Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia coli in Italy
Abstract
1. Introduction
2. Results
2.1. Bacterial Isolates and Phenotypic Characterization
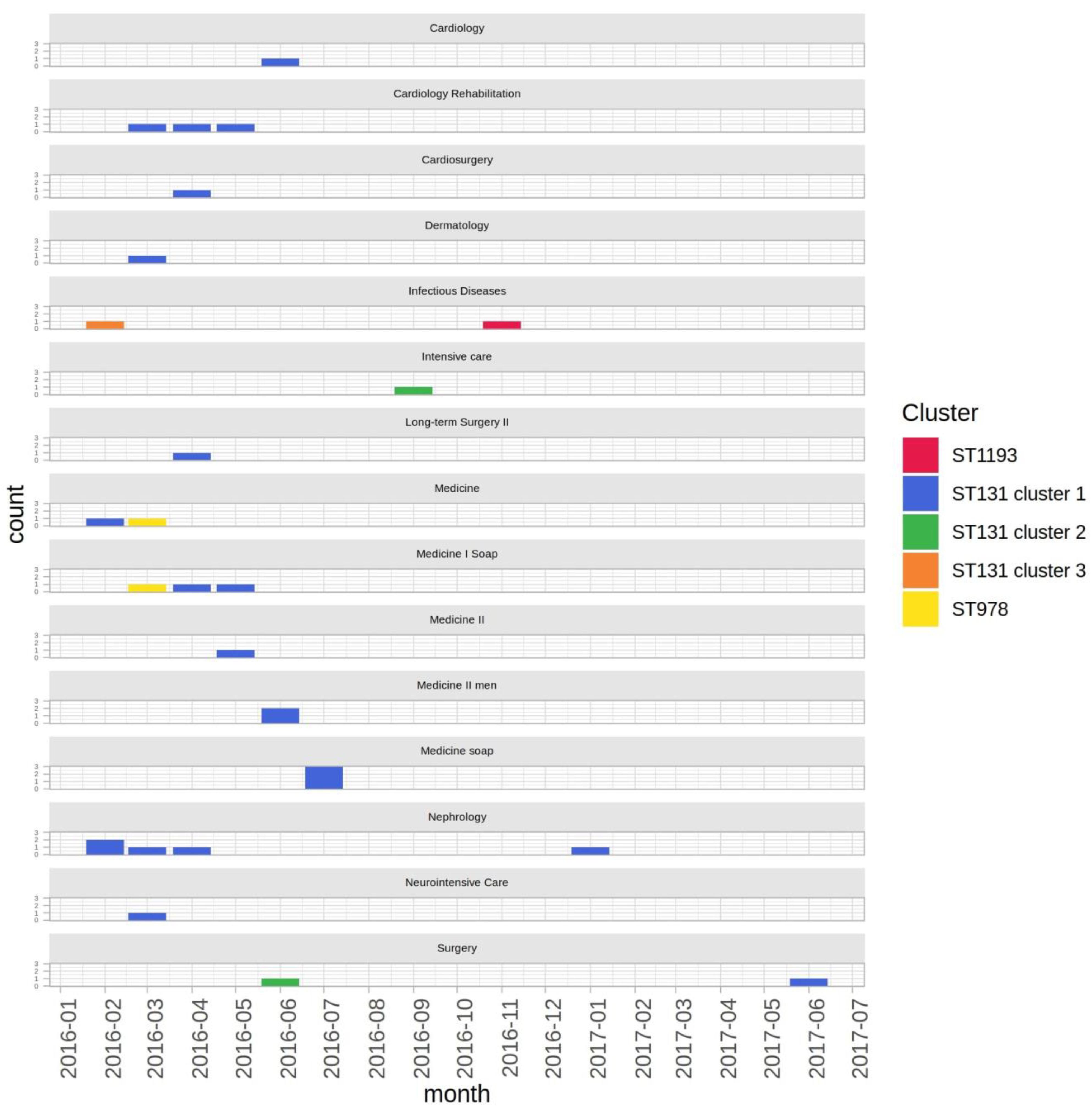
2.2. Epidemiological Context
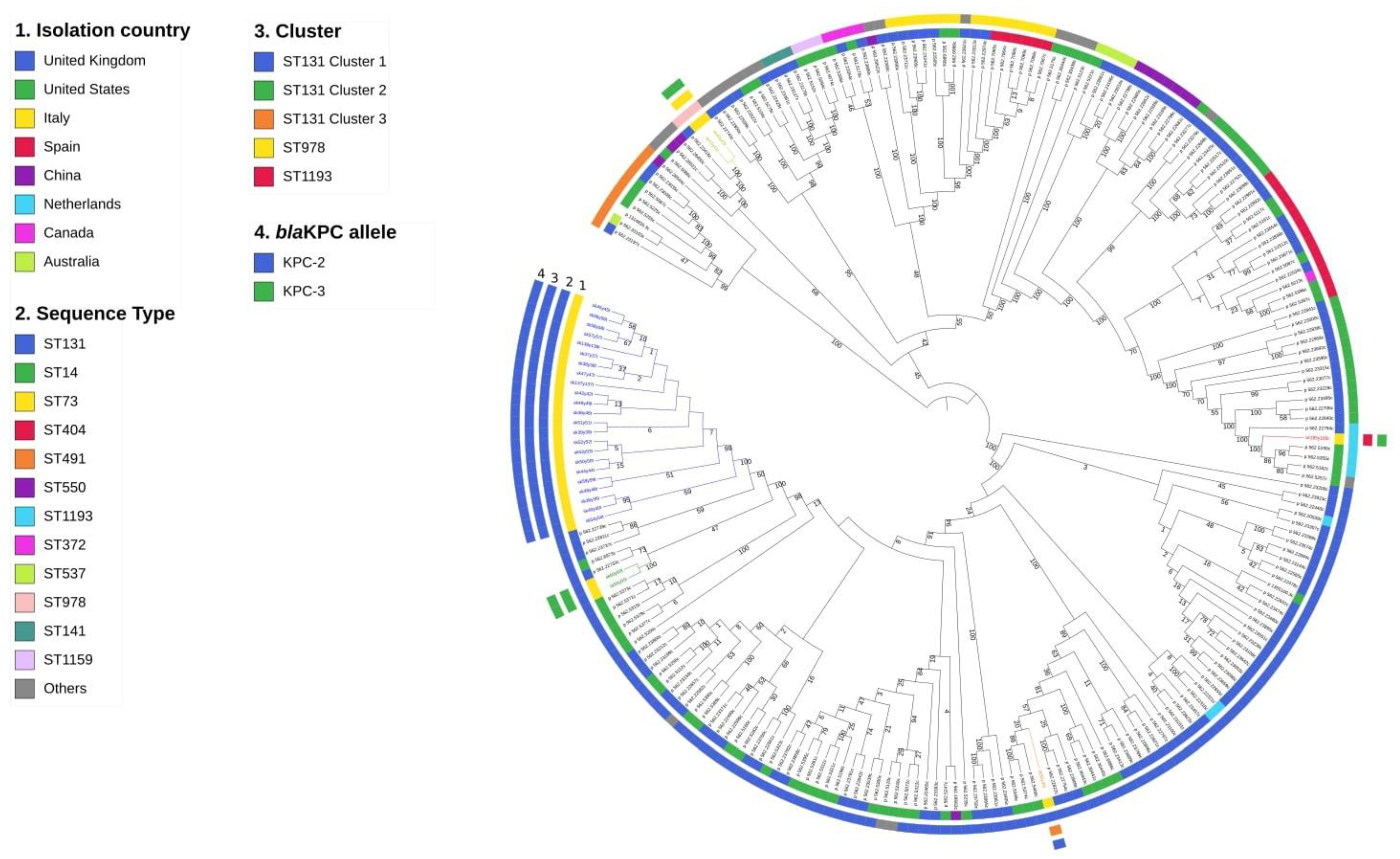
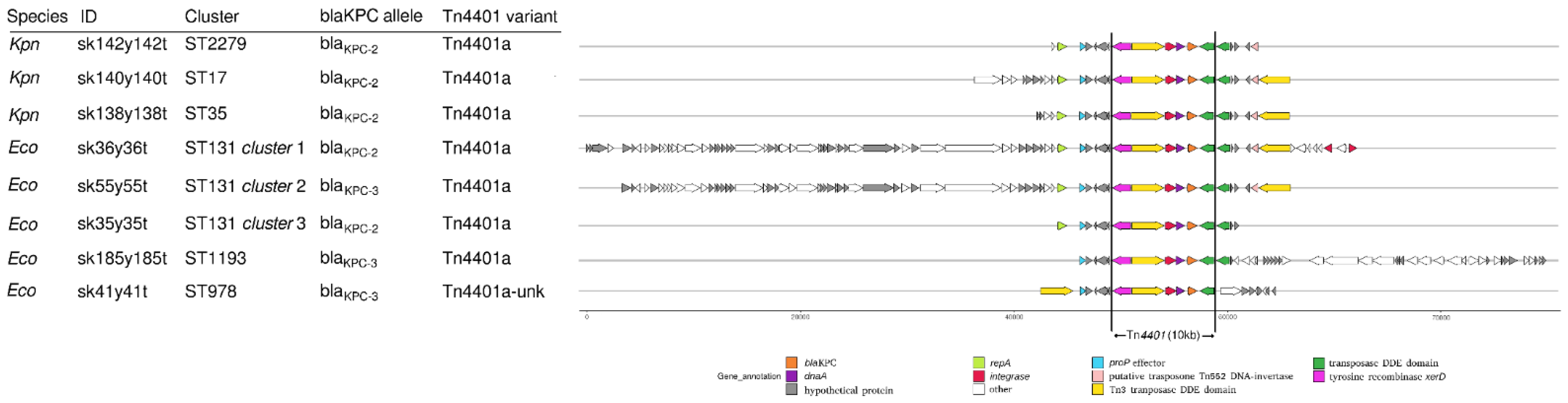
2.3. Whole-Genome Sequencing Characterization
3. Discussion
4. Materials and Methods
4.1. Epidemiological Context and Characterization of Bacterial Isolates
4.2. High Resolution Melting Assay
4.3. Whole-Genome Sequencing
4.4. CoreSNP Calling and Phylogenetic Analyses
4.5. Whole-Genome Sequencing-Based Typing
4.6. KPC-Harboring Contigs Comparison
4.7. Conjugation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brolund, A.; Lagerqvist, N.; Byfors, S.; Struelens, M.J.; Monnet, D.L.; Albiger, B.; Kohlenberg, A.; European Antimicrobial Resistance Genes Surveillance Network EURGen-Net Capacity Survey Group. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. Euro Surveill 2019, 24. [Google Scholar] [CrossRef]
- Decraene, V.; Phan, H.T.T.; George, R.; Wyllie, D.H.; Akinremi, O.; Aiken, Z.; Cleary, P.; Dodgson, A.; Pankhurst, L.; Crook, D.W.; et al. A Large, Refractory Nosocomial Outbreak of Klebsiella pneumoniae Carbapenemase-Producing Escherichia coli demonstrates carbapenemase gene outbreaks involving sink sites require novel approaches to infection control. Antimicrob. Agents Chemother. 2018, 62, e01689-18. [Google Scholar] [CrossRef] [PubMed]
- Stoesser, N.; Sheppard, A.E.; Peirano, G.; Anson, L.W.; Pankhurst, L.; Sebra, R.; Phan, H.T.T.; Kasarskis, A.; Mathers, A.J.; Peto, T.E.A.; et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci. Rep. 2017, 7, 5917. [Google Scholar] [CrossRef]
- Stoesser, N.; Sheppard, A.E.; Pankhurst, L.; De Maio, N.; Moore, C.E.; Sebra, R.; Turner, P.; Anson, L.W.; Kasarskis, A.; Batty, E.M.; et al. Evolutionary History of the Global Emergence of the Escherichia coli Epidemic Clone ST131. MBio 2016, 7, e02162. [Google Scholar] [CrossRef]
- Grundmann, H.; Glasner, C.; Albiger, B.; Aanensen, D.M.; Tomlinson, C.T.; Andrasević, A.T.; Cantón, R.; Carmeli, Y.; Friedrich, A.W.; Giske, C.G.; et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): A prospective, multinational study. Lancet Infect Dis. 2017, 17, 153–163. [Google Scholar] [CrossRef]
- Petrella, S.; Ziental-Gelus, N.; Mayer, C.; Renard, M.; Jarlier, V.; Sougakoff, W. Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France. Antimicrob. Agents Chemother. 2008, 52, 3725–3736. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Urban, C.; Bradford, P.A.; Tuckman, M.; Segal-Maurer, S.; Wehbeh, W.; Grenner, L.; Colon-Urban, R.; Mariano, N.; Rahal, J.J. Carbapenem-resistant Escherichia coli harboring Klebsiella pneumoniae carbapenemase beta-lactamases associated with long-term care facilities. Clin Infect Dis. 2008, 46, e127–e130. [Google Scholar] [CrossRef] [PubMed]
- Goren, M.G.; Navon-Venezia, S.; Chmelnitsky, I.; Carmeli, Y. Carbapenem-resistant KPC-2-producing Escherichia coli in a Tel Aviv Medical Center, 2005 to 2008. Antimicrob. Agents Chemother. 2010, 54, 2687–2691. [Google Scholar] [CrossRef]
- Naas, T.; Cuzon, G.; Gaillot, O.; Courcol, R.; Nordmann, P. When carbapenem-hydrolyzing beta-lactamase KPC meets Escherichia coli ST131 in France. Antimicrob. Agents Chemother. 2011, 55, 4933–4934. [Google Scholar] [CrossRef]
- Morris, D.; Boyle, F.; Ludden, C.; Condon, I.; Hale, J.; O’Connell, N.; Power, L.; Boo, T.W.; Dhanji, H.; Lavallee, C.; et al. Production of KPC-2 carbapenemase by an Escherichia coli clinical isolate belonging to the international ST131 clone. Antimicrob. Agents Chemother. 2011, 55, 4935–4936. [Google Scholar] [CrossRef]
- Giani, T.; Antonelli, A.; Caltagirone, M.; Mauri, C.; Nicchi, J.; Arena, F.; Nucleo, E.; Bracco, S.; Pantosti, A.; Luzzaro, F.; et al. Evolving beta-lactamase epidemiology in Enterobacteriaceae from Italian nationwide surveillance, October 2013: KPC-carbapenemase spreading among outpatients. Euro Surveill. 2017, 22, 30583. [Google Scholar] [CrossRef] [PubMed]
- Iacchini, S.; Sabbatucci, M.; Gagliotti, C.; Rossolini, G.M.; Moro, M.L.; Iannazzo, S.; D’Ancona, F.; Pezzotti, P.; Pantosti, A. Bloodstream infections due to carbapenemase-producing Enterobacteriaceae in Italy: Results from nationwide surveillance, 2014 to 2017. Euro Surveill. 2019, 24. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Caltagirone, M.; Bitar, I.; Nucleo, E.; Spalla, M.; Fogato, E.; D’Angelo, R.; Pagani, L.; Migliavacca, R. Emergence of Escherichia coli Sequence Type 131 (ST131) and ST3948 with KPC-2, KPC-3 and KPC-8 carbapenemases from a Long-Term Care and Rehabilitation Facility (LTCRF) in Northern Italy. Adv. Exp. Med. Biol. 2016, 901, 77–89. [Google Scholar] [CrossRef]
- Adler, A.; Miller-Roll, T.; Assous, M.V.; Geffen, Y.; Paikin, S.; Schwartz, D.; Weiner-Well, Y.; Hussein, K.; Cohen, R.; Carmeli, Y. A multicenter study of the clonal structure and resistance mechanism of KPC-producing Escherichia coli isolates in Israel. Clin. Microbiol. Infect. 2015, 21, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Mavroidi, A.; Miriagou, V.; Malli, E.; Stefos, A.; Dalekos, G.N.; Tzouvelekis, L.S.; Petinaki, E. Emergence of Escherichia coli sequence type 410 (ST410) with KPC-2 β-lactamase. Int. J. Antimicrob. Agents. 2012, 39, 247–250. [Google Scholar] [CrossRef]
- Landman, D.; Urban, C.; Bäcker, M.; Kelly, P.; Shah, N.; Babu, E.; Bratu, S.; Quale, J. Susceptibility profiles, molecular epidemiology, and detection of KPC-producing Escherichia coli isolates from the New York City vicinity. J. Clin. Microbiol. 2010, 48, 4604–4607. [Google Scholar] [CrossRef]
- Leung, V.; Loo, V.G.; Frenette, C.; Domingo, M.C.; Bourgault, A.M.; Mulvey, M.R.; Robson, H.G. First Canadian outbreak of Enterobacteriaceae-expressing Klebsiella pneumoniae carbapenemase type 3. Can. J. Infect. Dis. Med. Microbiol. 2012, 23, 117–120. [Google Scholar] [CrossRef]
- Accogli, M.; Giani, T.; Monaco, M.; Giufrè, M.; García-Fernández, A.; Conte, V.; D’Ancona, F.; Pantosti, A.; Rossolini, G.M.; Cerquetti, M. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J. Antimicrob. Chemother. 2014, 69, 2293–2296. [Google Scholar] [CrossRef][Green Version]
- Chavda, K.D.; Chen, L.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. Molecular Diversity and Plasmid Analysis of KPC-Producing Escherichia coli. Antimicrob. Agents Chemother. 2016, 60, 4073–4081. [Google Scholar] [CrossRef]
- Petty, N.K.; Ben Zakour, N.L.; Stanton-Cook, M.; Skippington, E.; Totsika, M.; Forde, B.M.; Phan, M.D.; Gomes Moriel, D.; Peters, K.M.; Davies, M.; et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proc. Natl. Acad. Sci. USA 2014, 111, 5694–5699. [Google Scholar] [CrossRef]
- Ben Zakour, N.L.; Alsheikh-Hussain, A.S.; Ashcroft, M.M.; Khanh Nhu, N.T.; Roberts, L.W.; Stanton-Cook, M.; Schembri, M.A.; Beatson, S.A. Erratum for Ben Zakour et al., Sequential Acquisition of Virulence and Fluoroquinolone Resistance Has Shaped the Evolution of Escherichia coli ST131. MBio 2016, 7, e00958-16. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 6.0; EUCAST: Basel, Switzerland, 2016; Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_6.0.pdf (accessed on 14 June 2021).
- Perini, M.; Piazza, A.; Panelli, S.; Di Carlo, D.; Corbella, M.; Gona, F.; Vailati, F.; Marone, P.; Cirillo, D.M.; Farina, C.; et al. EasyPrimer: User-friendly tool for pan-PCR/HRM primers design. Development of an HRM protocol on wzi gene for fast Klebsiella pneumoniae typing. Sci. Rep. 2020, 10, 1307. [Google Scholar] [CrossRef] [PubMed]
- Perini, M.; Batisti Biffignandi, G.; Di Carlo, D.; Pasala, A.R.; Piazza, A.; Panelli, S.; Zuccotti, G.V.; Comandatore, F. MeltingPlot, a user-friendly online tool for epidemiological investigation using High Resolution Melting data. BMC Bioinform. 2021, 22, 76. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Gona, F.; Comandatore, F.; Battaglia, S.; Piazza, A.; Trovato, A.; Lorenzin, G.; Cichero, P.; Biancardi, A.; Nizzero, P.; Moro, M.; et al. Comparison of core-genome MLST, coreSNP and PFGE methods for Klebsiella pneumoniae cluster analysis. Microb. Genom. 2020, 6, e000347. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 90. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Micobiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, A.E.; Stoesser, N.; German-Mesner, I.; Vegesana, K.; Sarah Walker, A.; Crook, D.W.; Mathers, A.J. TETyper: A bioinformatic pipeline for classifying variation and genetic contexts of transposable elements from short-read whole-genome sequencing data. Microb. Genom. 2018, 4, e000232. [Google Scholar] [CrossRef]
- Guy, L.; Kultima, J.R.; Andersson Siv, G.E. genoPlotR: Comparative gene and genome visualization in R. Bioinformatics 2010, 26, 2334–2335. [Google Scholar] [CrossRef]
| ID Strain | Antimicrobial Susceptibility (MIC, µg/mL) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | PTZ | CTX | CAZ | CZA | C/T | MER | IMI | ERT | AMK | GNT | AZT | CIP | TBR | TIG | SXT | COL | |
| sk46y46t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 4 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk36y36t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk35y35t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | >4 (R) | >4 (R) | >1 (R) | >4 (R) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk39y39t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk37y37t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 2/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk38y38t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk42y42t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | 16 (I) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk40y40t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk47y47t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | >8 (R) | >1 (R) | 16 (I) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk41y41t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 16/4 (R) | 8 (I) | 8 (I) | >1 (R) | >16 (R) | 4 (I) | >4 (R) | ≤0,06 (S) | >4 (R) | ≤1 (S) | ≤2/38 (S) | <0,25 (S) |
| sk43y43t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 32/4 (R) | 8 (I) | 8 (I) | >1 (R) | >16 (R) | 4 (I) | >4 (R) | ≤0,06 (S) | >4 (R) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk54y54t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | >8 (R) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk44y44t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk45y45t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk48y48t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk49y49t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | >8 (R) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | ≤2/38 (S) | <0,25 (S) |
| sk51y51t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk50y50t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk52y52t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | 4 (I) | >4 (R) | >1 (R) | 4 (I) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk53y53t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk56y56t | >32 (R) | >16 (R) | >32 (R) | 16 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk55y55t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 16/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | ≤2/38 (S) | 0,5 (S) |
| sk57y57t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | <0,25 (S) |
| sk58y58t | >32 (R) | >16 (R) | >32 (R) | 8 (R) | <0,5/4 (S) | 2/4 (R) | 4 (I) | 4 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk59y59t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk60y60t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 32/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 1 (S) |
| sk185y185t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk136y136t | >32 (R) | >16 (R) | >32 (R) | 32 (R) | <0,5/4 (S) | 2/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| sk137y137t | >32 (R) | >16 (R) | >32 (R) | >32 (R) | <0,5/4 (S) | 4/4 (R) | 8 (I) | 8 (I) | >1 (R) | <8 (S) | ≤2 (S) | >4 (R) | >1 (R) | ≤2 (S) | ≤1 (S) | >4/76 (R) | 0,5 (S) |
| ID Sample | Microorganism | Ward | Isolation Date | Material |
|---|---|---|---|---|
| sk35y35t | Escherichia coli | Infectious Diseases | 15 February 2016 | Rectal swab |
| sk36y36t | Escherichia coli | Nephrology | 16 February 2016 | Rectal swab |
| sk37y37t | Escherichia coli | Medicine | 26 February 2016 | Urine |
| sk38y38t | Escherichia coli | Nephrology | 29 February 2016 | Rectal swab |
| sk39y39t | Escherichia coli | Cardiology Rehabilitation | 1 March 2016 | Urine |
| sk40y40t | Escherichia coli | Neurointensive Care | 3 March 2016 | Purulent exudate |
| sk42y42t | Escherichia coli | Dermatology | 3 March 2016 | Urine |
| sk41y41t | Escherichia coli | Medicine I Soap | 8 March 2016 | Blood |
| sk43y43t | Escherichia coli | Medicine | 17 March 2016 | Rectal swab |
| sk44y44t | Escherichia coli | Nephrology | 23 March 2016 | Blood |
| sk45y45t | Escherichia coli | Cardiology Rehabilitation | 5 April 2016 | Urine |
| sk46y46t | Escherichia coli | Cardiosurgery | 18 April 2016 | Bronchial aspirate |
| sk47y47t | Escherichia coli | Medicine I Soap | 18 April 2016 | Urine |
| sk48y48t | Escherichia coli | Nephrology | 21 April 2016 | Surgical wound swab |
| sk49y49t | Escherichia coli | Long-term Surgery II | 24 April 2016 | Peritoneal fluid |
| sk50y50t | Escherichia coli | Medicine II | 10 May 2016 | Respiratory secretion |
| sk51y51t | Escherichia coli | Cardiology Rehabilitation | 13 May 2016 | Urine |
| sk52y52t | Escherichia coli | Medicine I Soap | 22 May 2016 | Urine |
| sk53y53t | Escherichia coli | Medicine II men | 11 June 2016 | Urine |
| sk54y54t | Escherichia coli | Medicine II men | 25 June 2016 | Urine |
| sk55y55t | Escherichia coli | Surgery | 29 June 2016 | Drainage fluid |
| sk56y56t | Escherichia coli | Cardiology | 30 June 2016 | Urine |
| sk57y57t | Escherichia coli | Medicine soap | 13 July 2016 | Urine |
| sk58y58t | Escherichia coli | Medicine soap | 13 July 2016 | Rectal swab |
| sk59y59t | Escherichia coli | Medicine soap | 14 July 2016 | Purulent exudate |
| sk60y60t | Escherichia coli | Intensive care | 30 September 2016 | Rectal swab |
| sk185y185t | Escherichia coli | Infectious Diseases | 14 November 2016 | Respiratory secretion |
| sk136y136t | Escherichia coli | Nephrology | 23 January 2017 | Rectal swab |
| sk137y137t | Escherichia coli | Surgery | 5 June 2017 | Rectal swab |
| sk138y138t | Klebsiella pneumoniae | Nephrology | 22 January 2016 | Urine |
| sk139y139t | Klebsiella pneumoniae | Cardiology | 16 February 2016 | Rectal swab |
| sk140y140t | Klebsiella pneumoniae | Rehabilitation Medicine | 25 February 2016 | Rectal swab |
| sk141y141t | Klebsiella pneumoniae | Urology | 25 February 2016 | Urine |
| sk142y142t | Klebsiella pneumoniae | Infectious Diseases | 22 March 2016 | Rectal swab |
| sk143y143t | Klebsiella pneumoniae | Medicine | 31 March 2016 | Rectal swab |
| ID Sample | Isolation Date | MLST | Cluster | Serotype | fimH | Resistance Genes | Virulence Genes | Plasmid Incompatibility Groups |
|---|---|---|---|---|---|---|---|---|
| sk35y35t | 15 February 2016 | 131 | ST131 cluster 3 | O25b:H4 | 30 | ampH, blaTEM-90, blaKPC-2,blaOXA-9, blaCTX-M-15, ampC2, catB4, blaOXA-1, aac(3)-IIa, parC1aAB, gyrA1AB | iha, sat, iss, cnf1, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk36y36t | 16 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-122, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk37y37t | 26 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-90, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk38y38t | 29 February 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-122, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk39y39t | 1 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-141, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk40y40t | 3 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-122, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk42y42t | 3 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk41y41t | 8 March 2016 | 978 | ST978 | O83:H27 | 2 | aac(6)-Ib, ampH, ampC2, blaKPC-3 | vat, pic, gad | IncX3_1__JN247852 |
| sk43y43t | 17 March 2016 | 978 | ST978 | O83:H27 | 2 | aac(6)-Ib, ampH, ampC2, blaKPC-3 | vat, pic, gad | IncX3_1__JN247852 |
| sk44y44t | 23 March 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-122, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk45y45t | 5 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk46y46t | 18 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-156, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk47y47t | 18 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-54, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk48y48t | 21 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-192, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk49y49t | 24 April 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, blaTEM-122, blaKPC-2, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk50y50t | 10 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-122, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk51y51t | 13 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk52y52t | 22 May 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-150, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk53y53t | 11 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-168, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk54y54t | 25 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, blaTEM-122, blaKPC-2, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk55y55t | 29 June 2016 | 131 | ST131 cluster 2 | O25b:H4 | 30 | ampH, blaTEM-79, blaKPC-3, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | sat, iss, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk56y56t | 30 June 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781, Col(BS512)_1_NC_010656 |
| sk57y57t | 13 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-168, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk58y58t | 13 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-141, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk59y59t | 14 July 2016 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-79, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk60y60t | 30 September 2016 | 131 | ST131 cluster 2 | O25b:H4 | 30 | ampH, blaTEM-79, blaKPC-3, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | sat, iss, gad | IncFIB(pQil)_1_pQil_JN233705 |
| sk185y185t | 14 November 2016 | 1193 | ST1193 | O75:H5 | 64 | qnrB1, blaTEM-198, blaKPC-3, ampC2, aac(3)-IId, dfrA14 | iha, sat, vat | IncFIB(pQil)_1_pQil_JN233705, Col(BS512)_1_NC_010656 |
| sk136y136t | 23 January 2017 | 131 | ST131 cluster 1 | O25b:H4 | 30 | strA, ampH, blaTEM-168, blaKPC-2, sulII, blaOXA-9, blaCTX-M-27, ampC2, aadA5, mphA, tetR, dfrA17, tetA, sulI, strB, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk137y137t | 5 June 2017 | 131 | ST131 cluster 1 | O25b:H4 | 30 | ampH, blaTEM-150, blaKPC-2, blaOXA-9, blaCTX-M-27, ampC2, parC1aAB, gyrA1AB | iha, sat, iss, senB, gad | IncFIB(pQil)_1_pQil_JN233705, Col156_1_NC_009781 |
| sk138y138t | 22 January 2016 | 35 | oqxBgb, blaKPC-2, blaTEM-79, blaOXA-9, oqxA, tetD, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk139y139t | 16 February 2016 | 3033 | strA, strB, oqxBgb, catB4, blaKPC-2, blaTEM-198, blaOXA-1, blaOXA-9, blaCTX-M-15, oqxA, dfrA14, sulII, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk140y140t | 25 February 2016 | 17 | oqxBgb, blaKPC-2, blaTEM-122, blaOXA-9, oqxA, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk141y141t | 25 February 2016 | 35 | oqxBgb, blaKPC-2, blaTEM-122, blaOXA-9, oqxA, tetD, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk142y142t | 22 March 2016 | 2279 | strA, strB, oqxBgb, blaKPC-2, blaTEM-198, blaOXA-9, aac(3)-Iia, blaCTX-M-15, oqxA, dfrA14, sulII, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 | ||||
| sk143y143t | 31 March 2016 | 17 | oqxBgb, blaKPC-2, blaTEM-122, blaOXA-9, oqxA, ampH | IncFIB(pQil)_1_pQil_JN233705, IncFIB(K)_1_Kpn3_JN233704 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piazza, A.; Principe, L.; Comandatore, F.; Perini, M.; Meroni, E.; Mattioni Marchetti, V.; Migliavacca, R.; Luzzaro, F. Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia coli in Italy. Antibiotics 2021, 10, 718. https://doi.org/10.3390/antibiotics10060718
Piazza A, Principe L, Comandatore F, Perini M, Meroni E, Mattioni Marchetti V, Migliavacca R, Luzzaro F. Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia coli in Italy. Antibiotics. 2021; 10(6):718. https://doi.org/10.3390/antibiotics10060718
Chicago/Turabian StylePiazza, Aurora, Luigi Principe, Francesco Comandatore, Matteo Perini, Elisa Meroni, Vittoria Mattioni Marchetti, Roberta Migliavacca, and Francesco Luzzaro. 2021. "Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia coli in Italy" Antibiotics 10, no. 6: 718. https://doi.org/10.3390/antibiotics10060718
APA StylePiazza, A., Principe, L., Comandatore, F., Perini, M., Meroni, E., Mattioni Marchetti, V., Migliavacca, R., & Luzzaro, F. (2021). Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia coli in Italy. Antibiotics, 10(6), 718. https://doi.org/10.3390/antibiotics10060718







