Penades, J.R.; Lasa, I. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 2003, 48, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Dunman, P.M.; Murphy, E.; Haney, S.; Palacios, D.; Tucker-Kellogg, G.; Wu, S.; Brown, E.L.; Zagursky, R.J.; Shlaes, D.; Projan, S.J. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 2001, 183, 7341–7353. [Google Scholar] [CrossRef] [Green Version]
- Beenken, K.E.; Blevins, J.S.; Smeltzer, M.S. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 2003, 71, 4206–4211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, L.H.; Cassat, J.E.; Shaw, L.N.; Beenken, K.E.; Smeltzer, M.S. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 2008, 3, e3361. [Google Scholar] [CrossRef]
- Cheung, A.L.; Projan, S.J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 1994, 176, 4168–4172. [Google Scholar] [CrossRef] [Green Version]
- Beenken, K.E.; Mrak, L.N.; Griffin, L.M.; Zielinska, A.K.; Shaw, L.N.; Rice, K.C.; Horswill, A.R.; Bayles, K.W.; Smeltzer, M.S. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5, e10790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.F.; Vuong, C.; Otto, M. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 2006, 296, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.P.; Bischoff, M.; Senn, M.M.; Berger-Bachi, B. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 2003, 71, 4167–4170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef] [Green Version]
- Kullik, I.; Giachino, P.; Fuchs, T. Deletion of the alternative sigma factor in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 1998, 180, 4814–4820. [Google Scholar] [CrossRef] [Green Version]
- Rachid, S.; Ohlsen, K.; Wallner, U.; Hacker, J.; Hecker, M.; Ziebuhr, W. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 2000, 182, 6824–6826. [Google Scholar] [CrossRef] [Green Version]
- Pane-Farre, J.; Jonas, B.; Forstner, K.; Engelmann, S.; Hecker, M. The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 2006, 296, 237–258. [Google Scholar] [CrossRef]
- Hu, C.; Xiong, N.; Zhang, Y.; Rayner, S.; Chen, S. Functional characterization of lipase in the pathogenesis of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 2012, 419, 617–620. [Google Scholar] [CrossRef]
- Xiong, N.; Hu, C.; Zhang, Y.; Chen, S. Interaction of sortase A and lipase 2 in the inhibition of Staphylococcus aureus biofilm formation. Arch. Microbiol. 2009, 191, 879. [Google Scholar] [CrossRef]
- Heukelekian, H.; Heller, A. Relation between food concentration and surface for bacterial growth. J. Bacteriol. 1940, 40, 547–558. [Google Scholar] [CrossRef] [Green Version]
- Maradit Kremers, H.; Larson, D.R.; Crowson, C.S.; Kremers, W.K.; Washington, R.E.; Steiner, C.A.; Jiranek, W.A.; Berry, D.J. Prevalence of total hip and knee replacement in the United States. J. Bone Jt. Surg. 2015, 97, 1386–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konopka, J.F.; Lee, Y.-Y.; Su, E.P.; McLawhorn, A.S. Quality-adjusted life years after hip and knee arthroplasty: Health-related quality of life after 12,782 joint replacements. JBJS Open Access 2018, 3, e0007. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.M.; Lau, E.; Watson, H.; Schmier, J.K.; Parvizi, J. Economic burden of periprosthetic joint infection in the United States. J. Arthroplast. 2012, 27, 61–65.e61. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.A.; Darouiche, R.O. Device-associated infections: A macroproblem that Starts with microadherence. Clin. Infect. Dis. 2001, 33, 1567–1572. [Google Scholar]
- Peel, T.N.; Cheng, A.C.; Buising, K.L.; Choong, P.F. Microbiological aetiology, epidemiology, and clinical profile of prosthetic joint infections: Are current antibiotic prophylaxis guidelines effective? Antimicrob. Agents Chemother. 2012, 56, 2386–2391. [Google Scholar] [CrossRef] [Green Version]
- Zimmerli, W.; Trampuz, A.; Ochsner, P.E. Prosthetic-joint infections. N. Engl. J. Med. 2004, 351, 1645–1654. [Google Scholar] [CrossRef] [Green Version]
- Del Pozo, J.L.; Patel, R. Clinical practice. Infection associated with prosthetic joints. N. Engl. J. Med. 2009, 361, 787–794. [Google Scholar] [CrossRef] [Green Version]
- Southwood, R.T.; Rice, J.L.; McDonald, P.J.; Hakendorf, P.H.; Rozenbilds, M.A. Infection in experimental hip arthroplasties. J. Bone Jt. Surg. 1985, 67, 229–231. [Google Scholar] [CrossRef]
- Kunutsor, S.K.; Whitehouse, M.R.; Blom, A.W.; Beswick, A.D. Re-infection outcomes following one- and two-stage surgical revision of infected hip prosthesis: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0139166. [Google Scholar] [CrossRef] [Green Version]
- Sierra, R.J.; Trousdale, R.T.; Pagnano, M.W. Above-the-knee amputation after a total knee replacement: Prevalence, etiology, and functional outcome. J. Bone Jt. Surg. 2003, 85, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ren, D. Quorum sensing inhibitors: A patent overview. Expert Opin. Ther. Pat. 2009, 19, 1581–1601. [Google Scholar] [CrossRef]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [Green Version]
- Krzyżek, P. Challenges and Limitations of Anti-quorum Sensing Therapies. Front. Microbiol. 2019, 10, 2473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirioni, O.; Giacometti, A.; Ghiselli, R.; Dell’Acqua, G.; Orlando, F.; Mocchegiani, F.; Silvestri, C.; Licci, A.; Saba, V.; Scalise, G.; et al. RNAIII-inhibiting peptide significantly reduces bacterial load and enhances the effect of antibiotics in the treatment of central venous catheter-associated Staphylococcus aureus infections. J. Infect. Dis. 2006, 193, 180–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anguita-Alonso, P.; Giacometti, A.; Cirioni, O.; Ghiselli, R.; Orlando, F.; Saba, V.; Scalise, G.; Sevo, M.; Tuzova, M.; Patel, R.; et al. RNAIII-inhibiting-peptide-loaded polymethylmethacrylate prevents in vivo Staphylococcus aureus biofilm formation. Antimicrob. Agents Chemother. 2007, 51, 2594–2596. [Google Scholar] [CrossRef] [Green Version]
- Balaban, N.; Cirioni, O.; Giacometti, A.; Ghiselli, R.; Braunstein, J.B.; Silvestri, C.; Mocchegiani, F.; Saba, V.; Scalise, G. Treatment of Staphylococcus aureus biofilm infection by the quorum-sensing inhibitor RIP. Antimicrob. Agents Chemother. 2007, 51, 2226–2229. [Google Scholar] [CrossRef] [Green Version]
- Cirioni, O.; Giacometti, A.; Ghiselli, R.; Dell’Acqua, G.; Gov, Y.; Kamysz, W.; Lukasiak, J.; Mocchegiani, F.; Orlando, F.; D’Amato, G.; et al. Prophylactic efficacy of topical temporin A and RNAIII-inhibiting peptide in a subcutaneous rat Pouch model of graft infection attributable to staphylococci with intermediate resistance to glycopeptides. Circulation 2003, 108, 767–771. [Google Scholar] [CrossRef] [Green Version]
- Gov, Y.; Bitler, A.; Dell’Acqua, G.; Torres, J.V.; Balaban, N. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: Structure and function analysis. Peptides 2001, 22, 1609–1620. [Google Scholar] [CrossRef]
- Balaban, N.; Goldkorn, T.; Nhan, R.T.; Dang, L.B.; Scott, S.; Ridgley, R.M.; Rasooly, A.; Wright, S.C.; Larrick, J.W.; Rasooly, R.; et al. Autoinducer of virulence as a target for vaccine and therapy against Staphylococcus aureus. Science 1998, 280, 438–440. [Google Scholar] [CrossRef]
- Balaban, N.; Goldkorn, T.; Gov, Y.; Hirshberg, M.; Koyfman, N.; Matthews, H.R.; Nhan, R.T.; Singh, B.; Uziel, O. Regulation of Staphylococcus aureus pathogenesis via target of RNAIII-activating protein (TRAP). J. Biol. Chem. 2001, 276, 2658–2667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, L.N.; Jonsson, M.; Singh, V.K.; Tarkowski, A.; Stewart, G.C. Inactivation of traP has no effect on the agr quorum-sensing system or virulence of Staphylococcus aureus. Infect. Immun. 2007, 75, 4519–4527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, G.J.; Novick, R.P. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 2004, 25, 1389–1403. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.; Gov, Y.; Bitler, A.; Boelaert, J.R. Prevention of Staphylococcus aureus biofilm on dialysis catheters and adherence to human cells. Kidney Int. 2003, 63, 340–345. [Google Scholar] [CrossRef] [Green Version]
- Kiran, M.D.; Adikesavan, N.V.; Cirioni, O.; Giacometti, A.; Silvestri, C.; Scalise, G.; Ghiselli, R.; Saba, V.; Orlando, F.; Shoham, M.; et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol. Pharmacol. 2008, 73, 1578–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [Green Version]
- Brackman, G.; Breyne, K.; De Rycke, R.; Vermote, A.; Van Nieuwerburgh, F.; Meyer, E.; Van Calenbergh, S.; Coenye, T. The quorum sensing inhibitor hamamelitannin increases antibiotic susceptibility of Staphylococcus aureus biofilms by affecting peptidoglycan biosynthesis and eDNA release. Sci. Rep. 2016, 6, 20321. [Google Scholar] [CrossRef]
- Vermote, A.; Brackman, G.; Risseeuw, M.D.; Vanhoutte, B.; Cos, P.; Van Hecke, K.; Breyne, K.; Meyer, E.; Coenye, T.; Van Calenbergh, S. Hamamelitannin analogues that modulate quorum sensing as potentiators of antibiotics against Staphylococcus aureus. Angew. Chem. Int. Ed. 2016, 55, 6551–6555. [Google Scholar] [CrossRef] [PubMed]
- Goerke, C.; Kümmel, M.; Dietz, K.; Wolz, C. Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J. Infect. Dis. 2003, 188, 250–256. [Google Scholar] [CrossRef] [Green Version]
- MDowell, P.; Affas, Z.; Reynolds, C.; Holden, M.T.; Wood, S.J.; Saint, S.; Cockayne, A.; Hill, P.J.; Dodd, C.E.; Bycroft, B.W.; et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol. Microbiol. 2001, 41, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayville, P.; Ji, G.; Beavis, R.; Yang, H.; Goger, M.; Novick, R.P.; Muir, T.W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl. Acad. Sci. USA 1999, 96, 1218–1223. [Google Scholar] [CrossRef] [Green Version]
- Lyon, G.J.; Mayville, P.; Muir, T.W.; Novick, R.P. Rational design of a global inhibitor of the virulence response in Staphylococcus aureus, based in part on localization of the site of inhibition to the receptor-histidine kinase, AgrC. Proc. Natl. Acad. Sci. USA 2000, 97, 13330–13335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, G.J.; Wright, J.S.; Muir, T.W.; Novick, R.P. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 2002, 41, 10095–10104. [Google Scholar] [CrossRef] [PubMed]
- Janda, K.D.; Kaufmann, G.F.; Park, J. Antibody-Mediated Disruption of Quorum Sensing in Bacteria. 2016. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2009055054&tab=PCTDESCRIPTION (accessed on 31 August 2021).
- Park, J.; Jagasia, R.; Kaufmann, G.F.; Mathison, J.C.; Ruiz, D.I.; Moss, J.A.; Meijler, M.M.; Ulevitch, R.J.; Janda, K.D. Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 2007, 14, 1119–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.; Fearon, U.; Veale, D.J.; Godson, C. Macrophages in synovial inflammation. Front. Immunol. 2011, 2, 52. [Google Scholar] [CrossRef] [Green Version]
- Thabit, A.K.; Fatani, D.F.; Bamakhrama, M.S.; Barnawi, O.A.; Basudan, L.O.; Alhejaili, S.F. Antibiotic penetration into bone and joints: An updated review. Int. J. Infect. Dis. 2019, 81, 128–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allarakhia, M. Open-source approaches for the repurposing of existing or failed candidate drugs: Learning from and applying the lessons across diseases. Drug Des. Dev. Ther. 2013, 7, 753–766. [Google Scholar] [CrossRef] [Green Version]
- Imperi, F.; Massai, F.; Ramachandran Pillai, C.; Longo, F.; Zennaro, E.; Rampioni, G.; Visca, P.; Leoni, L. New life for an old drug: The anthelmintic drug niclosamide inhibits Pseudomonas aeruginosa quorum sensing. Antimicrob. Agents Chemother. 2013, 57, 996–1005. [Google Scholar] [CrossRef] [Green Version]
- Aguinagalde, L.; Díez-Martínez, R.; Yuste, J.; Royo, I.; Gil, C.; Lasa, Í.; Martín-Fontecha, M.; Marín-Ramos, N.I.; Ardanuy, C.; Liñares, J.; et al. Auranofin efficacy against MDR Streptococcus pneumoniae and Staphylococcus aureus infections. J. Antimicrob. Chemother. 2015, 70, 2608–2617. [Google Scholar] [CrossRef] [Green Version]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.N.; Sobreira, T.J.P.; Hedrick, V.E.; Paul, L.N.; Seleem, M.N. Antibacterial activity and mechanism of action of auranofin against multi-drug resistant bacterial pathogens. Sci. Rep. 2016, 6, 22571. [Google Scholar] [CrossRef] [Green Version]
- She, P.; Zhou, L.; Li, S.; Liu, Y.; Xu, L.; Chen, L.; Luo, Z.; Wu, Y. Synergistic microbicidal effect of auranofin and antibiotics against planktonic and biofilm-encased S. aureus and E. faecalis. Front. Microbiol. 2019, 10, 2453. [Google Scholar] [CrossRef]
- Tharmalingam, N.; Ribeiro, N.Q.; da Silva, D.L.; Naik, M.T.; Cruz, L.I.; Kim, W.; Shen, S.; Dos Santos, J.D.; Ezikovich, K.; D’Agata, E.M.; et al. Auranofin is an effective agent against clinical isolates of Staphylococcus aureus. Future Med. Chem. 2019, 11, 1417–1425. [Google Scholar] [CrossRef]
- Kupferwasser, L.I.; Yeaman, M.R.; Shapiro, S.M.; Nast, C.C.; Sullam, P.M.; Filler, S.G.; Bayer, A.S. Acetylsalicylic acid reduces vegetation bacterial density, hematogenous bacterial dissemination, and frequency of embolic events in experimental Staphylococcus aureus endocarditis through antiplatelet and antibacterial effects. Circulation 1999, 99, 2791–2797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlacek, M.; Gemery, J.M.; Cheung, A.L.; Bayer, A.S.; Remillard, B.D. Aspirin treatment is associated with a significantly decreased risk of Staphylococcus aureus bacteremia in hemodialysis patients with tunneled catheters. Am. J. Kidney Dis. 2007, 49, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Kupferwasser, L.I.; Yeaman, M.R.; Nast, C.C.; Kupferwasser, D.; Xiong, Y.Q.; Palma, M.; Cheung, A.L.; Bayer, A.S. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Investig. 2003, 112, 222–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dotto, C.; Lombarte Serrat, A.; Cattelan, N.; Barbagelata, M.S.; Yantorno, O.M.; Sordelli, D.O.; Ehling-Schulz, M.; Grunert, T.; Buzzola, F.R. The active component of aspirin, salicylic acid, promotes Staphylococcus aureus biofilm formation in a PIA-dependent manner. Front. Microbiol. 2017, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Lade, H.; Park, J.H.; Chung, S.H.; Kim, I.H.; Kim, J.-M.; Joo, H.-S.; Kim, J.-S. Biofilm formation by Staphylococcus aureus clinical isolates is differentially affected by glucose and sodium chloride supplemented culture media. J. Clin. Med. 2019, 8, 1853. [Google Scholar] [CrossRef] [Green Version]
- Dotto, C.; Lombarte Serrat, A.; Ledesma, M.; Vay, C.; Ehling-Schulz, M.; Sordelli, D.O.; Grunert, T.; Buzzola, F. Salicylic acid stabilizes Staphylococcus aureus biofilm by impairing the agr quorum-sensing system. Sci. Rep. 2021, 11, 2953. [Google Scholar] [CrossRef]
- Springthorpe, B.; Bailey, A.; Barton, P.; Birkinshaw, T.N.; Bonnert, R.V.; Brown, R.C.; Chapman, D.; Dixon, J.; Guile, S.D.; Humphries, R.G.; et al. From ATP to AZD6140: The discovery of an orally active reversible P2Y12 receptor antagonist for the prevention of thrombosis. Bioorg. Med. Chem. Lett. 2007, 17, 6013–6018. [Google Scholar] [CrossRef]
- Wallentin, L.; Becker, R.C.; Budaj, A.; Cannon, C.P.; Emanuelsson, H.; Held, C.; Horrow, J.; Husted, S.; James, S.; Katus, H.; et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N. Engl. J. Med. 2009, 361, 1045–1057. [Google Scholar] [CrossRef]
- Storey, R.F.; James, S.K.; Siegbahn, A.; Varenhorst, C.; Held, C.; Ycas, J.; Husted, S.E.; Cannon, C.P.; Becker, R.C.; Steg, P.G.; et al. Lower mortality following pulmonary adverse events and sepsis with ticagrelor compared to clopidogrel in the PLATO study. Platelets 2014, 25, 517–525. [Google Scholar] [CrossRef]
- Sexton, T.R.; Zhang, G.; Macaulay, T.E.; Callahan, L.A.; Charnigo, R.; Vsevolozhskaya, O.A.; Li, Z.; Smyth, S. Ticagrelor reduces thromboinflammatory markers inpatients with pneumonia. JACC Basic Transl. Sci. 2018, 3, 435–449. [Google Scholar] [CrossRef] [PubMed]
- Lancellotti, P.; Musumeci, L.; Jacques, N.; Servais, L.; Goffin, E.; Pirotte, B.; Oury, C. Antibacterial activity of ticagrelor in conventional antiplatelet dosages against antibiotic-resistant Gram-positive bacteria. JAMA Cardiol. 2019, 4, 596–599. [Google Scholar] [CrossRef] [Green Version]
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [CrossRef] [Green Version]
- Hannachi, N.; Grac, L.; Baudoin, J.P.; Fournier, P.E.; Habib, G.; Camoin-Jau, L. Effect of antiplatelet agents on platelet antistaphylococcal capacity: An in vitro study. Int. J. Antimicrob. Agents 2020, 55, 105890. [Google Scholar] [CrossRef] [PubMed]
- Graziano, T.S.; Cuzzullin, M.C.; Franco, G.C.; Schwartz-Filho, H.O.; de Andrade, E.D.; Groppo, F.C.; Cogo-Müller, K. Statins and antimicrobial effects: Simvastatin as a potential drug against Staphylococcus aureus biofilm. PLoS ONE 2015, 10, e0128098. [Google Scholar] [CrossRef] [PubMed]
- Thangamani, S.; Mohammad, H.; Abushahba, M.F.; Hamed, M.I.; Sobreira, T.J.; Hedrick, V.E.; Paul, L.N.; Seleem, M.N. Exploring simvastatin, an antihyperlipidemic drug, as a potential topical antibacterial agent. Sci. Rep. 2015, 5, 16407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, B.L.; Sohnle, P.G. Effect of thioridazine on experimental cutaneous staphylococcal infections. In Vivo 2014, 28, 33–38. [Google Scholar]
- Thorsing, M.; Klitgaard, J.K.; Atilano, M.L.; Skov, M.N.; Kolmos, H.J.; Filipe, S.R.; Kallipolitis, B.H. Thioridazine induces major changes in global gene expression and cell wall composition in methicillin-resistant Staphylococcus aureus USA300. PLoS ONE 2013, 8, e64518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pant, N.; Eisen, D.P. Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections. Antibiotics 2021, 10, 1060. https://doi.org/10.3390/antibiotics10091060
Pant N, Eisen DP. Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections. Antibiotics. 2021; 10(9):1060. https://doi.org/10.3390/antibiotics10091060
Chicago/Turabian StylePant, Narayan, and Damon P. Eisen. 2021. "Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections" Antibiotics 10, no. 9: 1060. https://doi.org/10.3390/antibiotics10091060
APA StylePant, N., & Eisen, D. P. (2021). Non-Antimicrobial Adjuvant Strategies to Tackle Biofilm-Related Staphylococcus aureus Prosthetic Joint Infections. Antibiotics, 10(9), 1060. https://doi.org/10.3390/antibiotics10091060






