Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach
Abstract
:1. Introduction
2. Results
2.1. Isolates Characteristics
2.2. Conjugation Efficiency of blaCTX-M-15 Gene among Isolates of Human, Animals and the Environment
2.3. Transferrable Resistance of Non-Beta-Lactam Phenotype among Isolates of Human, Animal and the Environment
2.4. Replicon Types of Plasmids Carrying blaCTX-M-15
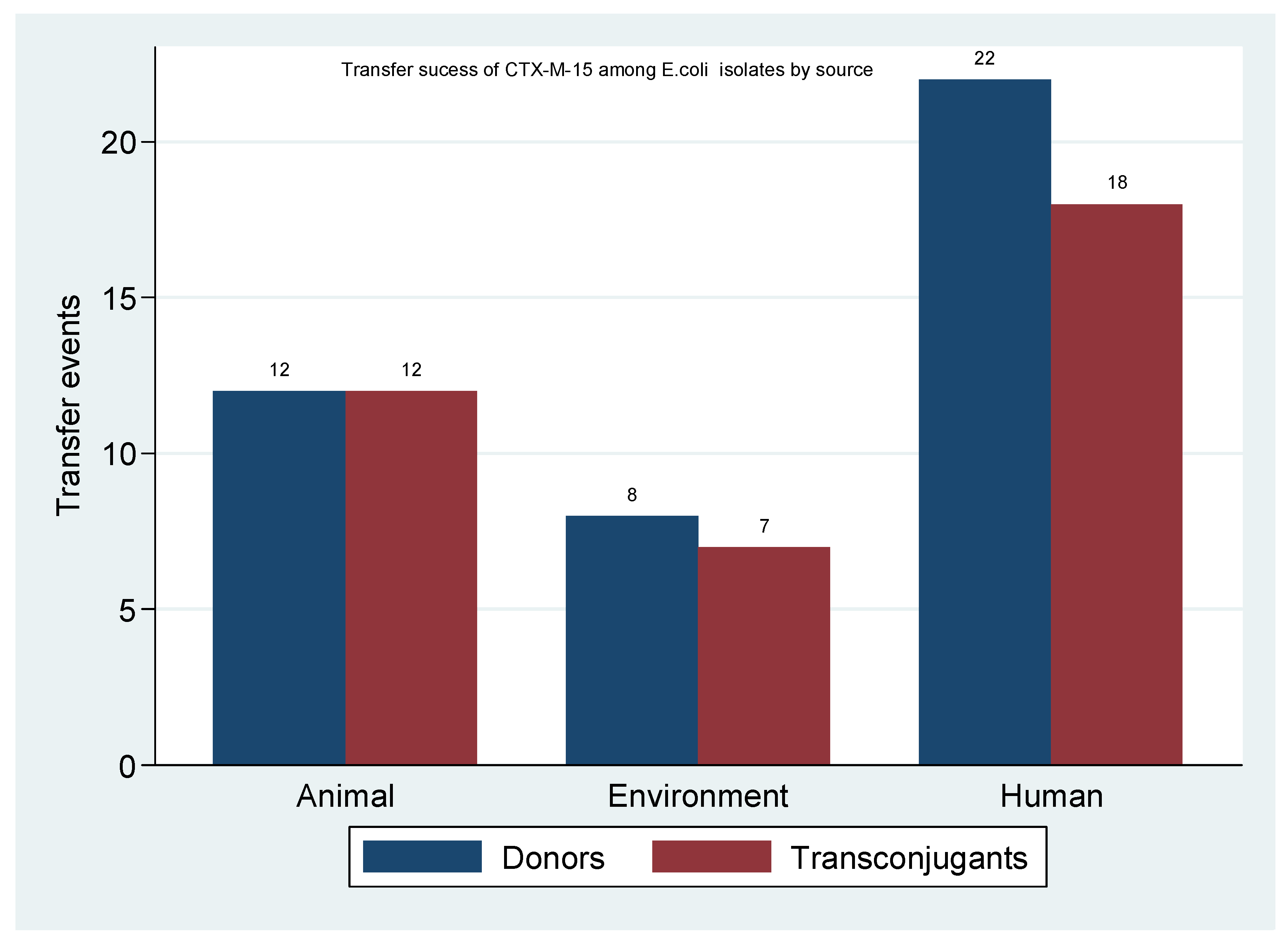
2.5. Transfer Success of blaCTX-M-15 among Escherichia coli Isolates
3. Discussion
4. Materials and Methods
4.1. Study Isolates
4.2. Antibiotic Susceptibility Testing
4.3. Conjugation Experiment
4.4. Genomic Extraction of Donor and Transconjugants DNA
4.5. PCR Based Replicon Typing
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| CREATES | Centre for Research, Agricultural advancement, Teaching Excellence and Sustainability in Food and Nutritional Security |
| CUHAS | Catholic University of Health and Allied Sciences |
| DNA | Deoxyribonucleic acid |
| EDTA | Ethylene Diaminetetracetic acid |
| ESBL | Extended spectrum beta-lactamase |
| GIT | Gastrointestinal tract |
| LB | Luria Bertani |
| MGE | Mobile genetic element |
| NaCl | Sodium Chloride |
| NaN3 | Sodium azide |
| PCR | Polymerase chain reaction |
| TAE | Tris acetate EDTA |
References
- Barlow, M. What antimicrobial resistance has taught us about horizontal gene transfer. Horiz. Gene Transf. 2009, 397–411. [Google Scholar] [CrossRef]
- Bonnet, R. Growing group of extended-spectrum β-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lahlaoui, H.; Khalifa, A.B.H.; Moussa, M.B. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL). Med. Et Mal. Infect. 2014, 44, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Hirai, I.; Fukui, N.; Taguchi, M.; Yamauchi, K.; Nakamura, T.; Okano, S.; Yamamoto, Y. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int. J. Antimicrob. Agents 2013, 42, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Novais, Â.; Carattoli, A.; Poirel, L.; Peixe, L.; Baquero, F.; Cantón, R.; Nordmann, P. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 2008, 14, 195. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.F.; Liras, P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 1989, 43, 173–206. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Moe, L.A.; Rodbumrer, J.; Gaarder, A.; Handelsman, J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009, 3, 243–251. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [Green Version]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [Green Version]
- Naseer, U.; Sundsfjord, A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 2011, 17, 83–97. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting plasmids to limit acquisition and transmission of antimicrobial resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Lopatkin, A.J.; Meredith, H.R.; Srimani, J.K.; Pfeiffer, C.; Durrett, R.; You, L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 2017, 8, 1689. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Rozen, D.E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 2006, 4, 556–562. [Google Scholar] [CrossRef] [PubMed]
- Benz, F.; Huisman, J.S.; Bakkeren, E.; Herter, J.A.; Stadler, T.; Ackermann, M.; Diard, M.; Egli, A.; Hall, A.R.; Hardt, W.-D. Plasmid-and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo. ISME J. 2021, 15, 862–878. [Google Scholar] [CrossRef]
- Yang, Q.E.; Agouri, S.R.; Tyrrell, J.M.; Walsh, T.R. Heavy metal resistance genes are associated with blaNDM-1-and blaCTX-M-15-carrying Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, e02642-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Ramírez, K.C.; Reyes-Gallegos, R.I.; Chávez-Jacobo, V.M.; Díaz-Magaña, A.; Meza-Carmen, V.; Ramírez-Díaz, M. A plasmid-encoded mobile genetic element from Pseudomonas aeruginosa that confers heavy metal resistance and virulence. Plasmid 2018, 98, 15–21. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.-J.; Lavigne, J.-P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Mshana, S.E.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T.; Imirzalioglu, C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect. Dis. 2013, 13, 466. [Google Scholar] [CrossRef] [Green Version]
- Mshana, S.E.; Falgenhauer, L.; Mirambo, M.M.; Mushi, M.F.; Moremi, N.; Julius, R.; Seni, J.; Imirzalioglu, C.; Matee, M.; Chakraborty, T. Predictors of bl a CTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect. Dis. 2016, 16, 187. [Google Scholar] [CrossRef] [Green Version]
- Mshana, S.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring blaCTX-M-15 in a tertiary hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef] [Green Version]
- Seni, J.; Falgenhauer, L.; Simeo, N.; Mirambo, M.M.; Imirzalioglu, C.; Matee, M.; Rweyemamu, M.; Chakraborty, T.; Mshana, S.E. Multiple ESBL-producing Escherichia coli sequence types carrying quinolone and aminoglycoside resistance genes circulating in companion and domestic farm animals in Mwanza, Tanzania, harbor commonly occurring plasmids. Front. Microbiol. 2016, 7, 142. [Google Scholar] [CrossRef]
- Moremi, N.; Manda, E.V.; Falgenhauer, L.; Ghosh, H.; Imirzalioglu, C.; Matee, M.; Chakraborty, T.; Mshana, S.E. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol. 2016, 7, 1862. [Google Scholar] [CrossRef] [Green Version]
- Mshana, S.E.; Gerwing, L.; Minde, M.; Hain, T.; Domann, E.; Lyamuya, E.; Chakraborty, T.; Imirzalioglu, C. Outbreak of a novel Enterobacter sp. carrying blaCTX-M-15 in a neonatal unit of a tertiary care hospital in Tanzania. Int. J. Antimicrob. Agents 2011, 38, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Seni, J.; Falgenhauer, L.; Simeo, N.; Mirambo, M.; Imirzalioglu, C.; Matee, M.; Rweyemamu, M.; Chakraborty, T.; Mshana, S.E. Preliminary insights into the occurrence of similar clones of extended-spectrum beta-lactamase-producing bacteria in humans, animals and the environment in Tanzania: A systematic review and meta-analysis between 2005 and 2016. Zoonoses Public Health 2018, 65, 1–10. [Google Scholar] [CrossRef]
- Hosuru Subramanya, S.; Bairy, I.; Nayak, N.; Amberpet, R.; Padukone, S.; Metok, Y.; Bhatta, D.R.; Sathian, B. Detection and characterization of ESBL-producing Enterobacteriaceae from the gut of healthy chickens, Gallus gallus domesticus in rural Nepal: Dominance of CTX-M-15-non-ST131 Escherichia coli clones. PLoS ONE 2020, 15, e0227725. [Google Scholar] [CrossRef]
- Mshana, S.E.; Kamugisha, E.; Mirambo, M.; Chakraborty, T.; Lyamuya, E. Prevalence of multiresistant gram-negative organisms in a tertiary hospital in Mwanza, Tanzania. BMC Res. Notes 2009, 2, 49. [Google Scholar] [CrossRef] [Green Version]
- Obeng-Nkrumah, N.; Labi, A.-K.; Blankson, H.; Awuah-Mensah, G.; Oduro-Mensah, D.; Anum, J.; Teye, J.; Kwashie, S.D.; Bako, E.; Ayeh-Kumi, P.F. Household cockroaches carry CTX-M-15-, OXA-48-and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019, 19, 272. [Google Scholar] [CrossRef]
- Lyimo, B.; Buza, J.; Subbiah, M.; Temba, S.; Kipasika, H.; Smith, W.; Call, D.R. IncF plasmids are commonly carried by antibiotic resistant Escherichia coli isolated from drinking water sources in northern Tanzania. Int. J. Microbiol. 2016, 2016, 3103672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gekenidis, M.-T.; Rigotti, S.; Hummerjohann, J.; Walsh, F.; Drissner, D. Long-Term Persistence of blaCTX-M-15 in Soil and Lettuce after Introducing Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli via Manure or Water. Microorganisms 2020, 8, 1646. [Google Scholar] [CrossRef]
- Zurfluh, K.; Glier, M.; Hächler, H.; Stephan, R. Replicon typing of plasmids carrying blaCTX-M-15 among Enterobacteriaceae isolated at the environment, livestock and human interface. Sci. Total Environ. 2015, 521, 75–78. [Google Scholar] [CrossRef] [PubMed]
- San Millan, A.; Peña-Miller, R.; Toll-Riera, M.; Halbert, Z.V.; McLean, A.R.; Cooper, B.S.; MacLean, R.C. Positive selection and compensatory adaptation interact to stabilize non-transmissible plasmids. Nat. Commun. 2014, 5, 5208. [Google Scholar] [CrossRef] [Green Version]
- Dionisio, F.; Conceicao, I.C.; Marques, A.C.R.; Fernandes, L.; Gordo, I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 2005, 1, 250–252. [Google Scholar] [CrossRef] [Green Version]
- Ragupathi, N.K.D.; Sethuvel, D.P.M.; Gajendran, R.; Anandan, S.; Walia, K.; Veeraraghavan, B. Horizontal transfer of antimicrobial resistance determinants among enteric pathogens through bacterial conjugation. Curr. Microbiol. 2019, 76, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [Green Version]
- Yousfi, M.; Mairi, A.; Touati, A.; Hassissene, L.; Brasme, L.; Guillard, T.; De, C.C. Extended spectrum β-lactamase and plasmid mediated quinolone resistance in Escherichia coli fecal isolates from healthy companion animals in Algeria. J. Infect. Chemother. 2016, 22, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Rafaï, C.; Frank, T.; Manirakiza, A.; Gaudeuille, A.; Mbecko, J.-R.; Nghario, L.; Serdouma, E.; Tekpa, B.; Garin, B.; Breurec, S. Dissemination of IncF-type plasmids in multiresistant CTX-M-15-producing Enterobacteriaceae isolates from surgical-site infections in Bangui, Central African Republic. BMC Microbiol. 2015, 15, 15. [Google Scholar] [CrossRef] [Green Version]
- Kiiru, J.; Butaye, P.; Goddeeris, B.M.; Kariuki, S. Analysis for prevalence and physical linkages amongst integrons, ISE cp 1, IS CR 1, Tn 21 and Tn 7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992–2011). BMC Microbiol. 2013, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Mshana, S.E.; Imirzalioglu, C.; Hossain, H.; Hain, T.; Domann, E. Chakraborty, T. Conjugative IncFI plasmids carrying CTX-M-15 among Escherichia coli ESBL producing isolates at a University hospital in Germany. BMC Infect. Dis. 2009, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Maherault, A.-C.; Kemble, H.; Magnan, M.; Gachet, B.; Roche, D.; Le Nagard, H.; Tenaillon, O.; Denamur, E.; Branger, C.; Landraud, L. Advantage of the F2: A1: B-IncF pandemic plasmid over IncC plasmids in in vitro acquisition and evolution of blaCTX-M gene-bearing plasmids in Escherichia coli. Antimicrob. Agents Chemother. 2019, 63, e01130-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.Y.; Ko, K.S.; Kang, C.–I.; Chung, D.R.; Peck, K.R.; Song, J.-H. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: Diverse clones and clonal dissemination. Int. J. Antimicrob. Agents 2011, 38, 160–163. [Google Scholar] [CrossRef]
- Dionisio, F.; Zilhão, R.; Gama, J.A. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid 2019, 102, 29–36. [Google Scholar] [CrossRef]
- Johnson, T.J.; Wannemuehler, Y.M.; Johnson, S.J.; Logue, C.M.; White, D.G.; Doetkott, C.; Nolan, L.K. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl. Environ. Microbiol. 2007, 73, 1976–1983. [Google Scholar] [CrossRef] [Green Version]
- Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Standard M100: Wayne, PA, USA, 2018. [Google Scholar]
- Jacoby, G.A.; Han, P. Detection of extended-spectrum beta-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 1996, 34, 908–911. [Google Scholar] [CrossRef] [Green Version]
- Casquet, J.; Thebaud, C.; Gillespie, R.G. Chelex without boiling, a rapid and easy technique to obtain stable amplifiable DNA from small amounts of ethanol-stored spiders. Mol. Ecol. Resour. 2012, 12, 136–141. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.; Threlfall, E. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]


| Sample Origin | Sample Type | Frequency n% | Species | Species n (%) | Total n (%) |
|---|---|---|---|---|---|
| Human | Human | 22 (43.14) | E. coli | 22 (43.1) | 22 (43.14) |
| Goat | 1 (1.96) | E. coli | 1 (1.96) | ||
| Animal | Pig | 3 (5.88) | E. coli | 3 (5.88) | |
| Dog | 6 (11.76) | E. coli | 6 (11.76) | 12 (23.52) | |
| Chicken | 2 (3.92) | E. coli | 2 (3.92) | ||
| Environment | Soil | 6 (11.76) | E. coli | 6 (11.76) | |
| E. coli | 2 (3.92) | ||||
| Fish | 11 (21.57) | K. pneumoniae | 3 (5.88) | 17 (33.32) | |
| C. braakii | 2 (3.92) | ||||
| E. cloacae | 4 (7.84) | ||||
| Total (n) | 51 (100) | 51 (100) |
| Sample ID | Source | Species | Conjugation Frequency |
|---|---|---|---|
| CN1 | Fish | E. cloacae | 8.2 × 10−5 |
| CN2 | Fish | E. cloacae | 2.3 × 10−4 |
| CN3 | Fish | E. cloacae | 5.2 × 10−5 |
| CN4 | Fish | E. cloacae | NIL |
| CN5 | Fish | C. braakii | 7.5 × 10−6 |
| CN6 | Fish | E. coli | 7.6 × 10−3 |
| CN7 | Fish | E. coli | NIL |
| CN8 | Fish | K. pneumoniae | 2.0 × 10−5 |
| CN9 | Fish | K. pneumoniae | 4.2 × 10−4 |
| CN10 | Fish | K. pneumoniae | 3.3 × 10−5 |
| CN11 | Fish | C. braakii | 9.4 × 10−4 |
| CN12 | Pig | E. coli | 4.7 × 10−5 |
| CN13 | Pig | E. coli | 2.6 × 10−6 |
| CN14 | Pig | E. coli | 9.8 × 10−5 |
| CN15 | Local chicken | E. coli | 4.7 × 10−5 |
| CN16 | Local chicken | E. coli | 8.4 × 10−7 |
| CN17 | Goat | E. coli | 4.1 × 10−6 |
| CN18 | Dog | E. coli | 2.1 × 10−5 |
| CN19 | Dog | E. coli | 1.2 × 10−7 |
| CN20 | Dog | E. coli | 5.0 × 10−5 |
| CN21 | Dog | E. coli | 1.1 × 10−6 |
| CN22 | Dog | E. coli | 6.0 × 10−4 |
| CN23 | Dog | E. coli | 9.6 × 10−6 |
| CN24 | Environment | E. coli | 1.5 × 10−9 |
| CN25 | Environment | E. coli | 2.6 × 10−7 |
| CN26 | Environment | E. coli | 3.5 × 10−6 |
| CN27 | Environment | E. coli | 2.9 × 10−7 |
| CN28 | Environment | E. coli | 6.1 × 10−6 |
| CN29 | Environment | E. coli | 7.2 × 10−3 |
| CN30 | Human | E. coli | 1.0 × 10−3 |
| CN31 | Human | E. coli | 4.7 × 10−4 |
| CN32 | Human | E. coli | 2.1 × 10−4 |
| CN33 | Human | E. coli | 4.0 × 10−5 |
| CN34 | Human | E. coli | 5.4 × 10−5 |
| CN35 | Human | E. coli | 4.8 × 10−1 |
| CN36 | Human | E. coli | 1.7 × 10−4 |
| CN37 | Human | E. coli | 3.5 × 10−7 |
| CN38 | Human | E. coli | 8.1 × 10−5 |
| CN39 | Human | E. coli | 1.2 × 10−5 |
| CN40 | Human | E. coli | 2.7 × 10−5 |
| CN41 | Human | E. coli | 2.4 × 10−7 |
| CN42 | Human | E. coli | NIL |
| CN43 | Human | E. coli | 5.5 × 10−6 |
| CN44 | Human | E. coli | 4.4 × 10−6 |
| CN45 | Human | E. coli | 2.9 × 10−6 |
| CN46 | Human | E. coli | NIL |
| CN47 | Human | E. coli | 2.1 × 10−5 |
| CN48 | Human | E. coli | 1.2 × 10−4 |
| CN49 | Human | E. coli | 1.1 × 10−7 |
| CN50 | Human | E. coli | NIL |
| CN51 | Human | E. coli | NIL |
| Sample No. | Source | Species | Donor’s Non-B-lactam Resistance Phenotype |
|---|---|---|---|
| CN1 | Fish | E. cloacae | SXT *, CIP *,CN *,TE * |
| CN2 | Fish | E. cloacae | CIP, SXT, CN, TE |
| CN3 | Fish | E. cloacae | CIP *, SXT *, TE *, CN* |
| CN4 | Fish | E. cloacae | CIP, CN, TE, SXT |
| CN5 | Fish | C. braakii | CIP *, SXT *, CN *, TE * |
| CN6 | Fish | E. coli | CIP, SXT, CN, TE |
| CN7 | Fish | E. coli | CIP, TE |
| CN8 | Fish | K. pneumoniae | CIP *, SXT *, CN *, TE * |
| CN9 | Fish | K. pneumoniae | CIP *, SXT *, CN *, TE * |
| CN10 | Fish | K. pneumoniae | CIP, SXT, CN, TE |
| CN11 | Fish | C. braakii | CIP, SXT, CN, TE * |
| CN12 | Pig | E. coli | CIP *, SXT *, TE * |
| CN13 | Pig | E. coli | TE, CIP, CN |
| CN14 | Pig | E. coli | CIP *, SXT *, TE *, CN * |
| CN15 | Local chicken | E. coli | CIP, SXT, CN, TE |
| CN16 | Local chicken | E. coli | CIP, SXT, CN, TE |
| CN17 | Goat | E. coli | SXT, TE *, CN, CIP * |
| CN18 | Dog | E. coli | SXT |
| CN19 | Dog | E. coli | SXT *, CIP *, TE, CN |
| CN20 | Dog | E. coli | CIP *, SXT *, TE * |
| CN21 | Dog | E. coli | CIP *, SXT *, TE *, CN * |
| CN22 | Dog | E. coli | CIP *, CN *, TE *, SXT * |
| CN23 | Dog | E. coli | SXT, TE, CN, CIP |
| CN24 | Environment | E. coli | SXT *, CIP *, TE * |
| CN25 | Environment | E. coli | SXT, TE, CIP |
| CN26 | Environment | E. coli | CIP *, SXT *, TE* |
| CN27 | Environment | E. coli | CIP * |
| CN28 | Environment | E. coli | CIP *, SXT *, CN *, TE * |
| CN29 | Environment | E. coli | CN, CIP *, SXT *, TE * |
| CN30 | Human | E. coli | TE *, CIP *, CN, SXT * |
| CN31 | Human | E. coli | CIP *, SXT * |
| CN32 | Human | E. coli | SXT *, CIP * |
| CN33 | Human | E. coli | TE *, CN *, CIP *, SXT * |
| CN34 | Human | E. coli | SXT *, TE *, CN *, CIP |
| CN35 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN36 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN37 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN38 | Human | E. coli | SXT *, TE *, CIP*, CN * |
| CN39 | Human | E. coli | SXT, TE, CIP *, CN * |
| CN40 | Human | E. coli | SXT *, TE * |
| CN41 | Human | E. coli | SXT, TE *, CIP *, CN |
| CN42 | Human | E. coli | SXT, CIP, CN, TE |
| CN43 | Human | E. coli | CN *, CIP *, SXT *, TE * |
| CN44 | Human | E. coli | SXT, TE, CIP |
| CN45 | Human | E. coli | SXT, TE, CIP, CN |
| CN46 | Human | E. coli | TE, SXT |
| CN47 | Human | E. coli | SXT *, TE *, CIP, CN |
| CN48 | Human | E. coli | SXT *, TE *, CIP, CN |
| CN49 | Human | E. coli | CIP *, CN *, SXT *, TE * |
| CN50 | Human | E. coli | SXT, TE |
| CN51 | Human | E. coli | CN, CIP, SXT, TE |
| Sample Source | Conjugation Efficiency | Conjugation Range | Donor’s Plasmid Replicon | Transconjugant Replicon Type |
|---|---|---|---|---|
| Human | 1.2 × 10−4 | FIB | FIA | |
| Human | 8.1 × 10−5 | FIA, FIB | FIB | |
| Dog | 5.0 × 10−5 | FIB | FIB | |
| Human | 5.4 × 10−5 | 10−6–10−3 | FIB | FIB |
| Human | 2.1 × 10−4 | FIB | FIB | |
| Environment | 7.2 × 10−3 | FIB | FIB | |
| Dog | 1.1 × 10−6 | FIB | FIB | |
| Human | 1.7 × 10−4 | FIB | FIB | |
| Dog | 9.6 × 10−6 | no rep | FIB | |
| Dog | 2.1 × 10−5 | no rep | FIB | |
| Human | 1.2 × 10−5 | 10−7–10−4 | no rep | FIB |
| Human | 4.7 × 10−4 | no rep | FIB | |
| Environment | 2.9 × 10−7 | no rep | FIB | |
| Fish | 2.3 × 10−4 | no rep | FIB | |
| Fish | NIL | FIA, Y | NA | |
| Human | NIL | no rep | NA | |
| Human | NIL | 0 | no rep | NA |
| Human | NIL | no rep | NA | |
| Human | NIL | no rep | NA | |
| Fish | NIL | no rep | NA | |
| Fish | 4.2 × 10−4 | A/C, FIA | no rep | |
| Pig | 2.6 × 10−6 | FIA | no rep | |
| Human | 5.5 × 10−6 | FIA | no rep | |
| Dog | 6.0 × 10−4 | FIA | no rep | |
| Pig | 9.8 × 10−5 | FIA | no rep | |
| Human | 2.9 × 10−6 | FIA | no rep | |
| Human | 4.0 × 10−5 | FIA | no rep | |
| Human | 4.8 × 10−1 | 10−9–10−1 | FIA | no rep |
| Dog | 1.2 × 10−7 | FIB | no rep | |
| Human | 3.5 × 10−7 | FIB | no rep | |
| Environment | 1.5 × 10−9 | FIB | no rep | |
| Environment | 2.6 × 10−7 | FIB | no rep | |
| Human | 4.4 × 10−6 | FIB | no rep | |
| Environment | 3.5 × 10−6 | FIB | no rep | |
| Human | 2.1 × 10−5 | FIB | no rep | |
| Fish | 7.5 × 10−6 | no rep | no rep | |
| Fish | 9.4 × 10−4 | no rep | no rep | |
| Human | 2.7 × 10−5 | no rep | no rep | |
| Local chicken | 4.7 × 10−5 | no rep | no rep | |
| Pig | 4.7 × 10−5 | no rep | no rep | |
| Human | 2.4 × 10−7 | no rep | no rep | |
| Fish | 3.3 × 10−5 | no rep | no rep | |
| Fish | 2.0 × 10−5 | 10−7–10−3 | no rep | no rep |
| Fish | 7.6 × 10−3 | no rep | no rep | |
| Human | 1.1 × 10−7 | no rep | no rep | |
| Fish | 5.2 × 10−5 | no rep | no rep | |
| Goat | 4.1 × 10−6 | no rep | no rep | |
| Environment | 6.1 × 10−6 | no rep | no rep | |
| Local chicken | 8.4 × 10−7 | no rep | no rep | |
| Human | 1.0 × 10−3 | no rep | no rep | |
| Fish | 8.2 × 10−5 | no rep | no rep |
| Source | E. coli Donors n | E. coli Transconjugants n (%) |
|---|---|---|
| Human | 22 | 18 (81.8) |
| Animal | 12 | 12 (100.0) |
| Environment | 8 | 7 (87.5) |
| Total | 42 | 37(88.1) |
| Primer Panel/Target | Direction | Primer Sequence | Annealing Temp (°C) | Amplicon Size (bp) |
|---|---|---|---|---|
| Panel 1 | ||||
| B/O | F | 5′-gcggtccggaaagccagaaaac-3′ | 60 | 159 |
| R | 5′-tctgcgttccgccaagttcga-3′ | |||
| FIC | F | 5′-gtgaactggcagatgaggaagg-3′ | 60 | 262 |
| R | 5′-ttctcctcgtcgccaaactagat-3′ | |||
| A/C | F | 5′-gagaaccaaagacaaagacctgga3′ | 60 | 465 |
| R | 5′-acgacaaacctgaattgcctcctt-3′ | |||
| P | F | 5′ctatggccctgcaaacgcgccagaaa3′ | 60 | 534 |
| R | 5′-tcacgcgccagggcgcagcc-3′ | |||
| T | F | 5′-ttggcctgtttgtgcctaaaccat-3′ | 60 | 750 |
| R | 5′-cgttgattacacttagctttggac-3′ | |||
| Panel 2 | ||||
| K/B | F | 5′-gcggtccggaaagccagaaaac-3′ | 60 | 160 |
| R | 5′-tctttcacgagcccgccaaa-3 | |||
| W | F | 5′-cctaagaacaacaaagcccccg-3′ | 60 | 242 |
| R | 5′-ggtgcgcggcatagaaccgt-3′ | |||
| FIIS | F | 5′-ctgtcgtaagctgatggc-3′ | 60 | 270 |
| R | 5′-ctctgccacaaacttcagc-3′ | |||
| FIA | F | 5′-ccatgctggttctagagaaggtg-3′ | 60 | 462 |
| R | 5′-gtatatccttactggcttccgcag-3′ | |||
| FIB | F | 5′-ggagttctgacacacgattttctg-3′ | 60 | 702 |
| 5′-ctcccgtcgcttcagggcatt-3′ | ||||
| Y | F | 5′-aattcaaacaacactgtgcagcctg-3′ | 60 | 765 |
| R | 5′-gcgagaatggacgattacaaaacttt-3′ | |||
| Panel 3 | ||||
| I1 | F | 5′-cgaaagccggacggcagaa-3′ | 60 | 139 |
| R | 5′-tcgtcgttccgccaagttcgt-3′ | |||
| FrepB | F | 5′-tgatcgtttaaggaattttg-3′ | 60 | 270 |
| R | 5′-gaagatcagtcacaccatcc-3′ | |||
| X | F | 5′-aaccttagaggctatttaagttgctgat-3′ | 60 | 376 |
| R | 5′-tgagagtcaatttttatctcatgttttagc3′ | |||
| HI1 | F | 5′-ggagcgatggattacttcagtac-3′ | 60 | 471 |
| R | 5′-tgccgtttcacctcgtgagta-3′ | |||
| N | F | 5′-gtctaacgagcttaccgaag-3′ | 60 | 559 |
| R | 5′-gtttcaactctgccaagttc-3′ | |||
| HI2 | F | 5′-tttctcctgagtcacctgttaacac-3′ | 60 | 644 |
| R | 5′-ggctcactaccgttgtcatcct-3′ | |||
| L/M | F | 5′-ggatgaaaactatcagcatctgaag-3′ | 60 | 785 |
| R | 5′-ctgcaggggcgattctttagg-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Minja, C.A.; Shirima, G.; Mshana, S.E. Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach. Antibiotics 2021, 10, 836. https://doi.org/10.3390/antibiotics10070836
Minja CA, Shirima G, Mshana SE. Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach. Antibiotics. 2021; 10(7):836. https://doi.org/10.3390/antibiotics10070836
Chicago/Turabian StyleMinja, Caroline A., Gabriel Shirima, and Stephen E. Mshana. 2021. "Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach" Antibiotics 10, no. 7: 836. https://doi.org/10.3390/antibiotics10070836
APA StyleMinja, C. A., Shirima, G., & Mshana, S. E. (2021). Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach. Antibiotics, 10(7), 836. https://doi.org/10.3390/antibiotics10070836






