Absence of Host-Specific Genes in Canine and Human Staphylococcus pseudintermedius as Inferred from Comparative Genomics
Abstract
:1. Introduction
2. Results
2.1. MSSP-Infections Determinants
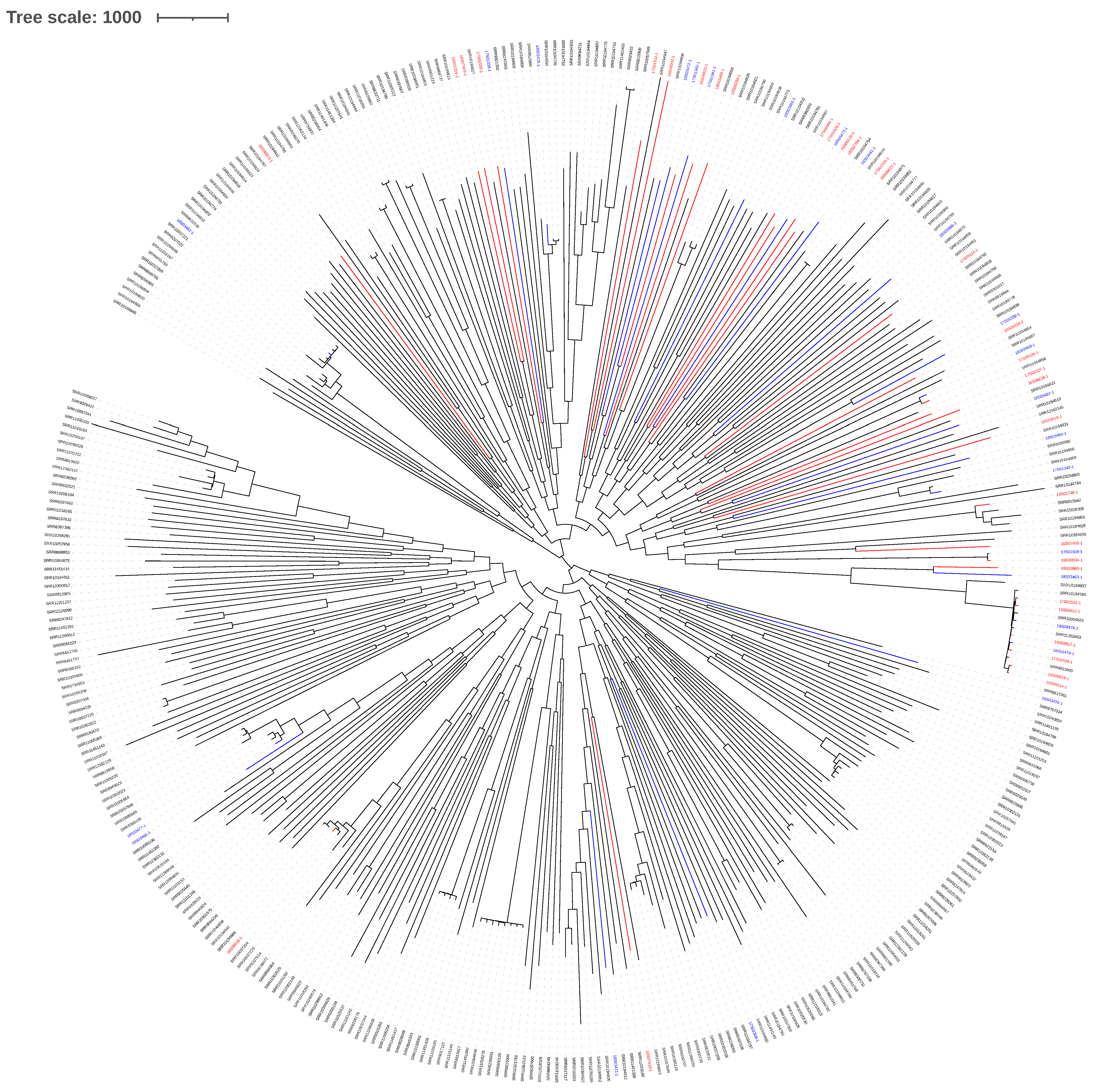
2.2. S. pseudintermedius Phylogeny
2.3. Antimicrobial Resistance Genes
2.4. Host-Associated Genes
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Genome Analysis
4.3. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Somayaji, R.; Rubin, J.E.; Priyantha, M.A.; Church, D. Exploring Staphylococcus pseudintermedius: An emerging zoonotic pathogen? Future Microbiol. 2016, 11, 1371–1374. [Google Scholar] [CrossRef] [Green Version]
- Börjesson, S.; Gómez-Sanz, E.; Ekström, K.; Torres, C.; Grönlund, U. Staphylococcus pseudintermedius can be misdiagnosed as staphylococcus aureus in humans with dog bite wounds. Eur. J. Clin. Microbiol. Infect. Dis. 2014. [Google Scholar] [CrossRef]
- Viau, R.; Hujer, A.M.; Hujer, K.M.; Bonomo, R.A.; Jump, R.L.P. Are Staphylococcus intermedius infections in humans cases of mistaken identity? A case series and literature review: Table 1. Open Forum Infect. Dis. 2015, 2, ofv110. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Murray, A.; Bendall, R.; Gaze, W.; Zhang, L.; Vos, M. Improved detection of Staphylococcus intermedius group in a routine diagnostic laboratory. J. Clin. Microbiol. 2015, 53, 961–963. [Google Scholar] [CrossRef] [Green Version]
- Nisa, S.; Bercker, C.; Midwinter, A.C.; Bruce, I.; Graham, C.F.; Venter, P.; Bell, A.; French, N.P.; Benschop, J.; Bailey, K.M.; et al. Combining MALDI-TOF and genomics in the study of methicillin resistant and multidrug resistant Staphylococcus pseudintermedius in New Zealand. Sci. Rep. 2019, 9, 1271. [Google Scholar] [CrossRef] [Green Version]
- Somayaji, R.; Priyantha, M.A.R.; Rubin, J.E.; Church, D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: Report of 24 cases. Diagn. Microbiol. Infect. Dis. 2016, 85, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; Rezusta, A.; Ferrer, I.; Pérez-Laguna, V.; Zarazaga, M.; Ruiz-Ripa, L.; Revillo, M.J.; Torres, C. Staphylococcus pseudintermedius human infection cases in Spain: Dog-to-human transmission. Vector Borne Zoonotic Dis. 2017, 17, 268–270. [Google Scholar] [CrossRef]
- Laarhoven, L.M.; de Heus, P.; van Luijn, J.; Duim, B.; Wagenaar, J.A.; van Duijkeren, E. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS ONE 2011, 6, e27788. [Google Scholar] [CrossRef] [PubMed]
- Starlander, G.; Börjesson, S.; Grönlund-Andersson, U.; Tellgren-Roth, C.; Melhus, Å. Cluster of infections caused by methicillin-resistant Staphylococcus pseudintermedius in humans in a tertiary hospital. J. Clin. Microbiol. 2014, 52, 3118–3120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matuszewska, M.; Murray, G.G.R.; Harrison, E.M.; Holmes, M.A.; Weinert, L.A. The evolutionary genomics of host specificity in staphylococcus aureus. Trends Microbiol. 2020, 28, 465–477. [Google Scholar] [CrossRef]
- Latronico, F.; Moodley, A.; Nielsen, S.S.; Guardabassi, L. Enhanced adherence of methicillin-resistant Staphylococcus pseudintermedius sequence type 71 to canine and human corneocytes. Vet. Res. 2014, 45, 70. [Google Scholar] [CrossRef] [Green Version]
- Little, S.V.; Bryan, L.K.; Hillhouse, A.E.; Konganti, K.; Lawhon, S.D. Whole-genome sequences of Staphylococcus pseudintermedius isolates from canine and human bacteremia infections. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [Green Version]
- Haenni, M.; el Garch, F.; Miossec, C.; Madec, J.-Y.Y.; Hocquet, D.; Valot, B. High Genetic Diversity among Methicillin-Susceptible Staphylococcus Pseudintermedius in Dogs in Europe. J. Glob. Antimicrob. Resist. 2020, 21, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Ceric, O.; Guag, J.; Nemser, S.; Borenstein, S.; Slavic, D.; Lippert, S.; McDowell, R.; Krishnamurthy, A.; Korosec, S.; et al. Genomics accurately predicts antimicrobial resistance in Staphylococcus pseudintermedius collected as part of Vet-LIRN resistance monitoring. Vet. Microbiol. 2021, 254, 109006. [Google Scholar] [CrossRef]
- Kang, J.H.; Hwang, C.Y. First detection of multiresistance PRE25-like elements from enterococcus spp. in Staphylococcus pseudintermedius isolated from canine pyoderma. J. Glob. Antimicrob. Resist. 2020, 20, 304–308. [Google Scholar] [CrossRef]
- Wladyka, B.; Piejko, M.; Bzowska, M.; Pieta, P.; Krzysik, M.; Mazurek, Ł.; Guevara-Lora, I.; Bukowski, M.; Sabat, A.J.; Friedrich, A.W.; et al. A peptide factor secreted by Staphylococcus pseudintermedius exhibits properties of both bacteriocins and virulence factors. Sci. Rep. 2015, 5, 14569. [Google Scholar] [CrossRef]
- Wegener, A.; Broens, E.M.; Zomer, A.; Spaninks, M.; Wagenaar, J.A.; Duim, B. Comparative genomics of phenotypic antimicrobial resistances in methicillin-resistant Staphylococcus pseudintermedius of canine origin. Vet. Microbiol. 2018, 225, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Brooks, M.R.; Padilla-Vélez, L.; Khan, T.A.; Qureshi, A.A.; Pieper, J.B.; Maddox, C.W.; Alam, M.T. Prophage-mediated disruption of genetic competence in Staphylococcus Pseudintermedius. mSystems 2020, 5. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.C.; Vitry, P.; Prystopiuk, V.; Garcia, B.; Höök, M.; Schoenebeck, J.; Geoghegan, J.A.; Dufrêne, Y.F.; Ross Fitzgerald, J. Host-specialized fibrinogen-binding by a bacterial surface protein promotes biofilm formation and innate immune evasion. PLoS Pathog. 2019, 15, e1007816. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, A.J.; Harrison, E.M.; Stanczak-Mrozek, K.; Leggett, B.; Waller, A.; Holmes, M.A.; Lloyd, D.H.; Lindsay, J.A.; Loeffler, A. Genomic insights into the rapid emergence and evolution of MDR in Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 2014, 70, 997–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannoehr, J.; ben Zakour, N.L.; Waller, A.S.; Guardabassi, L.; Thoday, K.L.; van den Broek, A.H.M.; Fitzgerald, J.R. Population genetic structure of the Staphylococcus intermedius group: Insights into Agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 2007, 189, 8685–8692. [Google Scholar] [CrossRef] [Green Version]
- Bannoehr, J.; Zakour, N.L.B.; Reglinski, M.; Inglis, N.F.; Prabhakaran, S.; Fossum, E.; Smith, D.G.; Wilson, G.J.; Cartwright, R.A.; Haas, J.; et al. Genomic and surface proteomic analysis of the canine pathogen Staphylococcus pseudintermedius reveals proteins that mediate adherence to the extracellular matrix. Infect. Immun. 2011, 79, 3074–3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, J.A.; Vehkala, M.; Välimäki, N.; Harris, S.R.; Chewapreecha, C.; Croucher, N.J.; Marttinen, P.; Davies, M.R.; Steer, A.C.; Tong, S.Y.C.; et al. Sequence element enrichment analysis to determine the genetic basis of bacterial phenotypes. Nat. Commun. 2016, 7, 12797. [Google Scholar] [CrossRef]
- Koop, G.; Vrieling, M.; Storisteanu, D.M.L.; Lok, L.S.C.; Monie, T.; van Wigcheren, G.; Raisen, C.; Ba, X.; Gleadall, N.; Hadjirin, N.; et al. Identification of LukPQ, a novel, equid-adapted leukocidin of Staphylococcus aureus. Sci. Rep. 2017, 7, 40660. [Google Scholar] [CrossRef]
- Abouelkhair, M.A.; Bemis, D.A.; Kania, S.A. Characterization of Recombinant Wild-Type and Nontoxigenic Protein A from Staphylococcus pseudintermedius. Virulence 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, M.C.F.; Ahrenfeldt, J.; Cisneros, J.L.B.; Jurtz, V.; Larsen, M.V.; Hasman, H.; Aarestrup, F.M.; Lund, O. A bacterial analysis platform: An integrated system for analysing bacterial whole genome sequencing data for clinical diagnostics and surveillance. PLoS ONE 2016, 11, e0157718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (ITOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote Pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid scoring of genes in microbial pan-genome-wide association studies with scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Resistance | Gene | Human Isolates (n = 32) | Canine Isolates (n = 25) |
|---|---|---|---|
| β-lactam | blaZ | 25 (78%) | 23 (92%) |
| aminoglycoside | ant6-Ia, aph(3′)-III | 14 (44%) | 14 (56%) |
| aac(6′)-Ie-aph(2”)-Ia | 0 | 2 (8%) | |
| chloramphenicol | cat(pC221) | 13 (41%) | 9 (36%) |
| macrolide | erm(B) | 13 (41%) | 10 (40%) |
| tetracycline | tet(M) | 6 (19%) | 11 (44%) |
| lincosamide | Inu(A) | 0 | 1 (4%) |
| folate inhibitor | dfrG | 0 | 1 (4%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wegener, A.; Broens, E.M.; van der Graaf-van Bloois, L.; Zomer, A.L.; Visser, C.E.; van Zeijl, J.; van der Meer, C.; Kusters, J.G.; Friedrich, A.W.; Kampinga, G.A.; et al. Absence of Host-Specific Genes in Canine and Human Staphylococcus pseudintermedius as Inferred from Comparative Genomics. Antibiotics 2021, 10, 854. https://doi.org/10.3390/antibiotics10070854
Wegener A, Broens EM, van der Graaf-van Bloois L, Zomer AL, Visser CE, van Zeijl J, van der Meer C, Kusters JG, Friedrich AW, Kampinga GA, et al. Absence of Host-Specific Genes in Canine and Human Staphylococcus pseudintermedius as Inferred from Comparative Genomics. Antibiotics. 2021; 10(7):854. https://doi.org/10.3390/antibiotics10070854
Chicago/Turabian StyleWegener, Alice, Els M. Broens, Linda van der Graaf-van Bloois, Aldert L. Zomer, Caroline E. Visser, Jan van Zeijl, Coby van der Meer, Johannes G. Kusters, Alex W. Friedrich, Greetje A. Kampinga, and et al. 2021. "Absence of Host-Specific Genes in Canine and Human Staphylococcus pseudintermedius as Inferred from Comparative Genomics" Antibiotics 10, no. 7: 854. https://doi.org/10.3390/antibiotics10070854
APA StyleWegener, A., Broens, E. M., van der Graaf-van Bloois, L., Zomer, A. L., Visser, C. E., van Zeijl, J., van der Meer, C., Kusters, J. G., Friedrich, A. W., Kampinga, G. A., Sips, G. J., Smeets, L., van Kerckhoven, M. E. J., Timmerman, A. J., Wagenaar, J. A., & Duim, B. (2021). Absence of Host-Specific Genes in Canine and Human Staphylococcus pseudintermedius as Inferred from Comparative Genomics. Antibiotics, 10(7), 854. https://doi.org/10.3390/antibiotics10070854






