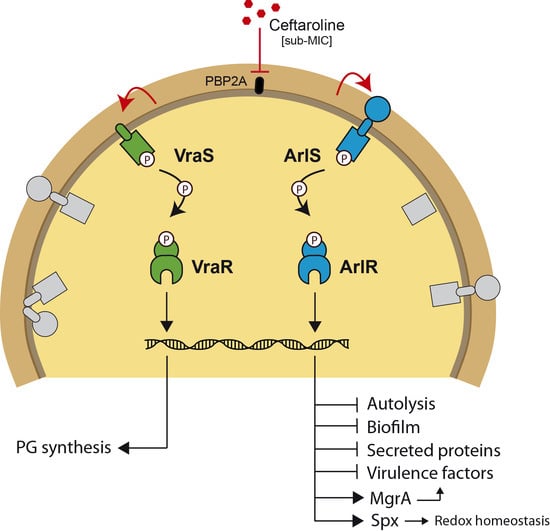
The Role of ArlRS and VraSR in Regulating Ceftaroline Hypersusceptibility in Methicillin-Resistant Staphylococcus aureus
Abstract
:1. Introduction
2. Results
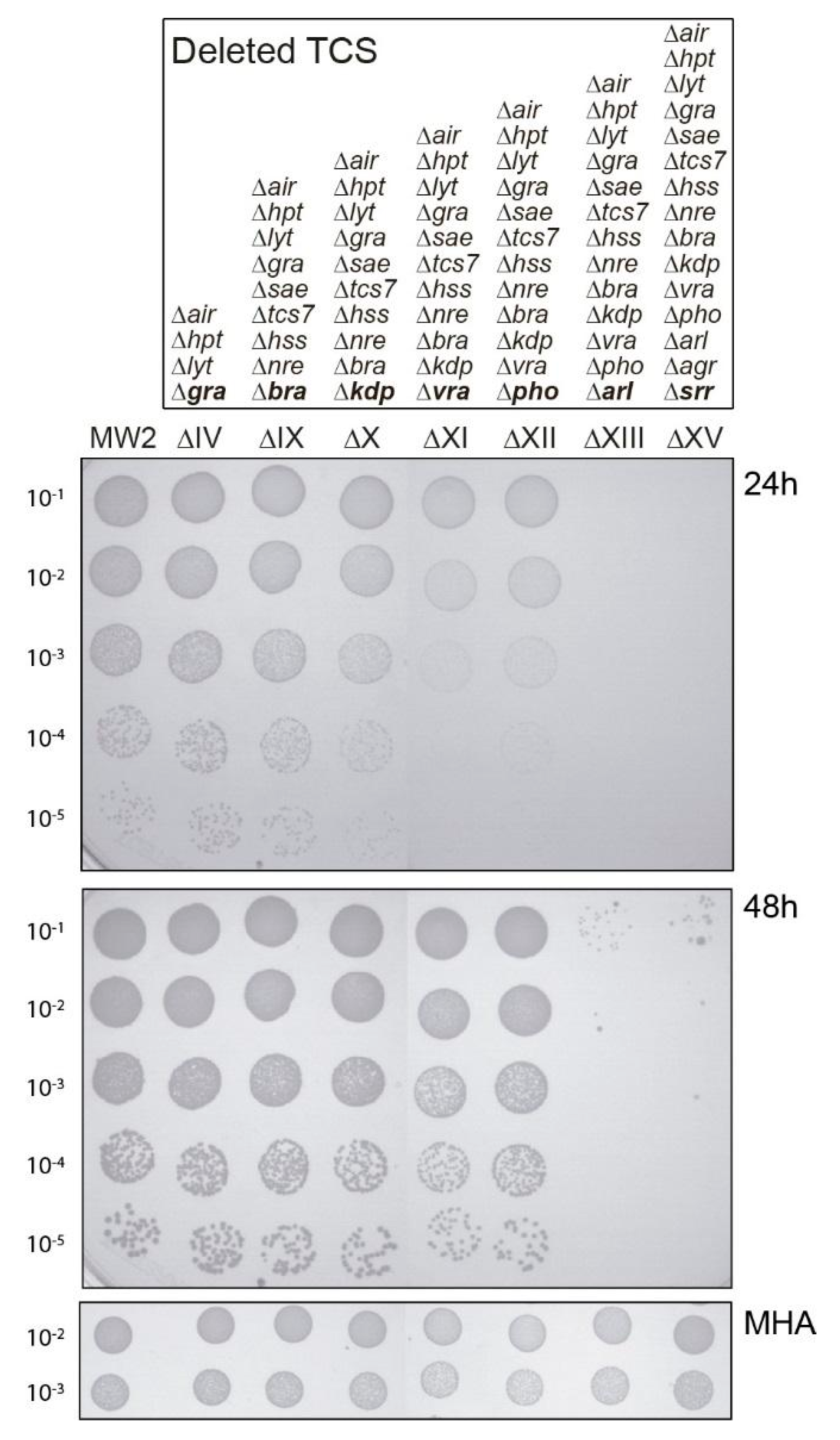
2.1. Both vraSR and arlRS TCSs Contribute to Sustain Bacterial Growth in the Presence of Sub-MIC Levels of Ceftaroline
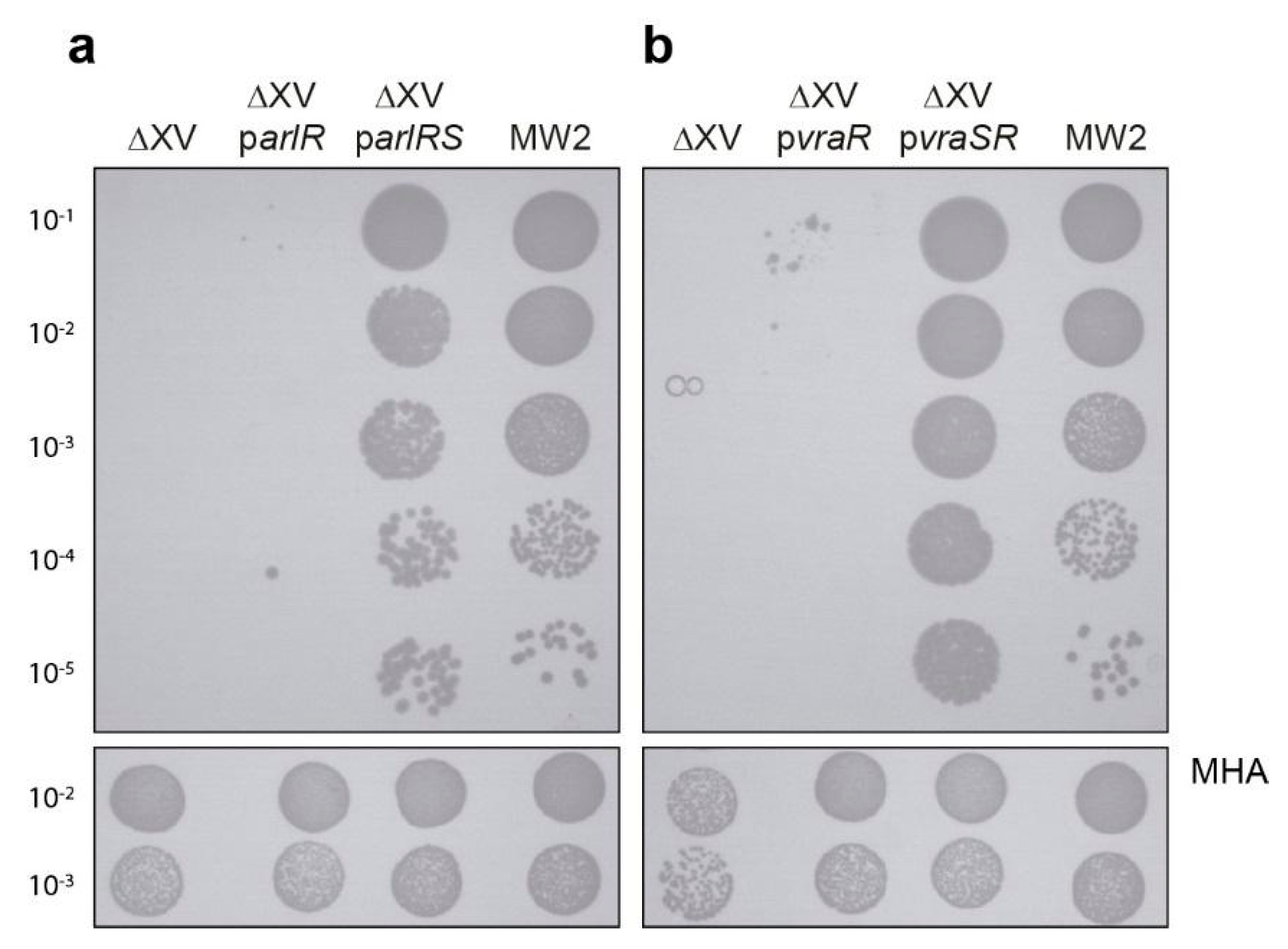
2.2. Analysis of arlRS and vraSR Single and Double Mutation
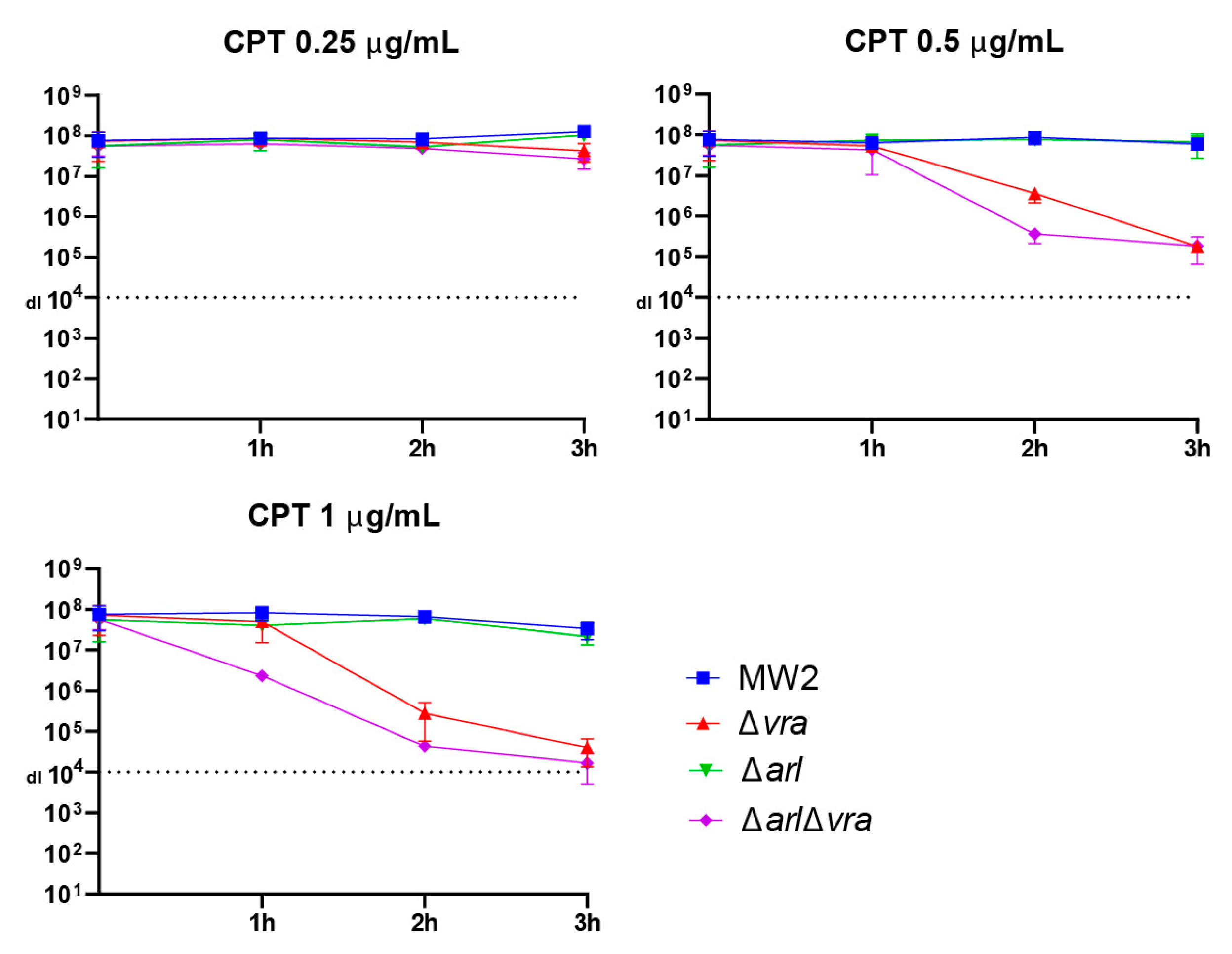
2.3. Analysis of Ceftaroline Sensitivity by Early Time Kill Assay
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Electrocompetent Staphylococcus Cells
4.3. Construction of MW2 Δarl Δvra Strain
4.4. Spot Test Assay
4.5. Antibiotic Susceptibility Tests
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Purrello, S.M.; Garau, J.; Giamarellos, E.; Mazzei, T.; Pea, F.; Soriano, A.; Stefani, S. Methicillin-resistant Staphylococcus aureus infections: A review of the currently available treatment options. J. Glob. Antimicrob. Resist. 2016, 7, 178–186. [Google Scholar] [CrossRef] [Green Version]
- Jacqueline, C.; Caillon, J.; Le Mabecque, V.; Miègeville, A.F.; Hamel, A.; Bugnon, D.; Ge, J.Y.; Potel, G. In vivo efficacy of ceftaroline (PPI-0903), a new broad-spectrum cephalosporin, compared with linezolid and vancomycin against methicillin-resistant and vancomycin-intermediate Staphylococcus aureus in a rabbit endocarditis model. Antimicrob. Agents Chemother. 2007, 51, 3397–3400. [Google Scholar] [CrossRef] [Green Version]
- Fishovitz, J.; Rojas-Altuve, A.; Otero, L.H.; Dawley, M.; Carrasco-López, C.; Chang, M.; Hermoso, J.A.; Mobashery, S. Disruption of allosteric response as an unprecedented mechanism of resistance to antibiotics. J. Am. Chem. Soc. 2014, 136, 9814–9817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.W.; Olsen, R.J.; Mehta, S.C.; Palzkill, T.; Cernoch, P.L.; Perez, K.K.; Musick, W.L.; Rosato, A.E.; Musser, M. PBP2a mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 2014, 58, 6668–6674. [Google Scholar] [CrossRef] [Green Version]
- Andrey, D.O.; François, P.; Manzano, C.; Bonetti, E.J.; Harbarth, S. Antimicrobial activity of ceftaroline against methicillin-resistant Staphylococcus aureus (MRSA) isolates collected in 2013–2014 at the Geneva University Hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Varela, M.C.; Roch, M.; Taglialegna, A.; Long, S.W.; Saavedra, M.O.; Rose, W.E.; Davis, J.J.; Hoffman, L.R.; Hernandez, R.E.; Rosato, R.R.; et al. Carbapenems drive the collateral resistance to ceftaroline in cystic fibrosis patients with MRSA. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Bouillet, S.; Stock, A.M. Structural basis of response regulator function. Annu. Rev. Microbiol. 2019, 73, 175–197. [Google Scholar] [CrossRef]
- Bai, J.; Zhu, X.; Zhao, K.; Yan, Y.; Xu, T.; Wang, J.; Huang, W.; Shi, L.; Shang, Y.; Lv, Z.; et al. The role of ArlRS in regulating oxacillin susceptibility in methicillin-resistant Staphylococcus aureus indicates it is a potential target for antimicrobial resistance breakers. Emerg. Microbes Infect. 2019, 8, 503–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona, S.T.; Choy, M.; Hogan, A.M. Essential two-component systems regulating cell envelope functions: Opportunities for novel antibiotic therapies. J. Membr. Biol. 2017, 251, 75–89. [Google Scholar] [CrossRef]
- Kuroda, M.; Kuroda, H.; Oshima, T.; Takeuchi, F.; Mori, H.; Hiramatsu, K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 2003, 49, 807–821. [Google Scholar] [CrossRef]
- Rapun-Araiz, B.; Haag, A.F.; Solano, C.; Lasa, I. The impact of two-component sensorial network in staphylococcal speciation. Curr. Opin. Microbiol. 2020, 55, 40–47. [Google Scholar] [CrossRef]
- Villanueva, M.; García, B.; Valle, J.; Rapún, B.; Ruiz De Los Mozos, I.; Solano, C.; Martí, M.; Penadés, J.R.; Toledo-Arana, A.; Lasa, I. Sensory deprivation in Staphylococcus aureus. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Rapun-Araiz, B.; Haag, A.F.; De Cesare, V.; Gil, C.; Dorado-Morales, P.; Penades, J.R.; Lasa, I. Systematic Reconstruction of the Complete Two-Component Sensorial Network in Staphylococcus aureus. Msystems 2020, 5, 1–16. [Google Scholar] [CrossRef]
- Galbusera, E.; Renzoni, A.; Andrey, D.O.; Monod, A.; Barras, C.; Tortora, P.; Polissi, A.; Kelley, W.L. Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance. Antimicrob. Agents Chemother. 2011, 55, 1008–1020. [Google Scholar] [CrossRef] [Green Version]
- Roch, M.; Lelong, E.; Panasenko, O.O.; Sierra, R.; Renzoni, A.; Kelley, W.L. Thermosensitive PBP2a requires extracellular folding factors PrsA and HtrA1 for Staphylococcus aureus MRSA B-lactam resistance. Commun. Biol. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Crosby, H.A.; Tiwari, N.; Kwiecinski, J.M.; Xu, Z.; Dykstra, A.; Jenul, C.; Fuentes, E.J.; Horswill, A.R. The Staphylococcus aureus ArlRS two-component system regulates virulence factor expression through MgrA. Mol. Microbiol. 2020, 113, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Linzner, N.; Van Loi, V.; Fritsch, V.N.; Antelmann, H. Thiol-based redox switches in the major pathogen Staphylococcus aureus. Biol. Chem. 2021, 402, 333–361. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.R.; Bae, T.; Williams, W.A.; Duguid, E.M.; Rice, P.A.; Schneewind, O.; He, C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2006, 2, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, M.; Jousselin, A.; Baek, K.T.; Prados, J.; Andrey, D.O.; Renzoni, A.; Ingmer, H.; Frees, D.; Kelley, W.L. Rifampin resistance rpoB alleles or multicopy thioredoxin/thioredoxin reductase suppresses the lethality of disruption of the global stress regulator spx in Staphylococcus aureus. J. Bacteriol. 2016, 198, 2719–2731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pamp, S.J.; Frees, D.; Engelmann, S.; Hecker, M.; Ingmer, H. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2006, 188, 4861–4870. [Google Scholar] [CrossRef] [Green Version]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Eguchi, Y.; Watanabe, T.; Okamoto, S.; Doi, A.; Utsumi, R. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 2010, 13, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Anton, A.I.; Barry, P.; Alfonso, B.; Fang, Y.; Novick, R.P. Novel cassette-based shuttle vector system for Gram-positive bacteria. Appl. Environ. Microbiol. 2004, 70, 6076–6085. [Google Scholar] [CrossRef] [Green Version]
- Schenk, S.; Laddaga, R.A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 1992, 94, 133–138. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.G.O.; Penade, R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Débarbouillé, M.; Penadés, J.R.; Lasa, I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef] [Green Version]
- Renzoni, A.; Andrey, D.O.; Jousselin, A.; Barras, C.; Monod, A.; Vaudaux, P.; Lew, D.; Kelley, W.L. Whole genome sequencing and complete genetic analysis reveals novel pathways to glycopeptide resistance in Staphylococcus aureus. PLoS ONE 2011, 6, e21577. [Google Scholar] [CrossRef]

| Strains | ECF a with Ceftaroline at 0.25 µg/mL | Modal MIC (Range) | Modal MBC (Range) |
|---|---|---|---|
| MW2 | ≈1 | 0.5 (0.5–1) | 1 |
| Δarl | ≈1 | 0.5 | 1 (0.5–1) |
| Δvra | 2 × 10−2 | 0.5 (0.5–1) | 0.5 |
| Δarl Δvra b | <10−8 | 0.5 (0.25–0.5) | 0.5 |
| ΔXV b | <10−8 | 0.25 (0.25–0.5) | 0.5 |
| ΔXV parlRS | 2 × 10−1 | 0.5 | 0.5 |
| ΔXV pvraSR | ≈1 | 1 (0.5–1) | 1 (0.5–1) |
| ATCC29213 | <10−8 | 0.5 (0.25–0.5) | 0.5 |
| Plasmids | Relevant Characteristics | Reference |
| pMAD::TCS12AD | pMAD plasmid containing the allele for deletion of the vraSR genes | [14] |
| parlRS | pCN51 plasmid expressing arlRS genes | [14] |
| pvraSR | pCN51 plasmid expressing vraSR genes | [14] |
| parlR | pCN51 plasmid expressing arlR gene | [14] |
| pvraR | pCN51 plasmid expressing vraR gene | [14] |
| Strains | Relevant Characteristics | Reference |
| ATCC29213 | Standard QC strain MSSA | |
| MW2 | Typical community-acquired strain of MRSA, which was isolated in 1998 in North Dakota, USA. bla+ ΔmecR1 mecI− mecR2− | [14] |
| ΔIV | MW2 ∆airSR, ∆hptSR, ∆lytSR, ∆graRS | [14] |
| ΔIX | MW2 ∆airSR, ∆hptSR, ∆lytSR, ∆graRS, ∆saeRS, ∆tcs7, ∆hssRS, ∆nreBC, ∆braRS | [14] |
| ΔX | MW2 ∆IX ∆kdpDE | [14] |
| ΔXI | MW2 ∆X ∆vraSR | [14] |
| ΔXII | MW2 ∆XI ∆phoPR | [14] |
| ΔXIII | MW2 ∆XII ∆arlRS | [14] |
| ΔXV | MW2 ∆XIII ∆agrCA, ∆srrAB | [14] |
| ΔXIV (srrAB) | MW2 ∆XIII ∆agrCA | [14] |
| ΔXIV (agrBDCA) | MW2 ∆XIII ∆srrAB | [14] |
| Δsrr | MW2 ∆srrAB | [14] |
| Δagr | MW2 ∆agrCA | [14] |
| Δarl | MW2 ∆arlRS | [14] |
| Δvra | MW2 ∆vraSR | [14] |
| Δarl Δvra | MW2 ∆arlRS ∆vraSR | This study |
| ΔXV parlRS | MW2 ∆XV ∆arl carrying pCN51::arlRS plasmid | [14] |
| ΔXV pvraSR | MW2 ∆XV ∆arl carrying pCN51::vrASR plasmid | [14] |
| ΔXV parlR | MW2 ∆XV carrying pCN51::arlR plasmid | [14] |
| ΔXV pvraR | MW2 ∆XV carrying pCN51::vraR plasmid | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villanueva, M.; Roch, M.; Lasa, I.; Renzoni, A.; Kelley, W.L. The Role of ArlRS and VraSR in Regulating Ceftaroline Hypersusceptibility in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 821. https://doi.org/10.3390/antibiotics10070821
Villanueva M, Roch M, Lasa I, Renzoni A, Kelley WL. The Role of ArlRS and VraSR in Regulating Ceftaroline Hypersusceptibility in Methicillin-Resistant Staphylococcus aureus. Antibiotics. 2021; 10(7):821. https://doi.org/10.3390/antibiotics10070821
Chicago/Turabian StyleVillanueva, Maite, Melanie Roch, Iñigo Lasa, Adriana Renzoni, and William L. Kelley. 2021. "The Role of ArlRS and VraSR in Regulating Ceftaroline Hypersusceptibility in Methicillin-Resistant Staphylococcus aureus" Antibiotics 10, no. 7: 821. https://doi.org/10.3390/antibiotics10070821
APA StyleVillanueva, M., Roch, M., Lasa, I., Renzoni, A., & Kelley, W. L. (2021). The Role of ArlRS and VraSR in Regulating Ceftaroline Hypersusceptibility in Methicillin-Resistant Staphylococcus aureus. Antibiotics, 10(7), 821. https://doi.org/10.3390/antibiotics10070821