Taxonomic and Functional Distribution of Bacterial Communities in Domestic and Hospital Wastewater System: Implications for Public and Environmental Health
Abstract
1. Introduction
2. Results
2.1. Physicochemical Analysis
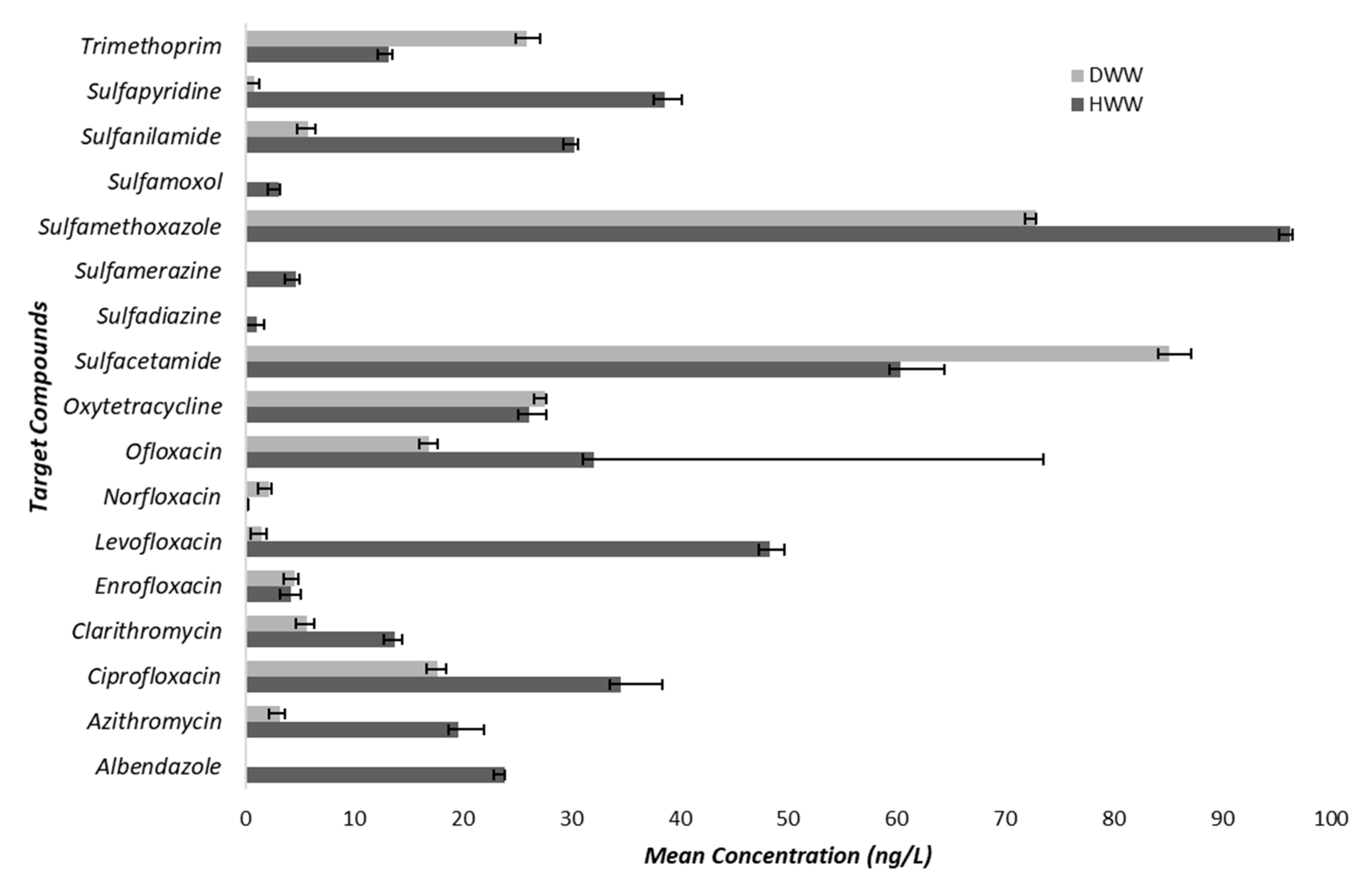
2.2. Antibiotics Concentration
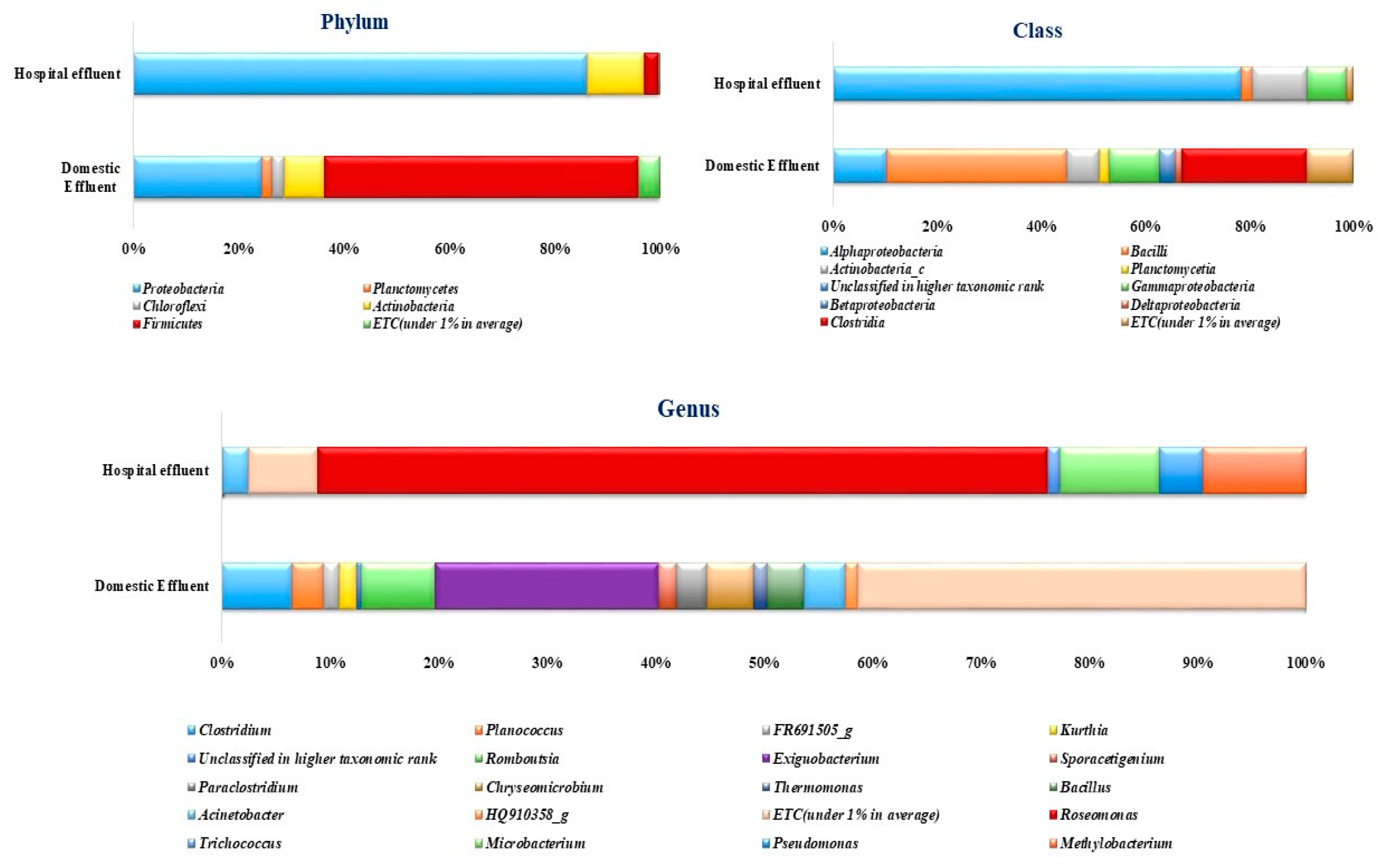
2.3. Diversity of Bacterial Communities
2.4. Functional Analysis
3. Discussion
4. Materials and Methods
4.1. Study Area and Sampling
4.2. Physico-Chemical Analysis of Wastewater Samples
4.3. Extraction and Quantification of Antibiotics
4.4. DNA Extraction, Library Preparation and Illumina Miseq High Throughput Sequencing
4.5. Sequence Data and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaur, R.; Yadav, B.; Tyagi, R.D. Microbiology of Hospital Wastewater. In Current developments in Biotechnology and bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 103–148. ISBN 9780128197226. [Google Scholar]
- Ramganesh, S.; Timothy, S.; Venkatachalam, S.; Kamika, I.; Nel, W.A.J. Industrial wastewaters harbor a unique diversity of bacterial communities revealed by high-throughput amplicon analysis. Ann. Microbiol. 2018, 68, 445–458. [Google Scholar]
- Asfaw, T.; Negash, L.; Kahsay, A.; Weldu, Y. Antibiotic Resistant Bacteria from Treated and Untreated Hospital Wastewater at Ayder Referral Hospital, Mekelle, North Ethiopia. Adv. Microbiol. 2017, 7, 871–886. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Chaudhury, R.R. Antibiotic Resistance in India: Drivers and Opportunities for Action. PLoS Med. 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Miyazaki, A.; Tung, R.; Taing, B.; Matsui, M.; Iwamoto, A.; Cox, S.E. Frequent unregulated use of antibiotics in rural Cambodian infants. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 401–407. [Google Scholar] [CrossRef]
- Sulis, G.; Batomen, B.; Kotwani, A.; Pai, M.; Gandra, S. Sales of antibiotics and hydroxychloroquine in India during the COVID-19 epidemic: An interrupted time series analysis. PLoS Med. 2021, 18, e1003682. [Google Scholar] [CrossRef]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Medicin. Chem. 2014, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Deters, B.; de la Bastide, A.; Korzen, M. Antibiotics Overuse and Bacterial Resistance. Ann. Microbiol. Res. 2019, 3, 93–99. [Google Scholar]
- Petrovich, M.L.; Ben Maamar, S.; Hartmann, E.M.; Murphy, B.T.; Poretsky, R.S.; Wells, G.F. Viral composition and context in metagenomes from biofilm and suspended growth municipal wastewater treatment plants. Microb. Biotechnol. 2019, 12, 1324–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef] [PubMed]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiologyopen 2021, 10, 1–11. [Google Scholar] [CrossRef]
- Kraemer, S.A.; Ramachandran, A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef]
- Le, T.H.; Ng, C.; Chen, H.; Yi, X.Z.; Koh, T.H.; Barkham, T.M.S.; Zhou, Z.; Gin, K.Y.H. Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrob. Agents Chemother. 2016, 60, 7449–7456. [Google Scholar] [CrossRef]
- Tsai, C.T.; Lai, J.S.; Lin, S.T. Quantification of pathogenic micro-organisms in the sludge from treated hospital wastewater. J. Appl. Microbiol. 1998, 85, 171–176. [Google Scholar] [CrossRef]
- Md. Mijanur, R.; Popy, D.; Rafshan, J.; Asma, T. Detection of multiple antibiotic-resistant bacteria from the hospital and non-hospital wastewater sources of a small town in Noakhali, Bangladesh. J. Appl. Biol. Biotechnol. 2021, 9, 59–65. [Google Scholar]
- Chagas, T.P.G.; Seki, L.M.; Cury, J.C.; Oliveira, J.A.L.; Dávila, A.M.R.; Silva, D.M.; Asensi, M.D. Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.; Choi, J. Bacterial communities and antibiotic resistance communities in a full-scale hospital wastewater treatment plant by high-throughput pyrosequencing. Water 2016, 8, 580. [Google Scholar] [CrossRef]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic resistance genes and bacterial community composition in a river influenced by a wastewater treatment plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Tiwari, B.; Sellamuthu, B.; Piché-Choquette, S.; Drogui, P.; Tyagi, R.D.; Vaudreuil, M.A.; Sauvé, S.; Buelna, G.; Dubé, R. The bacterial community structure of submerged membrane bioreactor treating synthetic hospital wastewater. Bioresour. Technol. 2019, 286, 121362. [Google Scholar] [CrossRef]
- Ekhaise, F.O.; Omavwoya, B.P. Influence of Hospital Wastewater Discharged from University of Benin Teaching Hospital (UBTH), Benin City on its Receiving Environment. Am. J. Agric. Environ. Sci. 2008, 4, 484–488. [Google Scholar]
- Hassoun-kheir, N.; Stabholz, Y.; Kreft, J.; De, R.; Romalde, J.L.; Nesme, J.; Sørensen, S.J.; Smets, B.F.; Graham, D.; Paul, M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Sci. Total Environ. 2020, 743, 140804. [Google Scholar] [CrossRef] [PubMed]
- Tylová, T.; Flieger, M.; Olšovská, J. Determination of antibiotics in influents and effluents of wastewater-treatment-plants in the Czech Republic-development and application of the SPE and a UHPLC-ToFMS method. Anal. Methods 2013, 5, 2110–2118. [Google Scholar] [CrossRef]
- Quoc Tuc, D.; Elodie, M.G.; Pierre, L.; Fabrice, A.; Marie-Jeanne, T.; Martine, B.; Joelle, E.; Marc, C. Fate of antibiotics from hospital and domestic sources in a sewage network. Sci. Total Environ. 2017, 575, 758–766. [Google Scholar]
- Gothwal, R.; Shashidhar, T. Antibiotic Pollution in the Environment: A Review. Clean-Soil Air Water 2015, 43, 479–489. [Google Scholar] [CrossRef]
- Karthikeyan, K.G.; Meyer, M.T. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 2006, 361, 196–207. [Google Scholar] [CrossRef]
- Chandra, S.; Prithvi, P.P.R.; Srija, K.; Jauhari, S.; Grover, A. Antimicrobial resistance: Call for rational antibiotics practice in India. J. Fam. Med. Prim. Care 2020, 9, 2192. [Google Scholar]
- WHO Global Tuberculosis Report 2020: Executive Summary; World Health Organization: Geneva, Switzerland, 2020.
- Finley, R.L.; Collignon, P.; Larsson, D.G.J.; McEwen, S.A.; Li, X.-Z.; Gaze, W.H.; Reid-Smith, R.; Timinouni, M.; Graham, D.W.; Topp, E. The scourge of antibiotic resistance: The important role of the environment. Clin. Infect. Dis. 2013, 57, 704–710. [Google Scholar] [CrossRef]
- Gideon Wyasu Determination of Bacteriological and some physicochemical properties of Hospital wastewater. Commun. Phys. Sci. 2019, 4, 141–150.
- Assress, H.A.; Selvarajan, R.; Nyoni, H.; Ntushelo, K.; Mamba, B.B.; Msagati, T.A.M. Diversity, Co-occurrence and Implications of Fungal Communities in Wastewater Treatment Plants. Sci. Rep. 2019, 9, 14056. [Google Scholar] [CrossRef]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, T.A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plants using high-throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef] [PubMed]
- Devries, S.L.; Zhang, P. Antibiotics and the Terrestrial Nitrogen Cycle: A Review. Curr. Pollut. Rep. 2016, 2, 51–67. [Google Scholar] [CrossRef]
- Jia, J.; Gomes-silva, G.; Plath, M.; Barbosa, B.; Ueiravieira, C.; Wang, Z. Ecotoxicology and Environmental Safety Shifts in bacterial communities and antibiotic resistance genes in surface water and gut microbiota of guppies (Poecilia reticulata) in the upper Rio. Ecotoxicol. Environ. Saf. 2021, 211, 111955. [Google Scholar] [CrossRef]
- Griffin, D.W.; Banks, K.; Gregg, K.; Shedler, S.; Walker, B.K. Antibiotic Resistance in Marine Microbial Communities Proximal to a Florida Sewage Outfall System. Antibiotics 2020, 9, 118. [Google Scholar] [CrossRef]
- Ochoa-Herrera, V.; Banihani, Q.; León, G.; Khatri, C.; Field, J.A.; Sierra-Alvarez, R. Toxicity of fluoride to microorganisms in biological wastewater treatment systems. Water Res. 2009, 43, 3177–3186. [Google Scholar] [CrossRef]
- Diwan, V.; Tamhankar, A.J.; Aggarwal, M.; Sen, S.; Khandal, R.K.; Lundborg, C.S. Detection of antibiotics in hospital effluents in India. Curr. Sci. 2009, 97, 12–15. [Google Scholar]
- Akiba, M.; Senba, H.; Otagiri, H.; Prabhasankar, V.P.; Taniyasu, S.; Yamashita, N.; Lee, K.; Yamamoto, T.; Tsutsui, T.; Ian, D.; et al. Impact of wastewater from different sources on the prevalence of antimicrobial-resistant Escherichia coli in sewage treatment plants in South India. Ecotoxicol. Environ. Saf. 2015, 115, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Verlicchi, P.; Al Aukidy, M.; Galletti, A.; Petrovic, M.; Barceló, D. Hospital effluent: Investigation of the concentrations and distribution of pharmaceuticals and environmental risk assessment. Sci. Total Environ. 2012, 430, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.H.M.L.M.; Gros, M.; Rodriguez-mozaz, S.; Delerue-matos, C.; Pena, A.; Barceló, D.; Montenegro, M.C.B.S.M. Contribution of hospital ef fl uents to the load of pharmaceuticals in urban wastewaters: Identi fi cation of ecologically relevant pharmaceuticals. Sci. Total Environ. 2013, 461–462, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Smilack, J.D. Trimethoprim-sulfamethoxazole. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 1999; Volume 74, pp. 730–734. [Google Scholar]
- Low, D.E. Fluoroquinolones for Treatment of Community-Acquired Pneumonia and Tuberculosis: Putting the Risk of Resistance into Perspective. Clin. Infect. Dis. 2009, 48, 1361–1363. [Google Scholar] [CrossRef][Green Version]
- Sharma, P.C.; Jain, A.; Jain, S. Fluoroquinolone Antibacterials: A review on Chemistry, Microbiology and Therapeutic prospects. Acta Pol. Pharm.-Drug Res. 2009, 66, 587–604. [Google Scholar]
- Szekeres, E.; Chiriac, C.M.; Baricz, A.; Szőke-Nagy, T.; Lung, I.; Soran, M.L.; Rudi, K.; Dragos, N.; Coman, C. Investigating antibiotics, antibiotic resistance genes, and microbial contaminants in groundwater in relation to the proximity of urban areas. Environ. Pollut. 2018, 236, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.L.; Andrei, A.S.; Rudi, K.; Balc�zar, J.L.; Dragos, N.; et al. Abundance of antibiotics, antibiotic resistance genes and bacterial community composition in wastewater effluents from different Romanian hospitals. Environ. Pollut. 2017, 225, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, J.; Zhao, Z.; Cao, Y.; Li, B. Hospital Wastewater as a Reservoir for Antibiotic Resistance Genes: A Meta-Analysis. Front. Public Health 2020, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Verburg, I.; Van Veelen, H.P.J.; Waar, K.; Rossen, J.W.A.; Friedrich, A.W.; Hern, L.; Garc, S.; Schmitt, H. Effects of Clinical Wastewater on the Bacterial Community Structure from Sewage to the Environment. Microorganisms 2021, 9, 718. [Google Scholar] [CrossRef] [PubMed]
- Ogwugwa, V.H.; Oyetibo, G.O.; Amund, O.O. Taxonomic profiling of bacteria and fungi in freshwater sewer receiving hospital wastewater. Environ. Res. 2021, 192, 110319. [Google Scholar] [CrossRef]
- Efstratiou, M.A.; Bountouni, M.; Kefalas, E. Spread of Antibiotic Resistance in Aquatic Environments: E. coli as a Case Study. Multidiscip. Digit. Publ. Inst. Proc. 2018, 2, 693. [Google Scholar] [CrossRef]
- Andrade, L.; Kelly, M.; Hynds, P.; Weatherill, J.; Majury, A.; Dwyer, J.O. Groundwater resources as a global reservoir for antimicrobial- resistant bacteria. Water Res. 2020, 170, 115360. [Google Scholar] [CrossRef] [PubMed]
- Berglund, B.; Khan, G.A.; Weisner, S.E.B.; Ehde, P.M.; Fick, J.; Lindgren, P.-E. Efficient removal of antibiotics in surface-flow constructed wetlands, with no observed impact on antibiotic resistance genes. Sci. Total Environ. 2014, 476, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, T.; Selvarajan, R.; Tekere, M. Targeted 16S rRNA amplicon analysis reveals the diversity of bacterial communities in carwash effluents. Int. Microbiol. 2019, 22, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Sekar, S.; Zintchem, A.A.E.A.; Keshri, J.; Kamika, I.; Momba, M.N.B. Bacterial profiling in brine samples of the Emalahleni water reclamation plant, South Africa, using 454-pyrosequencing method. FEMS Microbiol. Lett. 2014, 359, 55–63. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Ciesielski, S.; Korzeniewska, E.; Osińska, A. Environmental fate of Bacteroidetes, with particular emphasis on Bacteroides fragilis group bacteria and their specific antibiotic resistance genes, in activated sludge wastewater treatment plants. J. Hazard. Mater. 2020, 394, 122544. [Google Scholar] [CrossRef]
- Palanisamy, V.; Gajendiran, V.; Mani, K. Meta - analysis to identify the core microbiome in diverse wastewater. Int. J. Environ. Sci. Technol. 2021, 1–18. [Google Scholar] [CrossRef]
- Rubin, M.A.; Leff, L.G. Nutrients and other abiotic factors affecting bacterial communities in an Ohio River (USA). Microb. Ecol. 2007, 54, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N. Exiguobacterium. Benef. Microbes Agro-Ecol. 2020, 169–183. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Zhou, J.; Wu, H.; Li, D.; Cui, Y.; Lu, B. Exiguobacterium sp. A1b/GX59 isolated from a patient with community-acquired pneumonia and bacteremia: Genomic characterization and literature review. BMC Infect. Dis. 2017, 17, 1–7. [Google Scholar] [CrossRef]
- Romano-Bertrand, S.; Bourdier, A.; Aujoulat, F.; Michon, A.L.; Masnou, A.; Parer, S.; Marchandin, H.; Jumas-Bilak, E. Skin microbiota is the main reservoir of Roseomonas mucosa, an emerging opportunistic pathogen so far assumed to be environmental. Clin. Microbiol. Infect. 2016, 22, 737.e1–737.e7. [Google Scholar] [CrossRef] [PubMed]
- Dent, L.L.; Marshall, D.R.; Pratap, S.; Hulette, R.B. Multidrug resistant Acinetobacter baumannii: A descriptive study in a city hospital. BMC Infect. Dis. 2010, 10, 1–7. [Google Scholar] [CrossRef]
- Hu, Q.; Hu, Z.; Li, J.; Tian, B.; Xu, H.; Li, J. Detection of OXA-type carbapenemases and integrons among carbapenem-resistant Acinetobactor baumannii in a Teaching Hospital in China. J. Basic Microbiol. 2011, 51, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Kornelsen, V.; Kumar, A. Update on Multidrug Resistance Efflux Pumps in Acinetobacter spp. Antimicrob. Agents Chemother. 2021, 65, e00514-21. [Google Scholar] [CrossRef] [PubMed]
- Kizny Gordon, A.E.; Mathers, A.J.; Cheong, E.Y.L.; Gottlieb, T.; Kotay, S.; Walker, A.S.; Peto, T.E.A.; Crook, D.W.; Stoesser, N. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—A systematic review of the literature. Clin. Infect. Dis. 2017, 64, 1435–1444. [Google Scholar] [CrossRef]
- Alam, M.; Imran, M. Screening and Potential of Multi-drug Resistance in Gram-Negative Bacteria from Hospital Wastewater. Iran. J. Sci. Technol. Trans. A Sci. 2018, 42, 251–259. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Megahed, A.; Zeineldin, M.; Evans, K.; Maradiaga, N.; Blair, B.; Aldridge, B.; Lowe, J. Impacts of environmental complexity on respiratory and gut microbiome community structure and diversity in growing pigs. Sci. Rep. 2019, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.; Romanenko, M.; Piven, L.; Moseiko, V.; Lushchak, O.; Kryzhanovska, N.; Guryanov, V.; Koliada, A. Differences in the gut Firmicutes to Bacteroidetes ratio across age groups in healthy Ukrainian population. BMC Microbiol. 2020, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, B.; Drogui, P.; Tyagi, R.D. Multidrug-resistant genes and pathogenic bacteria in hospital wastewater. In Current Developments in Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2020; pp. 177–202. ISBN 9780128197226. [Google Scholar]
- Bonten, M.J.M.; Hayden, M.K.; Nathan, C.; van Voorhis, J.; Matushek, M.; Slaughter, S.; Rice, T.; Weinstein, R.A. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 1996, 348, 1615–1619. [Google Scholar] [CrossRef]
- Hocquet, D.; Muller, A.; Bertrand, X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016, 93, 395–402. [Google Scholar] [CrossRef]
- Das, B.; Verma, J.; Kumar, P.; Ghosh, A.; Ramamurthy, T. Antibiotic resistance in Vibrio cholerae: Understanding the ecology of resistance genes and mechanisms. Vaccine 2020, 38, A83–A92. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E.; Carnelutti, A.; Graziano, E.; Russo, A. Multidrug-resistant Klebsiella pneumoniae: Challenges for treatment, prevention and infection control. Expert Rev. Anti. Infect. Ther. 2018, 16, 749–761. [Google Scholar] [CrossRef]
- Mitchell, B.A.; Brown, M.H.; Skurray, R.A. QacA multidrug efflux pump from Staphylococcus aureus: Comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob. Agents Chemother. 1998, 42, 475–477. [Google Scholar] [CrossRef]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Elangovan, N.S.; Dharmendirakumar, M. Assessment of Groundwater Quality along the Cooum River, Chennai, Tamil Nadu, India. J. Chem. 2013, 2013. [Google Scholar] [CrossRef]
- Baird, R.B.; Rice, E.W.; Posavec, S. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association, American Water Works Association, Water Environment Federation: Alexandria, VA, USA, 2017. [Google Scholar]
- Mhuka, V.; Dube, S.; Nindi, M.M. Occurrence of pharmaceutical and personal care products (PPCPs) in wastewater and receiving waters in South Africa using LC-OrbitrapTM MS. Emerg. Contam. 2020, 6, 250–258. [Google Scholar] [CrossRef]
- Chen, C.; Khaleel, S.S.; Huang, H.; Wu, C.H. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol. Med. 2014, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Good, I.J.; Toulmin, G.H. The number of new species, and the increase in population coverage, when a sample is increased. Biometrika 1956, 43, 45–63. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 672295. [Google Scholar] [CrossRef]
- Sibanda, T.; Ramganesh, S. Taxonomic and functional analyses reveal existence of virulence and antibiotic resistance genes in beach sand bacterial populations. Arch. Microbiol. 2021, 203, 1753–1766. [Google Scholar] [CrossRef]
- R Core Team. R. A Language and Environment for Statistical Computing; Version 3.5.2; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]



| Domestic Effluent (DWW) | Hospital Effluent (HWW) | UNEP | ENVIS | |
|---|---|---|---|---|
| pH | 6.6 ± 0.07 | 7.2 ± 0.28 | 6.6–8.5 | 6.5–8.5 |
| DO (mg L−1) | 0.534 ± 0.50 | 0.94 ± 0.42 | - | - |
| Conductivity (μS/cm) | 902 ± 2.21 | 628 ± 6.36 | 380 | - |
| Salinity (SAL) | 0.45 ± 0.07 | 0.3 ± 0.00 | - | - |
| NH3-N | 0.745 ± 0.008 | 0.13 ± 0.04 | 0.21 | - |
| DOC (mg L−1) | 50.39 ± 0.62 | 67.35 ± 0.21 | - | - |
| TDS (mg L−1) | 587.5 ± 0.71 | 431.5 ± 18.68 | - | 1500 |
| Cl (mg L−1) | 19.64 ± 3.75 | 32.95 ± 2.82 | 20 | - |
| Br− (mg L−1) | 0.45 ± 0.095 | ND | - | - |
| F− (mg L−1) | 0.42 ± 0.081 | 4.58 ± 0.52 | - | 1.5 |
| Nitrite (mg L−1) | 0.425 ± 0.130 | ND | - | 20 |
| Nitrate (mg L−1) | 0.31 ± 0.05 | ND | 0.16 | 50 |
| Sulfate (mg L−1) | 29.68 ± 1.48 | 19.58 ± 0.36 | 500 | 400 |
| Phosphate (mg L−1) | 5.32 ± 0.036 | 19.69 ± 0.57 | 4.5 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selvarajan, R.; Sibanda, T.; Pandian, J.; Mearns, K. Taxonomic and Functional Distribution of Bacterial Communities in Domestic and Hospital Wastewater System: Implications for Public and Environmental Health. Antibiotics 2021, 10, 1059. https://doi.org/10.3390/antibiotics10091059
Selvarajan R, Sibanda T, Pandian J, Mearns K. Taxonomic and Functional Distribution of Bacterial Communities in Domestic and Hospital Wastewater System: Implications for Public and Environmental Health. Antibiotics. 2021; 10(9):1059. https://doi.org/10.3390/antibiotics10091059
Chicago/Turabian StyleSelvarajan, Ramganesh, Timothy Sibanda, Jeevan Pandian, and Kevin Mearns. 2021. "Taxonomic and Functional Distribution of Bacterial Communities in Domestic and Hospital Wastewater System: Implications for Public and Environmental Health" Antibiotics 10, no. 9: 1059. https://doi.org/10.3390/antibiotics10091059
APA StyleSelvarajan, R., Sibanda, T., Pandian, J., & Mearns, K. (2021). Taxonomic and Functional Distribution of Bacterial Communities in Domestic and Hospital Wastewater System: Implications for Public and Environmental Health. Antibiotics, 10(9), 1059. https://doi.org/10.3390/antibiotics10091059









