Pseudomonas aeruginosa PAO 1 In Vitro Time–Kill Kinetics Using Single Phages and Phage Formulations—Modulating Death, Adaptation, and Resistance
Abstract
:1. Introduction
2. Results
2.1. Selection of Phages Based on Host Range
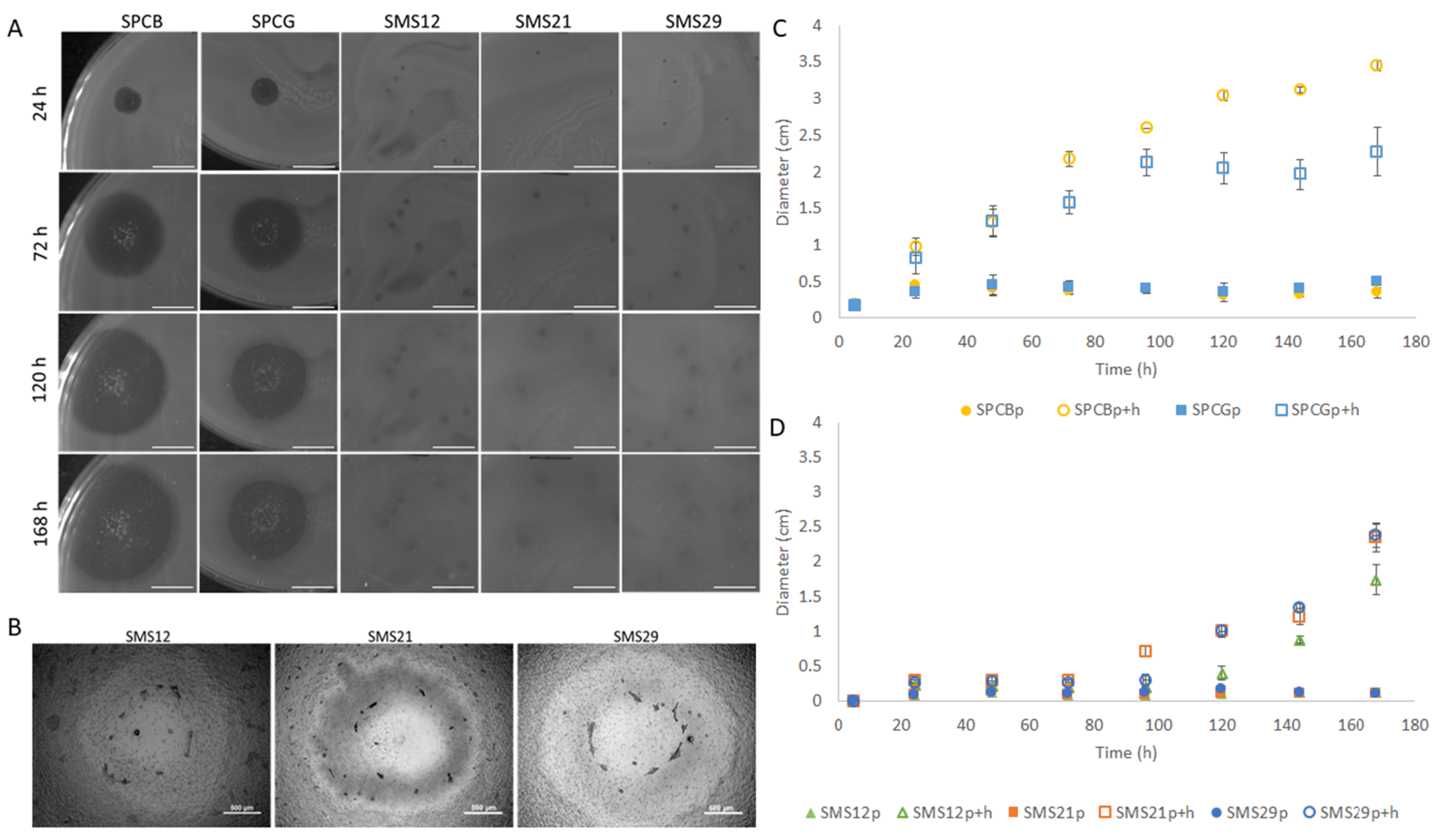
2.2. Virion Particle and Plaque Morphologies
2.3. One-Step Growth Characteristics
2.4. Phage Genomes and Comparative Analysis
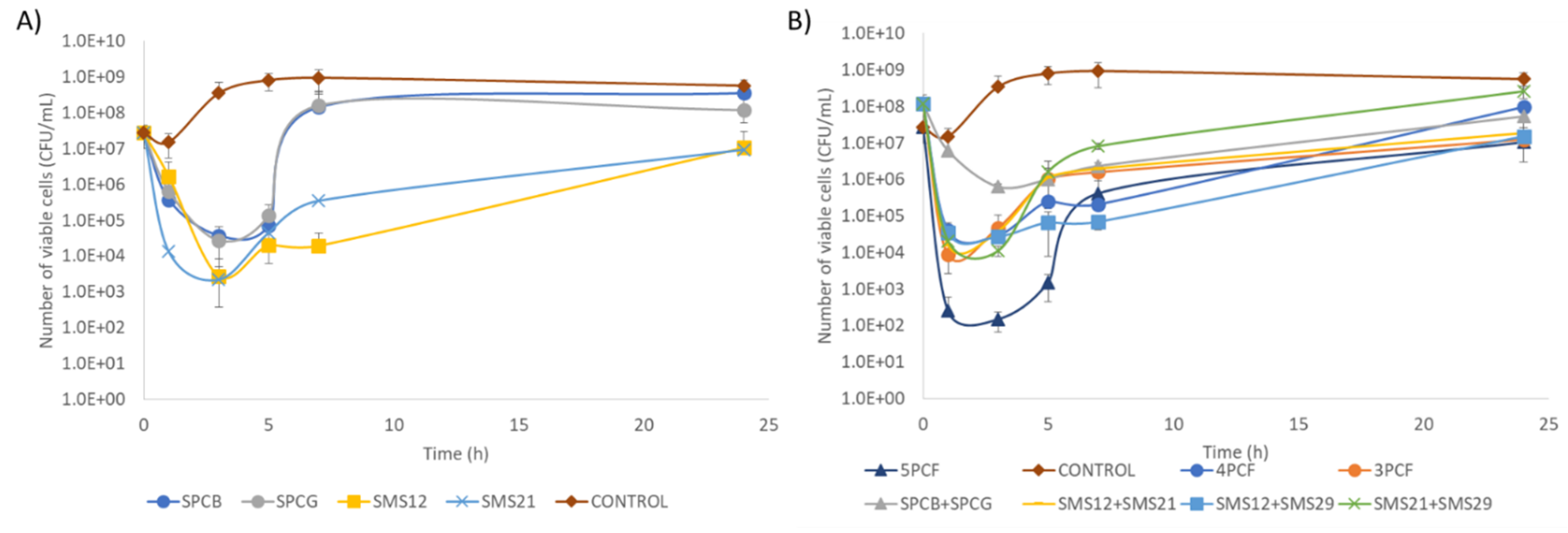
2.5. Time–Kill of Single Phage and Phage Cocktail Formulations
2.6. Assessment of the Survivor’s Susceptibility and Motility
3. Discussion
4. Materials and Methods
4.1. Bacteria, Phages, Growth Conditions
4.2. Phage Host Range Determination
4.3. Phage Propagation and Titration
4.4. Phage Plaque Morphology and Replication Characteristics
4.5. Transmission Electron Microscopy (TEM) Analysis of Phages
4.6. Phage DNA Extraction, Genome Sequencing, and Annotation
4.7. Time–Kill Experiments with Different Formulations
4.8. Characterization of the Motility Properties of Survivor Cells
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Hancock, R.E.W. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to manage Pseudomonas aeruginosa infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Serra, R.; Grande, R.; Butrico, L.; Rossi, A.; Settimio, U.F.; Caroleo, B.; Amato, B.; Gallelli, L.; De Franciscis, S. Chronic wound infections: The role of Pseudomonas aeruginosa and Staphylococcus aureus. Expert Rev. Anti. Infect. Ther. 2015, 13, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.R.; Isabella, V.M.; Lewis, K. Pseudomonas aeruginosa Biofilms in Disease. Microb. Ecol. 2014, 108, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Tümmler, B.; Kiewitz, C. Cystic fibrosis: An inherited susceptibility to bacterial respiratory infections. Mol. Med. Today 1999, 5, 351–358. [Google Scholar] [CrossRef]
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updates 2015, 21–22, 41–59. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Änggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; A preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- McCallin, S.; Sacher, J.C.; Zheng, J.; Chan, B.K. Current state of compassionate phage therapy. Viruses 2019, 11, 343. [Google Scholar] [CrossRef] [Green Version]
- McCallin, S.; Sarker, S.A.; Sultana, S.; Oechslin, F.; Brüssow, H. Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 2018, 20, 3278–3293. [Google Scholar] [CrossRef] [PubMed]
- Villarroel, J.; Larsen, M.V.; Kilstrup, M.; Nielsen, M. Metagenomic analysis of therapeutic PYO phage cocktails from 1997 to 2014. Viruses 2017, 9, 328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohler, T.; Curty, L.K.; Barja, F.; Van Delden, C.; Pechere, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef] [Green Version]
- Peterson, L.R. Bad Bugs, No Drugs: No ESCAPE Revisited. Clin. Infect. Dis. 2009, 49, 992–993. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Opar, A. Bad bugs need more drugs. Nat. Rev. Drug Discov. 2007, 6, 943–944. [Google Scholar] [CrossRef]
- Nadal Jimenez, P.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Ceyssens, P.J.; Glonti, T.; Kropinski, N.M.; Lavigne, R.; Chanishvili, N.; Kulakov, L.; Lashkhi, N.; Tediashvili, M.; Merabishvili, M. Phenotypic and genotypic variations within a single bacteriophage species. Virol. J. 2011, 8, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danis-Wlodarczyk, K.; Olszak, T.; Arabski, M.; Wasik, S.; Majkowska-Skrobek, G.; Augustyniak, D.; Gula, G.; Briers, Y.; Jang, H.B.; Vandenheuvel, D.; et al. Characterization of the newly isolated lytic bacteriophages KTN6 and KT28 and their efficacy against Pseudomonas aeruginosa biofilm. PLoS ONE 2015, 10, e0127603. [Google Scholar] [CrossRef]
- Murray, T.S.; Ledizet, M.; Kazmierczak, B.I. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 2010, 59, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Garbe, J.; Wesche, A.; Bunk, B.; Kazmierczak, M.; Selezska, K.; Rohde, C.; Sikorski, J.; Rohde, M.; Jahn, D.; Schobert, M. Characterization of JG024, a Pseudomonas aeruginosa PB1-like broad host range phage under simulated infection conditions. BMC Microbiol. 2010, 10, 301. [Google Scholar] [CrossRef] [Green Version]
- Trinh, J.T.; Székely, T.; Shao, Q.; Balázsi, G.; Zeng, L. Cell fate decisions emerge as phages cooperate or compete inside their host. Nat. Commun. 2017, 8, 14341. [Google Scholar] [CrossRef]
- Refardt, D. Within-host competition determines reproductive success of temperate bacteriophages. ISME J. 2011, 5, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Wayne, R.; Neilands, J.B. Evidence for common binding sites for ferrichrome compounds and bacteriophage phi 80 in the cell envelope of Escherichia coli. J. Bacteriol. 1975, 121, 497–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyman, P.; Abedon, S.T. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 2010, 70, 217–248. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Friman, V.P.; Hiltunen, T.; Jalasvuori, M.; Lindstedt, C.; Laanto, E.; Örmälä, A.M.; Laakso, J.; Mappes, J.; Bamford, J.K.H. High temperature and bacteriophages can indirectly select for bacterial pathogenicity in environmental reservoirs. PLoS ONE 2011, 6, e17651. [Google Scholar] [CrossRef]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A method for generation phage cocktail with great therapeutic potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a cocktail of three bacteriophages for biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanji, Y.; Shimada, T.; Yoichi, M.; Miyanaga, K.; Hori, K.; Unno, H. Toward rational control of Escherichia coli O157:H7 by a phage cocktail. Appl. Microbiol. Biotechnol. 2004, 64, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Chandler, C.E.; Horspool, A.M.; Hill, P.J.; Wozniak, D.J.; Schertzer, J.W.; Rasko, D.A.; Ernsta, R.K. Genomic and phenotypic diversity among ten laboratory isolates of Pseudomonas aeruginosa PAO1. J. Bacteriol. 2019, 201, e00595-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andres, D.; Hanke, C.; Baxa, U.; Seul, A.; Barbirz, S.; Seckler, R. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J. Biol. Chem. 2010, 286, 4165–4172. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Kim, S.; Park, B.; Ryu, S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 2014, 80, 1026–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabsch, W.; Ma, L.; Wiley, G.; Najar, F.Z.; Kaserer, W.; Schuerch, D.W.; Klebba, J.E.; Roe, B.A.; Laverde Gomez, J.A.; Schallmey, M.; et al. FepA- and TonB-dependent bacteriophage H8: Receptor binding and genomic sequence. J. Bacteriol. 2007, 189, 5658–5674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Washizaki, A.; Yonesaki, T.; Otsuka, Y. Characterization of the interactions between Escherichia coli receptors, LPS and OmpC, and bacteriophage T4 long tail fibers. Microbiologyopen 2016, 5, 1003–1015. [Google Scholar] [CrossRef] [Green Version]
- McCutcheon, J.G.; Peters, D.L.; Dennis, J.J. Identification and characterization of type IV pili as the cellular receptor of broad host range Stenotrophomonas maltophilia bacteriophages DLP1 and DLP2. Viruses 2018, 10, 338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, F.; Helm, R.F.; Broadway, K.M.; Scharf, B.E. More than rotating flagella: Lipopolysaccharide as a secondary receptor for flagellotropic phage 7-7-1. J. Bacteriol. 2018, 200, e00363-18. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, A.; Preuße, M.; Bruchmann, S.; Pawar, V.; Grahl, N.; Pils, M.C.; Nolan, L.M.; Filloux, A.; Weiss, S.; Häussler, S. Importance of flagella in acute and chronic Pseudomonas aeruginosa infections. Environ. Microbiol. 2019, 21, 883–897. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.-Y.; Guo, S.; Li, Z.; Zhao, Z.; Kojima, S.; Homma, M.; Wang, P.; Lo, C.-J.; Bai, F. Dynamic production and loss of flagellar filaments during the bacterial life cycle. bioRxiv 2019, 767319. [Google Scholar] [CrossRef] [Green Version]
- Feldman, M.; Bryan, R.; Rajan, S.; Scheffler, L.; Brunnert, S.; Tang, H.; Prince, A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 1998, 66, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Patankar, Y.R.; Lovewell, R.R.; Poynter, M.E.; Jyot, J.; Kazmierczak, B.I.; Berwin, B. Flagellar motility is a key determinant of the magnitude of the inflammasome response to Pseudomonas aeruginosa. Infect. Immun. 2013, 81, 2043–2052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, T.S.; Kazmierczak, B.I. FlhF Is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6995–7004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abeyrathne, P.D.; Daniels, C.; Poon, K.K.H.; Matewish, M.J.; Lam, J.S. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 2005, 187, 3002–3012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, A.; Sousa, J.C.; Silva, A.C.; Melo, L.D.R.; Sillankorva, S. Chestnut honey and bacteriophage application to control Pseudomonas aeruginosa and Escherichia coli biofilms: Evaluation in an ex vivo wound model. Front. Microbiol. 2018, 9, 1725. [Google Scholar] [CrossRef] [Green Version]
- Sillankorva, S.; Neubauer, P.; Azeredo, J. Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens. BMC Biotechnol. 2008, 8, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milho, C.; Andrade, M.; Vilas Boas, D.; Alves, D.; Sillankorva, S. Antimicrobial assessment of phage therapy using a porcine model of biofilm infection. Int. J. Pharm. 2019, 557, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russel, D. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Adams, M. Bacteriophages; Interscience Publishers: New York, NY, USA, 1959; ISBN 9781588296825. [Google Scholar]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Söding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Käll, L.; Sonnhammer, E.L.L. Reliability of transmembrane predictions in whole-genome data. FEBS Lett. 2002, 532, 415–418. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Krogh, A.; Sonnhammer, E.L.L. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004, 338, 1027–1036. [Google Scholar] [CrossRef]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, S. Promoter2.0: For the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Naville, M.; Ghuillot-Gaudeffroy, A.; Marchais, A.; Gautheret, D. ARNold: A web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 2011, 8, 11–13. [Google Scholar] [CrossRef] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Wang, Y.; Coleman-Derr, D.; Chen, G.; Gu, Y.Q. OrthoVenn: A web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2015, 43, W78–W84. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Barry, A.L.; Craig, W.A.; Nadler, H.; Reller, L.B.; Sanders, C.C.; Swenson, J.M. M26-A Methods for Determining Bactericidal Activity of Antimicrobial Agents, Approved Guideline, 1st ed.; Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 1999. [Google Scholar]
- Moons, P.; Faster, D.; Aertsen, A. Lysogenic conversion and phage resistance development in phage exposed Escherichia coli biofilms. Viruses 2013, 5, 150–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vater, S.M.; Weiße, S.; Maleschlijski, S.; Lotz, C.; Koschitzki, F.; Schwartz, T.; Obst, U.; Rosenhahn, A. Swimming behavior of Pseudomonas aeruginosa studied by holographic 3D tracking. PLoS ONE 2014, 9, e87765. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.G.; Kuchma, S.L.; O’Toole, G.A. Plate-based assay for swarming motility in Pseudomonas aeruginosa. In Pseudomonas Methods and Protocols. Methods in Molecular Biology (Methods and Protocols); Humana Press: New York, NY, USA, 2014; Volume 1149. [Google Scholar] [CrossRef]





| ID | Source | Year | Sex (M/F) * | Age (Years) | Antibiotic | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK | TOB | CN | ATM | FEP | CAZ | CIP | CS | IMI | MRP | PRL | TZP | TC | TTC | |||||
| I500546 | Cutaneous | 2013 | M | 82 | 8 | ≤1 | ≤1 | 8 | ≤2 | 4 | 0.25 | ≤1 | 2 | 2 | 8 | ≤2 | 8 | 4 |
| I499131 | Sputum | 2013 | F | 59 | >16 | ≤1 | >4 | 8 | ≤2 | 4 | 0.25 | ≤1 | 2 | 4 | 4 | ≤2 | 8 | 4 |
| C364224 | Sputum | 2013 | M | 52 | 4 | ≤1 | ≤1 | 8 | ≤2 | 4 | 2 | ≤1 | 2 | 2 | 8 | ≤2 | 8 | ≤2 |
| U570696 | Urine | 2013 | M | 89 | 2 | ≤1 | ≤1 | 8 | ≤2 | 4 | 0.25 | ≤1 | 4 | >16 | 8 | 8 | 8 | 4 |
| U572569 | Urine | 2013 | M | 82 | >16 | >4 | >4 | 8 | ≤2 | 4 | 4 | ≤1 | 2 | >16 | 4 | >32 | 8 | ≤2 |
| I66897 | Blood | 2018 | M | 63 | 2 | ≤1 | ≤1 | 16 | ≤2 | 4 | 0.25 | ≤1 | 2 | 2 | 8 | ≤2 | 32 | 4 |
| I29074 | Blood | 2017 | M | 68 | 2 | ≤1 | ≤1 | 16 | ≤2 | 16 | 1 | ≤1 | 2 | 2 | 8 | 32 | 8 | 8 |
| I41151 | Blood | 2017 | M | 80 | 16 | 8 | 8 | 16 | ≤2 | 2 | 2 | ≤1 | 4 | 4 | 2 | ≤2 | 32 | 4 |
| H73832 | Blood | 2018 | M | 61 | 2 | ≤1 | ≤1 | 16 | ≤2 | 4 | 0.25 | ≤1 | 2 | 4 | 4 | ≤2 | >16 | 4 |
| U88885 | Blood | 2017 | F | 74 | 4 | ≤1 | ≤1 | 16 | ≤2 | 2 | 0.25 | ≤1 | 4 | 2 | 4 | ≤2 | 8 | 4 |
| I60026 | Blood | 2018 | F | 83 | 2 | ≤1 | ≤1 | 16 | ≤2 | 4 | 0.25 | ≤1 | 2 | 2 | 4 | ≤2 | 32 | 4 |
| I41152 | Blood | 2018 | F | 52 | 4 | ≤1 | ≤1 | 16 | ≤2 | 4 | 0.25 | ≤1 | 2 | 8 | 4 | ≤2 | 8 | 4 |
| I60584 | Blood | 2018 | F | 73 | 2 | ≤1 | ≤1 | 16 | ≤2 | 4 | 0.25 | ≤1 | 4 | 2 | 8 | ≤2 | 32 | 4 |
| I97824 | Urine | 2019 | M | 88 | 2 | ≤1 | 8 | 8 | ≤2 | 16 | 0.25 | ≤1 | 16 | 2 | 8 | ≤2 | 8 | 8 |
| I93488 | Sputum | 2009 | M | 74 | 32 | 8 | 8 | 8 | ≤2 | 16 | 1 | ≤1 | 16 | 8 | 8 | ≤2 | 8 | 8 |
| I92986 | Urine | 2019 | F | 73 | 32 | ≤1 | 16 | 8 | ≤2 | 2 | 2 | ≤1 | 2 | 2 | 8 | ≤2 | 8 | 8 |
| C80117 | Ear | 2009 | F | 3 | 16 | ≤1 | 8 | 8 | ≤2 | 4 | 0.25 | ≤1 | 2 | 2 | 4 | ≤2 | 8 | 4 |
| U14706 | Urine | 2009 | M | 90 | 8 | ≤1 | ≤1 | 8 | ≤2 | 4 | 1 | ≤1 | 4 | 4 | 8 | ≤2 | 8 | 4 |
| I202628 | Sputum | 2019 | M | 68 | 2 | ≤1 | ≤1 | 8 | ≤2 | 2 | 0.25 | ≤1 | 2 | 2 | 8 | ≤2 | 8 | 4 |
| MIC Breakpoint | R > 16 | R > 4 | R > 4 | R > 16 | R > 8 | R > 8 | R > 0.5 | R > 2 | R > 8 | R > 8 | R > 16 | R > 16 | R > 16 | R > 16 | ||||
 Isolates showing resistance to a particular antibiotic according to the MIC Breakpoint (EUCAST, http://www.eucast.org/clinical_breakpoints/, accessed 20 January 2020).
Isolates showing resistance to a particular antibiotic according to the MIC Breakpoint (EUCAST, http://www.eucast.org/clinical_breakpoints/, accessed 20 January 2020).| Isolation Source | Phage | Bacterial Isolates | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAO1 | I500546 | 1499131 | C364224 | U570696 | U572569 | I66897 | I29074 | I41151 | H73832 | U88885 | I60026 | I51359 | I60584 | I97824 | I93488 | I92986 | C80117 | U14706 | I202628 | Efficacy (%) * | ||
| SPC (Microgen) | SPCA | 55 | ||||||||||||||||||||
| SPCB | 55 | |||||||||||||||||||||
| SPCC | 45 | |||||||||||||||||||||
| SPCE | 45 | |||||||||||||||||||||
| SPCF | 55 | |||||||||||||||||||||
| SPCG | 50 | |||||||||||||||||||||
| Sewage (2013) | SMS9 | 45 | ||||||||||||||||||||
| SMS10 | 30 | |||||||||||||||||||||
| SMS11 | 20 | |||||||||||||||||||||
| Sewage (2019) | SMS12 | 35 | ||||||||||||||||||||
| SMS13 | 30 | |||||||||||||||||||||
| SMS14 | 45 | |||||||||||||||||||||
| SMS15 | 30 | |||||||||||||||||||||
| SMS16 | 55 | |||||||||||||||||||||
| SMS17 | 30 | |||||||||||||||||||||
| SMS18 | 20 | |||||||||||||||||||||
| SMS19 | 35 | |||||||||||||||||||||
| SMS20 | 40 | |||||||||||||||||||||
| SMS21 | 30 | |||||||||||||||||||||
| SMS22 | 45 | |||||||||||||||||||||
| SMS23 | 30 | |||||||||||||||||||||
| SMS24 | 35 | |||||||||||||||||||||
| SMS25 | 30 | |||||||||||||||||||||
| SMS26 | 40 | |||||||||||||||||||||
| SMS27 | 15 | |||||||||||||||||||||
| SMS28 | 45 | |||||||||||||||||||||
| SMS29 | 45 | |||||||||||||||||||||
| SMS30 | 45 | |||||||||||||||||||||
 lysis from within;
lysis from within;  lysis from without;
lysis from without;  no lysis detected;
no lysis detected;  phages selected; * percentage of isolates that are susceptible to a given phage.
phages selected; * percentage of isolates that are susceptible to a given phage.| Susceptibility Profile | Susceptibility of Surviving Cells (%) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern | SPCB | SPCG | SMS12 | SMS21 | SMS29 | Control | 5PCF | 4PCF | 3PCF | SPCB + SPCG | SMS12 + SMS21 | SMS12 + SMS29 | SMS21 + SMS29 | SPCB | SPCG | SMS12 | SMS21 | SMS29 |
| 1 | R | R | R | R | R | 10.0 | 14.3 | 25.0 | 33.3 | 7.5 | 80.0 | 25.0 | 19.1 | 6.3 | ||||
| 2 | R | R | R | R | S | 5.0 | 28.5 | 25.0 | 12.5 | |||||||||
| 3 | R | R | R | S | R | 5.0 | 14.3 | 11.1 | ||||||||||
| 4 | R | R | R | S | S | 10.0 | 22.2 | 15.0 | 6.2 | |||||||||
| 5 | R | R | S | R | R | 15.0 | 14.3 | 25.0 | 20.0 | 12.5 | ||||||||
| 6 | R | R | S | R | S | 12.5 | 12.5 | 4.8 | ||||||||||
| 7 | R | R | S | S | R | 11.1 | 5.5 | 12.5 | 25.0 | |||||||||
| 8 | R | R | S | S | S | 30.0 | 20.0 | 4.8 | 18.8 | |||||||||
| 9 | R | S | R | R | R | 5.0 | ||||||||||||
| 10 | R | S | S | R | S | 15.0 | 10.0 | 9.5 | 18.8 | |||||||||
| 11 | R | S | S | S | R | 14.3 | ||||||||||||
| 12 | R | S | S | S | S | 15.0 | 14.3 | 28.0 | 42.9 | 100.0 | 12.5 | |||||||
| 13 | S | R | R | R | R | 4.8 | ||||||||||||
| 14 | S | R | S | R | R | 12.5 | 12.5 | |||||||||||
| 15 | S | R | S | S | S | 30.0 | 22.2 | 12.5 | 14.3 | |||||||||
| 16 | S | S | S | R | S | 35.0 | 12.5 | 100.0 | 100.0 | |||||||||
| 17 | S | S | S | S | S | 40.0 | ||||||||||||
| R * | 79.6 | 64.7 | 35.3 | 52.9 | 47.0 | 100.0 | 100.0 | 85.7 | 100.0 | 100.0 | 66.5 | 100.0 | 87.5 | 80.9 | 0.0 | 12.5 | 100.0 | 0.0 |
| S * | 29.4 | 35.3 | 64.7 | 47.1 | 53.0 | 0.0 | 0.0 | 14.3 | 0.0 | 0.0 | 33.5 | 0.0 | 12.5 | 19.1 | 100.0 | 71.5 | 0.0 | 100.0 |
 surviving P. aeruginosa cells that became resistant;
surviving P. aeruginosa cells that became resistant;  surviving cells that remained susceptible to the phage used in the treatment. When cocktail formulations were used, the value is highlighted in green only if the survivors remained susceptible to all phages present in the specific formulation. * R and S refer to the percentage of survivors showing resistance (R) or susceptibility (S) to a particular phage. 5PCF (phages SPCB + SPCG + SMS12 + SMS21 + SMS29); 4PCF (SPCG + SMS12 + SMS21 + SMS29); 3PCF (SMS12 + SMS21 + SMS29).
surviving cells that remained susceptible to the phage used in the treatment. When cocktail formulations were used, the value is highlighted in green only if the survivors remained susceptible to all phages present in the specific formulation. * R and S refer to the percentage of survivors showing resistance (R) or susceptibility (S) to a particular phage. 5PCF (phages SPCB + SPCG + SMS12 + SMS21 + SMS29); 4PCF (SPCG + SMS12 + SMS21 + SMS29); 3PCF (SMS12 + SMS21 + SMS29).| Swimming (%) | Swarming (%) | ||||||
|---|---|---|---|---|---|---|---|
| Cells Surviving Specific Treatment | No | Reduced to Moderate * | Good † | No | Dendritic | Smooth Edge | Suppressor |
| Control | 100.0 | 100.0 | |||||
| SPCB | 48.6 | 11.4 | 40.0 | 50.0 | 37.5 | 12.5 | |
| SPCG | 50.0 | 9.1 | 40.9 | 8.4 | 58.3 | 33.3 | |
| SMS12 | 44.4 | 14.8 | 40.7 | 66.7 | 11.1 | 22.2 | |
| SMS21 | 12.5 | 87.5 | 22.2 | 66.7 | 11.1 | ||
| SMS29 | 20.0 | 10.0 | 70.0 | 62.5 | 25.0 | 12.5 | |
| 5PCF | 73.7 | 26.3 | 15.8 | 5.2 | 79.0 | ||
| 4PCF | 38.4 | 30.8 | 30.8 | 57.1 | 42.9 | ||
| 3PCF | 42.9 | 21.4 | 35.7 | 87.5 | 12.5 | ||
| SPCB + SPCG | 45.5 | 4.5 | 50.0 | 63.6 | 27.3 | 9.1 | |
| SMS12 + SMS21 | 71.4 | 14.3 | 14.3 | 71.4 | 28.6 | ||
| SMS12 + SMS29 | 55.6 | 33.3 | 11.1 | 28.6 | 14.3 | 57.1 | |
| SMS21 + SMS29 | 60.0 | 26.7 | 13.3 | 25.0 | 50.0 | 25.0 | |
 predominant swimming pattern;
predominant swimming pattern;  predominant swarming pattern.
predominant swarming pattern.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, A.M.; Faustino, A.; Pastrana, L.M.; Bañobre-López, M.; Sillankorva, S. Pseudomonas aeruginosa PAO 1 In Vitro Time–Kill Kinetics Using Single Phages and Phage Formulations—Modulating Death, Adaptation, and Resistance. Antibiotics 2021, 10, 877. https://doi.org/10.3390/antibiotics10070877
Pinto AM, Faustino A, Pastrana LM, Bañobre-López M, Sillankorva S. Pseudomonas aeruginosa PAO 1 In Vitro Time–Kill Kinetics Using Single Phages and Phage Formulations—Modulating Death, Adaptation, and Resistance. Antibiotics. 2021; 10(7):877. https://doi.org/10.3390/antibiotics10070877
Chicago/Turabian StylePinto, Ana Mafalda, Alberta Faustino, Lorenzo M. Pastrana, Manuel Bañobre-López, and Sanna Sillankorva. 2021. "Pseudomonas aeruginosa PAO 1 In Vitro Time–Kill Kinetics Using Single Phages and Phage Formulations—Modulating Death, Adaptation, and Resistance" Antibiotics 10, no. 7: 877. https://doi.org/10.3390/antibiotics10070877
APA StylePinto, A. M., Faustino, A., Pastrana, L. M., Bañobre-López, M., & Sillankorva, S. (2021). Pseudomonas aeruginosa PAO 1 In Vitro Time–Kill Kinetics Using Single Phages and Phage Formulations—Modulating Death, Adaptation, and Resistance. Antibiotics, 10(7), 877. https://doi.org/10.3390/antibiotics10070877








