Abstract
Resistance to the last-line antibiotics against invasive Gram-negative bacterial infection is a rising concern in public health. Multidrug resistant (MDR) Acinetobacter baumannii Aci46 can resist colistin and carbapenems with a minimum inhibitory concentration of 512 µg/mL as determined by microdilution method and shows no zone of inhibition by disk diffusion method. These phenotypic characteristics prompted us to further investigate the genotypic characteristics of Aci46. Next generation sequencing was applied in this study to obtain whole genome data. We determined that Aci46 belongs to Pasture ST2 and is phylogenetically clustered with international clone (IC) II as the predominant strain in Thailand. Interestingly, Aci46 is identical to Oxford ST1962 that previously has never been isolated in Thailand. Two plasmids were identified (pAci46a and pAci46b), neither of which harbors any antibiotic resistance genes but pAci46a carries a conjugational system (type 4 secretion system or T4SS). Comparative genomics with other polymyxin and carbapenem-resistant A. baumannii strains (AC30 and R14) identified shared features such as CzcCBA, encoding a cobalt/zinc/cadmium efflux RND transporter, as well as a drug transporter with a possible role in colistin and/or carbapenem resistance in A. baumannii. Single nucleotide polymorphism (SNP) analyses against MDR ACICU strain showed three novel mutations i.e., Glu229Asp, Pro200Leu, and Ala138Thr, in the polymyxin resistance component, PmrB. Overall, this study focused on Aci46 whole genome data analysis, its correlation with antibiotic resistance phenotypes, and the presence of potential virulence associated factors.
1. Introduction
Acinetobacter baumannii is an opportunistic pathogenic bacterium that causes nosocomial infections in immunocompromised patients, especially patients treated in the intensive care unit (ICU) [1,2]. A. baumannii infections usually occur following: trauma, surgery, catheterization, or endotracheal intubation [3]. Moreover, this bacterium is well known for its multidrug resistant (MDR) characteristics, defined as resistance to at least one agent in three or more antibiotic categories [4], and as a nosocomial ESKAPE pathogen, a group including: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa, and Enterobacter species [5]. A. baumannii can resist almost all available antibiotics and it is possible for a strain to be pan drug resistant (PDR), which is defined as resistant to all agents in all antibiotic categories including last-resort antibiotics (carbapenems and polymyxins) [4].
Carbapenem-resistant A. baumannii or CRAB is considered by WHO (World Health Organization) as one of the leading threats to global human healthcare [6]. During the COVID-19 pandemic, CRAB infections have increased among COVID-19 patients who are subjected to long-term stays in the ICU [7,8]. The incidence of A. baumannii infection in Thailand is widely distributed in all regions of the country [9]. Clinical isolates of A. baumannii accounted for 15–16% of hospital-acquired bacteremia and CRAB comprises 70–88% of total A. baumannii clinical isolates [10]. Presence of the carbapenemase encoding genes, blaOXA-23 or blaOXA-51, in combination with insertion sequence elements is frequently found in CRAB [11].
Unfortunately, colistin-resistant A. baumannii strains that are either MDR or PDR have been reported worldwide [12,13]. Polymyxins, including polymyxin B and colistin, are alternative last-line drugs used against CRAB [14]. Cationic polymyxin molecules target the polyanionic portions of the outer membrane of the bacterial envelope, specifically lipid A in lipopolysaccharide (LPS) [15]. Polymyxins bind lipid A and disrupt the outer membrane, causing cytoplasm leakage [16]. In Thailand, colistin-resistant A. baumannii isolates are found in 35–44% of pneumonia patients [17,18]. Among the colistin-resistant A. baumannii strains, four known mechanisms have been identified: (i) modification of lipid A, (ii) loss of LPS, (iii) disruption of outer membrane asymmetry, and (iv) efflux pumps [19]. Modification of lipid A involves increased addition of phosphoethanolamine (PEtN) to lipid A resulting from a mutation in the PmrAB two-component system. Increased expression of pmrC, which encodes lipid A phosphoethanolamine transferase, enhances addition of PEtN to either the 4′-phosphate or 1′-phosphate of lipid A [20]. Inactivation or complete loss of LPS occurs as a result of mutations in LPS biosynthesis genes such as: lpxA, lpxC, and lpxD [21]. Mutations in vacJ, encoding an outer membrane lipoprotein, and pldA, encoding an outer membrane phospholipase, result in accumulation of phospholipid, disrupting LPS organization, membrane asymmetry, and colistin binding [22]. Efflux pumps have also been shown to play important roles in the osmotic stress response and colistin resistance, specifically the AdeRS two-component system, which regulates expression of the AdeABC [23] and EmrAB efflux pumps [24].
Our previous report characterized MDR A. baumannii Aci46 that was isolated from a pus sample from a Thai patient at Ramathibodi hospital [25]. This strain was reported as a CRAB, i.e., resistant to imipenem. In the current study, we further explored the Aci46 resistance profile against other antibiotics including the alternative last-line drug, colistin. Since the genotypic characteristics of the MDR Aci46 were still unknown, genome studies were applied to identify antibiotic resistance genes, sequence type, and international clonal (IC) group of Aci46. In addition, the Aci46 genome was compared with other MDR A. baumannii and drug sensitive A. baumannii to characterize the unique gene features of the MDR (or CRAB) Aci46.
2. Results and Discussion
2.1. Antibiotic Resistance Phenotypes of Aci46
A previous report has shown that Aci46 is susceptible to amikacin and resistant to cefoperazone-sulbactam, ceftazidime, ciprofloxacin, and imipenem [25]. To expand the antibiotic profile of Aci46, twenty drugs from eight classes (i.e., aminoglycosides, beta-lactams (and beta-lactam combined), carbapenems, quinolones, folate pathway blocks, phenicol, tetracycline, and colistin) were used in disk diffusion and microdilution assays. We found that Aci46 was resistant to all twenty drugs (Table 1, Figure S1 and Supplement Table S1) with a minimum inhibitory concentration (MIC) for colistin of 512 µg/mL (Figure 1). We have demonstrated that Aci46 is an XDR (extensively drug resistant) strain according to the definition described by Magiorakos et al. (non-susceptible to at least one agent in all but two antimicrobial categories specified for Acinetobacter spp.) [4].

Table 1.
Antibiotic susceptibility profiling of A. baumannii Aci46 by disk diffusion method.
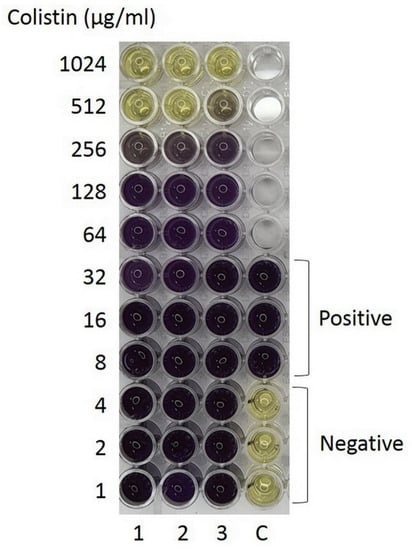
Figure 1.
Minimum inhibitory concentration (MIC) determination by microdilution assay using MTT staining. Aci46 viable cells and positive control are shown in purple while dead cells or negative control are shown in yellow. The experiments were tested with three biological replicates.
2.2. Whole Genome Sequencing Data
To further investigate the genetic makeup of XDR Aci46, the chromosome and plasmids of Aci46 were subjected to next-generation sequencing. The summarized genome data is shown in Table 2. The Aci46 genome size is 3,887,827 bp with a GC content of 38.87%. The number of predicted protein coding sequences, rRNA genes, and tRNA genes were 3754, 3, and 63, respectively. We also identified two plasmids from the whole genome data, namely pAci46a and pAci46b. The size of pAci46a was 70,873 bp with a GC content of 33.39% while pAci46b was 8808 bp with a GC content of 34.31%. The number of predicted protein coding sequences for pAci46a and pAci46b were 102 and 11, respectively. No rRNA or tRNA genes were present in either case.

Table 2.
Standard data of Aci46 genome features.
The microbial taxonomy of Aci46 was confirmed as A. baumannii at 100% identity based on variation of 54 genes encoding ribosomal protein subunits. Typing of Aci46 was classified by multi-locus sequence typing (MLST) using Oxford and Pasture schemes. The sequence type (ST) of Aci46 was ST1962 (gltA-1, gyrB-3, gdhB-189, recA-2, cpn60-2, gpi-140, rpoD-3) based on the Oxford scheme [26], while it belonged to ST2 (cpn60-2, fusA-2, gltA-2, pyrG-2, recA-2, rplB-2, rpoB-2) based on the Pasture scheme [27]. ST2 based on Pasture scheme is a predominant ST of CRAB found in Thailand and Southeast Asia [10,28]. However, ST1962 based on the Oxford scheme has never been reported in Thailand. ST1962 has been reported in the USA for only one strain (PubMLST database, to be published). From our previous report, we knew that Aci46 harbored class 1 integrase [25]. However, the class 1 integron can be transferred across two A. baumannii IC groups, IC I and IC II [29]. Phylogenetic analysis of the Aci46 genome compared with ten genomes of A. baumannii from three different IC’s, with the A. baylyi ADP1 genome as an outgroup to root the tree, revealed that Aci46 is more closely related to A. baumannii MDR-ZJ06 and belongs to IC II (Figure 2).

Figure 2.
Phylogenetic analysis of A. baumannii Aci46 and ten sequences of A. baumannii genomes for identification of international clonal group (IC). The phylogenetic tree was constructed by RAxML version 8.2.11 using 100 single-copy genes with bootstrap values set to 100 replicates. Selected strains belonging to IC I, IC II, and IC III are labeled. The accession numbers of the strains are as follows: AYE (CU459141.1), AB0057 (CP001182.2), AB307-0294 (CP001172), AC30 (ALXD00000000), ACICU (CP031380.1), MDR-ZJ06 (CP001937.2), R14 (PUDB01000000), ATCC17978 (CP000521.1), SDF (CU468230.2), and A. baylyi ADP1 (CR543861).
2.3. Antibiotic Resistance Gene, Efflux Pump, and Virulence Gene Predictions
Based on the phenotypic characteristics of the antibiotic resistance profile, a search for the presence of antibiotic resistance genes and genes encoding efflux pumps associated with the XDR phenotype in Aci46 was undertaken. ResFinder, CARD, and NDARO databases were used to predict the antibiotic resistance genes and efflux pumps present in Aci46. We found that Aci46 harbored sixteen resistance genes against eight classes of drugs (i.e., aminoglycosides, beta-lactams/carbapenems, beta-lactams/cephalosporins, colistin, fluoroquinolones, macrolides, tetracycline, and sulfonamide) and twenty-two genes belonging to five classes of drug transporters (i.e., RND (resistance-nodulation-division) efflux systems, MFS (major facilitator superfamily) family transporter, ABC (ATP-binding cassette) transporter, MATE (multidrug and toxic compound extrusion) family transporter, and SMR (small multidrug resistance)) (Table 3). The genes, blaOXA-23, blaOXA-66 or blaOXA-51-like, and oprD genes are present in Aci46 and they are known confer carbapenem-resistance [11]. These data correlate with presence of blaOXA-23 and blaOXA-51 in other CRAB isolates found in Thailand [30]. Moreover, class 1 and class 2 integrase genes and blaOXA-23 are often found in XDR A. baumannii [31,32].

Table 3.
List of predicted antibiotic resistance and drug transporter genes.
With regard to colistin resistance, we identified lpxA and lpxC in Aci46, these genes might play a role in loss of LPS and leading to colistin resistance [21]. In addition, we found four genes encoding efflux pumps (i.e., adeR, adeS, emrA, and emrB) and two genes with roles in lipid modification (i.e., pmrA and pmrB). AdeRS is a two-component system that regulates the expression of the AdeABC efflux pump, which is an RND efflux system [23]. The EmrAB efflux system belongs to the MFS family of transporters [24]. The PmrAB two-component system regulates PmrC expression. PmrC adds PEtN to lipid A [33]. In summary, the genotypic characteristics of Aci46 suggest that Aci46 resists colistin by way of lipid A modification, LPS loss, and AdeABC-mediated efflux.
Several virulence factors of A. baumannii have been identified by genome-based analysis [34]. The outer membrane protein, OmpA, which functions as a porin, is a key factor in virulence where it plays particular roles in cell invasion, development of cytotoxicity, and apoptosis [35]. Capsular polysaccharides and LPS are also virulence factors and contribute to serum resistance, biofilm formation, and escape from the host immune response [36]. A. baumannii uses combined strategies, namely, bacterial fitness and pathogenicity, to cause disease in humans [35]. PAI (pathogenicity islands), such as prophages and secretion systems, have also been implicated in virulence and pathogenicity [37,38]. In Aci46, we have identified pathogenicity islands comprising four prophages, one T4SS (type four secretion system), one T6SS (type six secretion system), and one ICE (integrative and conjugation element) (Table 4 and Supplement Table S2). No antibiotic resistance genes were found on the plasmids and prophages. Genotypic characteristics underlying the antibiotic resistance profile of Aci46 are found on its chromosome, and not on plasmids or other mobile genetic elements. T4SS is located in plasmid pAci46a, similar to pAC30c in A. baumannii AC30 and pAC29b in A. baumannii AC29 [2]. Generally, T4SS plays a role in the transfer antibiotic resistance genes via horizontal gene transfer [38]. Based on comparative genome analysis, T4SS loci are found in clinical isolates associated with hospital outbreaks [39]. The function of T4SS is still unclear in A. baumannii, but it might be implicated in pathogenicity or host–pathogen interaction [38,40]. Thus, pAci46a might play a role in pathogenesis instead of drug resistance. Moreover, we found attL (gtaataacaaagcaatcccgcagggttgcgacaaatagccctctaaatcgctctaattgcccctagattcaatttta) and attR (gtaataacaaagcaatcccgcagggttgcgacaaatagccctctaaatcgctctaattgcccctagattcaatttta) sites on pAci46a (or ICE region). It is possible that pAci46a could be a conjugative plasmid responsible for plasmid mobilization, similar to pAC30c and pAC29b [2]. T6SS injects toxic effectors into other bacteria in the same niche; therefore, it is an important factor for competitive killing and host colonization [34]. The plasmid, pAci46b, carries eleven genes encoding: one outer membrane receptor protein, one replication protein, and nine hypothetical proteins. The functions of pAci46b are still unclear.

Table 4.
Predicted pathogenicity islands in WGS of A. baumannii Aci46.
2.4. Comparative Pangenomic Analysis against Other A. baumannii Strains
In order to further explore the novel gene(s) that might be involved in the colistin and carbapenem-resistant phenotypes in Aci46, pangenome analysis was performed (Figure 3). Pangenome (all genes from all genomes) and core genes (present in all genomes) (Supplement Table S7) comprise 4605 and 2754 genes, respectively. We found that the number of strain-specific genes for Aci46, ACICU, ATCC17978, AC30, and R14 are 45, 193, 382, 91, and 145, respectively (Supplement Table S3). MDR (Aci46, ACICU, AC30, and R14) (Supplement Table S4), polymyxin and carbapenem resistance (renamed PCRAB) (Aci46, AC30, and R14), and colistin and carbapenem resistance (renamed CCRAB) (Aci46 and R14) groups contained 504, 34, and 8 accessory genes (i.e., genes present in specific genomes), respectively. We hypothesized that the accessory genes in PCRAB and/or CCRAB might be involved in resistance to polymyxins (polymyxin B and colistin). The thirty-four PCRAB specific genes encode: amidase, carbapenemase BlaOXA-23, CzcCBA cobalt/zinc/cadmium efflux RND transporter, twenty-three hypothetical proteins, lysophospholipid, nickel-cobalt-cadmium resistance protein, oxidoreductase, transcriptional regulator, and DNA-methyltransferase subunit M (Supplement Table S5). The eight CCRAB specific genes encode: one primosomal protein I and seven hypothetical proteins (Supplement Table S6). One of the PCRAB specific genes, blaOXA-23, is frequently reported in CRAB [41,42]. Interestingly, three genes (czcA, czcB, and czcC) encode CzcCBA cobalt/zinc/cadmium efflux RND transporters in PCRAB specific genes. CzcCBA cobalt/zinc/cadmium efflux RND transporters have functions in exporting cations (Co2+, Zn2+, and Cd2+) from the cytoplasm and confer heavy metal resistance [43,44], and are reported to be associated with the XDR phenotype in A. baumannii [45]. This efflux system might function in colistin resistance in PCRAB, including Aci46, by exporting colistin and polymyxin (cationic molecules). In the case of CCRAB specific genes, their functions are unknown.
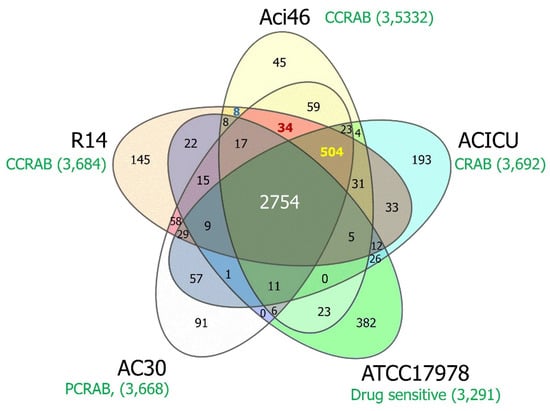
Figure 3.
Venn diagram of comparative pangenome analysis among A. baumannii strains. The yellow, blue, green, white, and orange represent Aci46, ACICU, ATCC17978, AC30, and R14, respectively, with the number of total genes and the character of drug resistance in each genome (shown in green). The overlapped areas show genes encoding shared protein families among strains. The number of genes in core genes (Aci46, ACICU, ATCC17978, AC30, and R14), MDR (Aci46, ACICU, AC30, and R14), PCRAB (Aci46, AC30, and R14), and CCRAB (Aci46 and R14) the specific groups are labeled in white, yellow, red, and blue, respectively. CRAB: carbapenem-resistant A. baumannii, CCRAB: colistin and carbapenem-resistant A. baumannii, PCRAB: polymyxins and carbapenem-resistant A. baumannii.
2.5. Pairwise SNP Analysis
Although we identified genes that were specific to PCRAB that encoded CzcCBA efflux pumps, known MDR genes were among the core genes (Supplement Table S7). Thus, in order to identify resistance-associated mutations in MDR and CCRAB strains, non-synonymous SNPs between Aci46 and ATCC17978 and between Aci46 and ACICU were identified. SNPs within known MDR genes from the core genes are listed in Table 5. Twenty genes show SNPs in Aci46 vs. ATCC17978 i.e., seven antibiotic resistance genes (blaADC-25, blaOXA-66, lpxA, lpxC, pmrB, gyrA, and gyrB) and thirteen drug transporter genes (adeA, adeB, adeF, adeG, adeH, adeJ, adeR, adeS, opmH, emrB, mdfA, macB, and abeS). The deduced amino acid sequences of these genes among the MDR strains were similar. For example, the deduced amino acid sequences of blaOXA-66 or blaOXA-51-like genes in Aci46, ACICU, AC30, and R14 showed conserved amino acids at Val36, Lys107, and Asn225 while ATCC17978 contained Glu36, Gln107, and Asp225 (Figure 4). In 2015, the Trp22Met mutation of blaOXA-51 was linked with carbapenem resistance function in A. baumannii [46]. This result suggested that amino acid sequences of antibiotic resistant and drug transporter proteins in MDR strains could be different from drug sensitive strains and might be linked to drug resistance level. For colistin resistance, we found three genes (blaADC-25, pmrB, and gyrB) that were mutated in Aci46 vs. ACICU. Of these only pmrB is related to colistin resistance. From comparisons of Aci46 vs. ACICU and Aci46 vs. ATCC17978, mutations in PmrB were detected in three positions: Ala138Thr, Pro200Leu, and Glu229Asp (Table 5). Known PmrB mutations that confer colistin resistance are Leu9-Gly12 deletion, Ala22Val, Ile232Thr, and Gln270Pro [47,48]. Hence, Ala138Thr, Pro200Leu, and Glu229Asp mutations in Aci46 PmrB might be novel mutations involved in colistin resistance.

Table 5.
List of non-synonymous SNPs in antibiotic resistance genes and efflux pumps from core gene group.

Figure 4.
Comparisons of deduced amino acid sequences of blaOXA-66 from five strains of A. baumannii by multiple sequence alignment.
3. Materials and Methods
3.1. Bacterial Strains
A. baumannii Aci46 was isolated from a male Thai patient treated at Ramathibodi hospital, Thailand [25]. This strain was isolated from a pus sample, identified by routine biochemical test, and confirmed by blaOXA51-like gene detection [25,49]. Aci46 was cultured on MHA (Mueller Hinton Agar, BD Difco, Eysins, Switzerland) and incubated at 37 °C for overnight.
3.2. Antibiotic Susceptibility Testing by Disk Diffusion and Microdilution
Antibiotic susceptibility was determined by disk diffusion method for 19 drugs (gentamicin, kanamycin, streptomycin, cephalothin, cefoxitin, cefotaxime, ceftazidime, ceftriaxone, ampicillin-clavulanic acid, imipenem, meropenem, ciprofloxacin, nalidixic acid, norfloxacin, trimethoprim, trimethoprim-sulfamethoxazole, ampicillin, chloramphenicol, and tetracycline) (Oxoid, Thermo Fisher Scientific, Waltham, MA, USA) and microdilution method for colistin. Aci46 was streaked on MHA and incubated overnight. Colonies were picked and resuspended in normal saline solution at 1 × 108 cfu/mL (OD600 = 0.08–0.12 or 0.5 McFarland). The cell suspension was spread on MHA using a cotton swab. Antibiotic disks were placed on the agar surface. After incubation at 37 °C for 20–24 h., the zones of inhibition were measured and the results were interpreted following the CLSI (Clinical and Laboratory Standard Institute) guideline [50]. For the microdilution method, cell suspensions of Aci46 were diluted in CAMHB (Cation-Adjusted Mueller Hinton Broth, BD Difco, Eysins, Switzerland) to 1 × 106 cfu/mL. Two-fold serial dilutions of colistin were prepared (1–1024 µg/mL) and mixed with 5 × 105 cfu/mL of Aci46 in 200 µL total volume. After incubation at 37 °C for 20–24 h., the minimum inhibitory concentration was observed and cell viability was measured by MTT-based staining [51]. Ten µL of 5 mg/mL MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (Invitrogen, Life Technologies, Carlsbad, CA, USA) in phosphate buffer saline was added in 100 µL of cell culture and incubated at 37 °C, 200 rpm, 1 h in the dark. One hundred µL of 10% SDS (sodium dodecyl sulfate, Merck, Darmstadt, Germany) and 50% DMSO (dimethyl sulfoxide, Sigma-Aldrich, St. Louis, MO, USA) was added and continually incubated at 37 °C, 200 rpm, 2 h in the dark. The absorbance of formazan dissolution was detected at 570 nm using a microplate reader (Azure Ao Absorbance Microplate Reader, Azure Biosystems, Dublin, CA, USA). Relative optical density at 570 nm was calculated by dividing OD570 of drug-containing wells with the OD570 of drug-free wells [52]. The cut-off for no detection was the relative OD570 of 0.1. Viable cells under MTT staining can also be observed by the naked eye i.e., color change from yellow to purple. Colistin resistance was determined by CLSI guideline (MIC ≥ 4 µg/mL; resistant) [50]. Escherichia coli ATCC25922 was selected to be a control strain for disk diffusion and microdilution methods.
3.3. Genomic DNA Extraction and Whole Genome Sequencing
Whole genomic DNA of Aci46 was extracted using a modified Marmur procedure [53]. Briefly, Aci46 cells were harvested from 3 mL of cell culture in CAMHB and resuspended in EDTA-saline (0.01 M EDTA and 0.15 M NaCl, pH 8.0). Thirty µL of 110 mg/mL lysozyme and 10 µL of 20 mg/mL RNase A were added and incubated at 37 °C for 2 h. After incubation, 80 µL of 20% SDS and 10 µL of 5 mg/mL proteinase K were added and incubated again at 65 °C, 30 min. Then, 5 M NaCl was added at 0.5 volume followed by phenol-chloroform extraction. The upper liquid phase was transferred to a new 1.5 mL microcentrifuge tube. A 0.25 volume of 5 M NaCl and a 0.1 volume of 3 M sodium acetate were added and mixed well. Ice-cold absolute ethanol was added at 2 volumes and inverted gently. The DNA pellet was hooked and transferred into a new 1.5 mL microcentrifuge tube, air dried, and resuspended in DNase-RNase-free water. Quality and quantity of DNA were measured by UV spectrophotometer (OD260/OD280 and OD260/OD230 ratio) (DeNovix DS-11 FX+ spectrophotometer, DeNovix, Wilmington, DE, USA), Qubit dsDNA BR assay kit (Invitrogen, Life Technologies, Carlsbad, CA, USA), and 1% agarose gel electrophoresis (Bio-rad, Hercules, CA, USA). One hundred ng of extracted DNA was used for library preparation using TruSeq Nano DNA Kit (Illumina, San Diego, CA, USA) followed by pair-end sequencing on Illumina HiSeq platform (Illumina, San Diego, CA, USA).
3.4. Genome Assembly, Annotation, and Pathogenicity Island Prediction
Raw sequence data of Aci46 were trimmed by Trim Galore version 0.6.3 [54] and the quality was checked using FastQC version 0.11.8 [55]. Trimmed reads were assembled using SPAdes version 3.12.0 [56], corrected assembly error by Pilon version 1.23 [57], and calculated genome coverage by SAMTools version 1.3 [58] in PATRIC (Pathosystems Resource Integration Center) version 3.6.9 [59]. The quality of de novo assembled contigs was assessed by QUAST version 5.0.2 [60] and visualized using Bandage version 0.8.1 [61]. Coding sequences and functional genes were annotated using RASTtk (Rapid Annotation using Subsystem Technology toolkit) [62]. Antibiotic resistance genes were predicted using ResFinder version 4.1 [63], CARD (Comprehensive Antibiotic Resistance Database) [64], and NDARO (National Database of Antibiotic Resistant Organisms) [65] databases. Pathogenicity islands (type 4 secretion system and type 6 secretion system) and prophages were predicted using VRprofile version 2.0 [66] and PHASTER (PHAge Search Tool Enhanced Release) [67], respectively. In plasmid analysis, trimmed reads were used for searching and assembling plasmid sequences using plasmidSPAdes version 3.12.0 [68] in PATRIC version 3.6.9 server [59]. Quality control, annotation, and pathogenicity island predictions of plasmids were assessed using the same tools as with genomic analysis.
3.5. MLST and Phylogenetic Analysis
Identification of A. baumannii Aci46 was confirmed by rMLST (ribosomal multilocus sequence typing) [69] in PubMLST server [70]. The ST (sequence typing) of Aci46 was identified by MLST (multilocus sequence typing) according to the Pasture scheme (based on seven housekeeping genes cpn60, gdhB, gltA, gpi, gyrB, recA, and rpoD) [27] and Oxford scheme (based on seven housekeeping genes cpn60, fusA, gltA, pyrG, recA, rplB, and rpoB) [26] in PubMLST server [70]. The IC (international clonal) group of Aci46 was determined by phylogenetic relationship analysis using RAxML version 8.2.11 [71] based on 100 single-copy genes selected by PATRIC PGFams program [72] in PATRIC version 3.6.9 server [59]. The phylogenetic tree was visualized by FigTree [73]. Genomes of A. baumannii from IC I, IC II, IC III groups were used to identify the IC of Aci46. IC I group included A. baumannii strain AYE (CU459141.1) [74], AB0057 (CP001182.2) [75], and AB307-0294 (CP001172) [75]. IC II group included A. baumannii strain AC30 (ALXD00000000) [2], ACICU (CP031380.1) [76], MDR-ZJ06 (CP001937.2) [77], and R14 (PUDB01000000) [78]. IC III included A. baumannii strain ATCC17978 (CP000521.1) [79] and SDF (CU468230.2) [74]. The outgroup strain was A. baylyi ADP1 (CR543861) [80].
3.6. Comparative Pangenome and Pairwise SNP Analysis
Genomic data from five strains comprised of colistin and carbapenem-resistant Aci46, polymyxin B and carbapenem-resistant AC30 (ALXD00000000) [2], colistin and carbapenem-resistant R14 (PUDB01000000) [78], wild-type or drug sensitive ATCC17978 (CP000521.1) [79], and colistin-sensitive and carbapenem-resistant ACICU (CP031380.1) [76] were compared using the Protein Family Sorter with PATRIC genus-specific families (PLfams) strategy in PATRIC server [59]. In pairwise SNP analysis, genome data of Aci46 was aligned with the ATCC17978 genome (CP000521.1) and ACICU (CP031380.1) using BWA-mem aligner [81] and SNP calling by FreeBayes [82]. The deduced amino acid sequences of blaOXA-66 from five genomes were compared by Clustal Omega in EMBL-EBI [83].
4. Conclusions
In summary, this study reported the genome data of colistin and carbapenem-resistant A. baumannii Aci46, which was isolated from a patient in a Thai hospital. The MLST genotype of Aci46 is Pasture ST2 which is a predominant ST found in Thailand and Oxford ST1962 which has never been reported in Thailand. The predicted antibiotic resistance genes (for example, blaOXA-23, blaOXA-66, and blaADC-25) are on the chromosome, not plasmids. Based on pangenome analysis, we found that the CzcCBA cobalt/zinc/cadmium efflux RND transporter might be involved in conferring resistance to colistin and/or carbapenem. From SNP analysis, we identified three points of non-synonymous mutations in pmrB (412G > A, 599C > T, and 687A > C) that change amino acid sequences. These amino acid changes, specifically Glu229Asp, Pro200Leu, and Ala138Thr may confer colistin resistance in MDR A. baumannii strains.
Supplementary Materials
Supplementary data are available online at https://www.mdpi.com/article/10.3390/antibiotics10091054/s1, Supplement Figure S1: The relative optical density at 570 nm for determination of minimum inhibitory concentration (MIC) by microdilution assay using MTT staining, Supplement Table S1: Guideline for interpretation of disk diffusion results, Supplement Table S2: Details of pathogenicity islands, Supplement Table S3: List of strain-specific genes, Supplement Table S4: List of multidrug resistant-specific genes (Aci46-ACICU-AC30-R14), Supplement Table S5: List of polymyxins and carbapenem-resistant-specific genes (Aci46-AC30-R14), Supplement Table S6: List of colistin and carbapenem-resistant-specific genes (Aci46-R14), Supplement Table S7: List of core genes (present in all genomes).
Author Contributions
N.T., S.C., S.S. and P.D. conceptualized the study; N.T. designed the research, tested antibiotic resistant profiling, analyzed genomic sequences, and wrote the paper; P.D. collected bacterial samples and identified A. baumannii Aci46. All authors have read and agreed to the published version of the manuscript.
Funding
The research project was partially supported by Postdoctoral fellowship award from Mahidol University, grant number MU-PD_2020_9.
Institutional Review Board Statement
Ethical review and approval were waived for this study, because the isolate used in this study was obtained from a collection of isolates that has already been published.
Informed Consent Statement
Informed consent was obtained from all subjects involved in a previous study.
Data Availability Statement
The whole genome and plasmid sequences of A. baumannii Aci46 have been deposited at DDBJ/ENA/GenBank under the BioProject ID PRJNA739068.
Acknowledgments
The research project was partially supported by Postdoctoral fellowship award from Mahidol University, grant number MU-PD_2020_9. We thank James M. Dubbs for editing the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, H.; Thangaraj, P.; Chakrabarti, A. Acinetobacter baumannii: A Brief Account of Mechanisms of Multidrug Resistance and Current and Future Therapeutic Management. J. Clin. Diagn. Res. 2013, 7, 2602–2605. [Google Scholar] [CrossRef] [PubMed]
- Lean, S.S.; Yeo, C.C.; Suhaili, Z.; Thong, K.L. Comparative Genomics of Two ST 195 Carbapenem-Resistant Acinetobacter baumannii with Different Susceptibility to Polymyxin Revealed Underlying Resistance Mechanism. Front. Microbiol. 2015, 6, 1445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, T.; Marchaim, D.; Johnson, P.C.; Awali, R.A.; Doshi, H.; Chalana, I.; Davis, N.; Zhao, J.J.; Pogue, J.M.; Parmar, S.; et al. Risk factors and outcomes for patients with bloodstream infection due to Acinetobacter baumannii-calcoaceticus complex. Antimicrob. Agents Chemother. 2014, 58, 4630–4635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet. Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Gottesman, T.; Fedorowsky, R.; Yerushalmi, R.; Lellouche, J.; Nutman, A. An outbreak of carbapenem-resistant Acinetobacter baumannii in a COVID-19 dedicated hospital. Infect. Prev. Pract. 2021, 3, 100113. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February-July 2020. MMWR Morb. Mortal Wkly Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Khuntayaporn, P.; Kanathum, P.; Houngsaitong, J.; Montakantikul, P.; Thirapanmethee, K.; Chomnawang, M.T. Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: A nationwide study. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 19. [Google Scholar] [CrossRef]
- Loraine, J.; Heinz, E.; Soontarach, R.; Blackwell, G.A.; Stabler, R.A.; Voravuthikunchai, S.P.; Srimanote, P.; Kiratisin, P.; Thomson, N.R.; Taylor, P.W. Genomic and Phenotypic Analyses of Acinetobacter baumannii Isolates from Three Tertiary Care Hospitals in Thailand. Front. Microbiol. 2020, 11, 548. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, D.A.; Biagi, M.; Tan, X.; Qasmieh, S.; Bulman, Z.P.; Wenzler, E. Multidrug Resistant Acinetobacter baumannii: Resistance by Any Other Name Would Still be Hard to Treat. Curr. Infect. Dis. Rep. 2019, 21, 46. [Google Scholar] [CrossRef]
- Karyne, R.; Curty Lechuga, G.; Almeida Souza, A.L.; Rangel da Silva Carvalho, J.P.; Simoes Villas Boas, M.H.; De Simone, S.G. Pan-Drug Resistant Acinetobacter baumannii, but Not Other Strains, Are Resistant to the Bee Venom Peptide Mellitin. Antibiotics 2020, 9, 178. [Google Scholar] [CrossRef] [Green Version]
- Huttner, B.; Jones, M.; Rubin, M.A.; Neuhauser, M.M.; Gundlapalli, A.; Samore, M. Drugs of last resort? The use of polymyxins and tigecycline at US Veterans Affairs medical centers, 2005–2010. PLoS ONE 2012, 7, e36649. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Roberts, K.D.; Nation, R.L.; Thompson, P.E.; Li, J. Pharmacology of polymyxins: New insights into an ‘old’ class of antibiotics. Future Microbiol. 2013, 8, 711–724. [Google Scholar] [CrossRef] [Green Version]
- Trimble, M.J.; Mlynarcik, P.; Kolar, M.; Hancock, R.E. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef] [Green Version]
- Thet, K.T.; Lunha, K.; Srisrattakarn, A.; Lulitanond, A.; Tavichakorntrakool, R.; Kuwatjanakul, W.; Charoensri, N.; Chanawong, A. Colistin heteroresistance in carbapenem-resistant Acinetobacter baumannii clinical isolates from a Thai university hospital. World J. Microbiol. Biotechnol. 2020, 36, 102. [Google Scholar] [CrossRef] [PubMed]
- Lertsrisatit, Y.; Santimaleeworagun, W.; Thunyaharn, S.; Traipattanakul, J. In vitro activity of colistin mono- and combination therapy against colistin-resistant Acinetobacter baumannii, mechanism of resistance, and clinical outcomes of patients infected with colistin-resistant A. baumannii at a Thai university hospital. Infect. Drug. Resist. 2017, 10, 437–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, G.J.; Domingues, S. Interplay between Colistin Resistance, Virulence and Fitness in Acinetobacter baumannii. Antibiotics 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beceiro, A.; Llobet, E.; Aranda, J.; Bengoechea, J.A.; Doumith, M.; Hornsey, M.; Dhanji, H.; Chart, H.; Bou, G.; Livermore, D.M.; et al. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 2011, 55, 3370–3379. [Google Scholar] [CrossRef] [Green Version]
- Moffatt Jennifer, H.; Harper, M.; Harrison, P.; Hale John, D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox Andrew, D.; et al. Colistin Resistance in Acinetobacter baumannii Is Mediated by Complete Loss of Lipopolysaccharide Production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thi Khanh Nhu, N.; Riordan, D.W.; Do Hoang Nhu, T.; Thanh, D.P.; Thwaites, G.; Huong Lan, N.P.; Wren, B.W.; Baker, S.; Stabler, R.A. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016, 6, 28291. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.; Hasdemir, U.; Aksu, B.; Altinkanat Gelmez, G.; Soyletir, G. Alterations in AdeS and AdeR regulatory proteins in 1-(1-naphthylmethyl)-piperazine responsive colistin resistance of Acinetobacter baumannii. J. Chemother. 2020, 32, 286–293. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Lan, C.Y. Contribution of EmrAB efflux pumps to colistin resistance in Acinetobacter baumannii. J. Microbiol. 2017, 55, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Kansakar, P.; Dorji, D.; Chongtrakool, P.; Mingmongkolchai, S.; Mokmake, B.; Dubbs, P. Local dissemination of multidrug-resistant Acinetobacter baumannii clones in a Thai hospital. Microb. Drug. Resist. 2011, 17, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodriguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef] [Green Version]
- Wareth, G.; Linde, J.; Nguyen, N.H.; Nguyen, T.N.M.; Sprague, L.D.; Pletz, M.W.; Neubauer, H. WGS-Based Analysis of Carbapenem-Resistant Acinetobacter baumannii in Vietnam and Molecular Characterization of Antimicrobial Determinants and MLST in Southeast Asia. Antibiotics 2021, 10, 563. [Google Scholar] [CrossRef]
- Liu, C.C.; Tang, C.Y.; Chang, K.C.; Kuo, H.Y.; Liou, M.L. A comparative study of class 1 integrons in Acinetobacter baumannii. Gene 2014, 544, 75–82. [Google Scholar] [CrossRef]
- Niumsup, P.R.; Boonkerd, N.; Tansawai, U.; Tiloklurs, M. Carbapenem-resistant Acinetobacter baumannii producing OXA-23 in Thailand. Jpn. J. Infect. Dis. 2009, 62, 152–154. [Google Scholar]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Teo, J.; Lim, T.P.; Hsu, L.Y.; Tan, T.Y.; Sasikala, S.; Hon, P.Y.; Kwa, A.L.; Apisarnthanarak, A. Extensively drug-resistant Acinetobacter baumannii in a Thai hospital: A molecular epidemiologic analysis and identification of bactericidal Polymyxin B-based combinations. Antimicrob. Resist. Infect. Control. 2015, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Adams Mark, D.; Nickel Gabrielle, C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs Michael, R.; Bonomo Robert, A. Resistance to Colistin in Acinetobacter baumannii Associated with Mutations in the PmrAB Two-Component System. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [Green Version]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [Green Version]
- McConnell, M.J.; Actis, L.; Pachon, J. Acinetobacter baumannii: Human infections, factors contributing to pathogenesis and animal models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, A.R.; Monteiro, R.; Azeredo, J. Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci. Rep. 2018, 8, 15346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhosseiny, N.M.; Attia, A.S. Acinetobacter: An emerging pathogen with a versatile secretome. Emerg. Microbes Infect. 2018, 7, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.C.; Kuo, H.Y.; Tang, C.Y.; Chang, K.C.; Liou, M.L. Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics 2014, 104, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmieri, M.; D’Andrea, M.M.; Pelegrin, A.C.; Perrot, N.; Mirande, C.; Blanc, B.; Legakis, N.; Goossens, H.; Rossolini, G.M.; van Belkum, A. Abundance of Colistin-Resistant, OXA-23- and ArmA-Producing Acinetobacter baumannii Belonging to International Clone 2 in Greece. Front. Microbiol. 2020, 11, 668. [Google Scholar] [CrossRef] [Green Version]
- Abdulzahra, A.T.; Khalil, M.A.F.; Elkhatib, W.F. First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. N. Microbes N. Infect. 2018, 26, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Nies, D.H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 1995, 177, 2707–2712. [Google Scholar] [CrossRef] [Green Version]
- Gheorghe, I.; Barbu, I.C.; Surleac, M.; Sarbu, I.; Popa, L.I.; Paraschiv, S.; Feng, Y.; Lazar, V.; Chifiriuc, M.C.; Otelea, D.; et al. Subtypes, resistance and virulence platforms in extended-drug resistant Acinetobacter baumannii Romanian isolates. Sci. Rep. 2021, 11, 13288. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Antunes, N.T.; Stewart, N.K.; Frase, H.; Toth, M.; Kantardjieff, K.A.; Vakulenko, S. Structural Basis for Enhancement of Carbapenemase Activity in the OXA-51 Family of Class D beta-Lactamases. ACS Chem. Biol. 2015, 10, 1791–1796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerson, S.; Betts Jonathan, W.; Lucaßen, K.; Nodari Carolina, S.; Wille, J.; Josten, M.; Göttig, S.; Nowak, J.; Stefanik, D.; Roca, I.; et al. Investigation of Novel pmrB and eptA Mutations in Isogenic Acinetobacter baumannii Isolates Associated with Colistin Resistance and Increased Virulence In Vivo. Antimicrob. Agents Chemother. 2019, 63, e01586-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, B.; Liu, H.; Jiang, Y.; Shao, L.; Yang, S.; Chen, D. New Mutations Involved in Colistin Resistance in Acinetobacter baumannii. mSphere 2020, 5, e00895-19. [Google Scholar] [CrossRef] [Green Version]
- Balows, A. Manual of Cinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Jorgenson, J.H., Pfaller, M.A., Yolken, R.H., Eds.; ASM Press: Washington, DC, USA, 2003; Volume 2, ISBN 1-555810255-4. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI. Supplement M100-ED31; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021; ISBN 978-1-68440-104-8. [Google Scholar]
- Benov, L. Effect of growth media on the MTT colorimetric assay in bacteria. PLoS ONE 2019, 14, e0219713. [Google Scholar] [CrossRef]
- Hundie, G.B.; Woldemeskel, D.; Gessesse, A. Evaluation of Direct Colorimetric MTT Assay for Rapid Detection of Rifampicin and Isoniazid Resistance in Mycobacterium tuberculosis. PLoS ONE 2016, 11, e0169188. [Google Scholar] [CrossRef]
- Salvà Serra, F.; Svensson Stadler, L.; Busquets, A.; Jaén-Luchoro, D.; Gomila, M. A Protocol for Extraction and Purification of High-Quality and Quantity Bacterial DNA Applicable for Genome Sequencing: A Modified Version of the Marmur Procedure. Protoc. Exch. 2018. [Google Scholar] [CrossRef]
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 27 June 2019).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 4 October 2018).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Brister, J.R.; Bolton, E.E.; Canese, K.; Comeau, D.C.; Funk, K.; Ketter, A.; Kim, S.; Kimchi, A.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020, 48, D9–D16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Tai, C.; Deng, Z.; Zhong, W.; He, Y.; Ou, H.Y. VRprofile: Gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief Bioinform. 2018, 19, 566–574. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling plasmids from whole genome sequencing data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Davis, J.J.; Gerdes, S.; Olsen, G.J.; Olson, R.; Pusch, G.D.; Shukla, M.; Vonstein, V.; Wattam, A.R.; Yoo, H. PATtyFams: Protein Families for the Microbial Genomes in the PATRIC Database. Front. Microbiol. 2016, 7, 118. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree, a Graphical Viewer of Phylogenetic Trees. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 November 2006).
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative analysis of Acinetobacters: Three genomes for three lifestyles. PLoS ONE 2008, 3, e1805. [Google Scholar] [CrossRef]
- Adams, M.D.; Goglin, K.; Molyneaux, N.; Hujer, K.M.; Lavender, H.; Jamison, J.J.; MacDonald, I.J.; Martin, K.M.; Russo, T.; Campagnari, A.A.; et al. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 2008, 190, 8053–8064. [Google Scholar] [CrossRef] [Green Version]
- Hamidian, M.; Wick, R.R.; Hartstein, R.M.; Judd, L.M.; Holt, K.E.; Hall, R.M. Insights from the revised complete genome sequences of Acinetobacter baumannii strains AB307-0294 and ACICU belonging to global clones 1 and 2. Microb. Genom. 2019, 5, e000298. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, T.; Yu, D.; Pi, B.; Yang, Q.; Zhou, J.; Hu, S.; Yu, Y. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 2011, 55, 4506–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mustapha, M.M.; Li, B.; Pacey, M.P.; Mettus, R.T.; McElheny, C.L.; Marshall, C.W.; Ernst, R.K.; Cooper, V.S.; Doi, Y. Phylogenomics of colistin-susceptible and resistant XDR Acinetobacter baumannii. J. Antimicrob. Chemother. 2018, 73, 2952–2959. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbe, V.; Vallenet, D.; Fonknechten, N.; Kreimeyer, A.; Oztas, S.; Labarre, L.; Cruveiller, S.; Robert, C.; Duprat, S.; Wincker, P.; et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004, 32, 5766–5779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).