In Vitro Selection of High-Level Beta-Lactam Resistance in Methicillin-Susceptible Staphylococcus aureus
Abstract
1. Introduction
2. Results
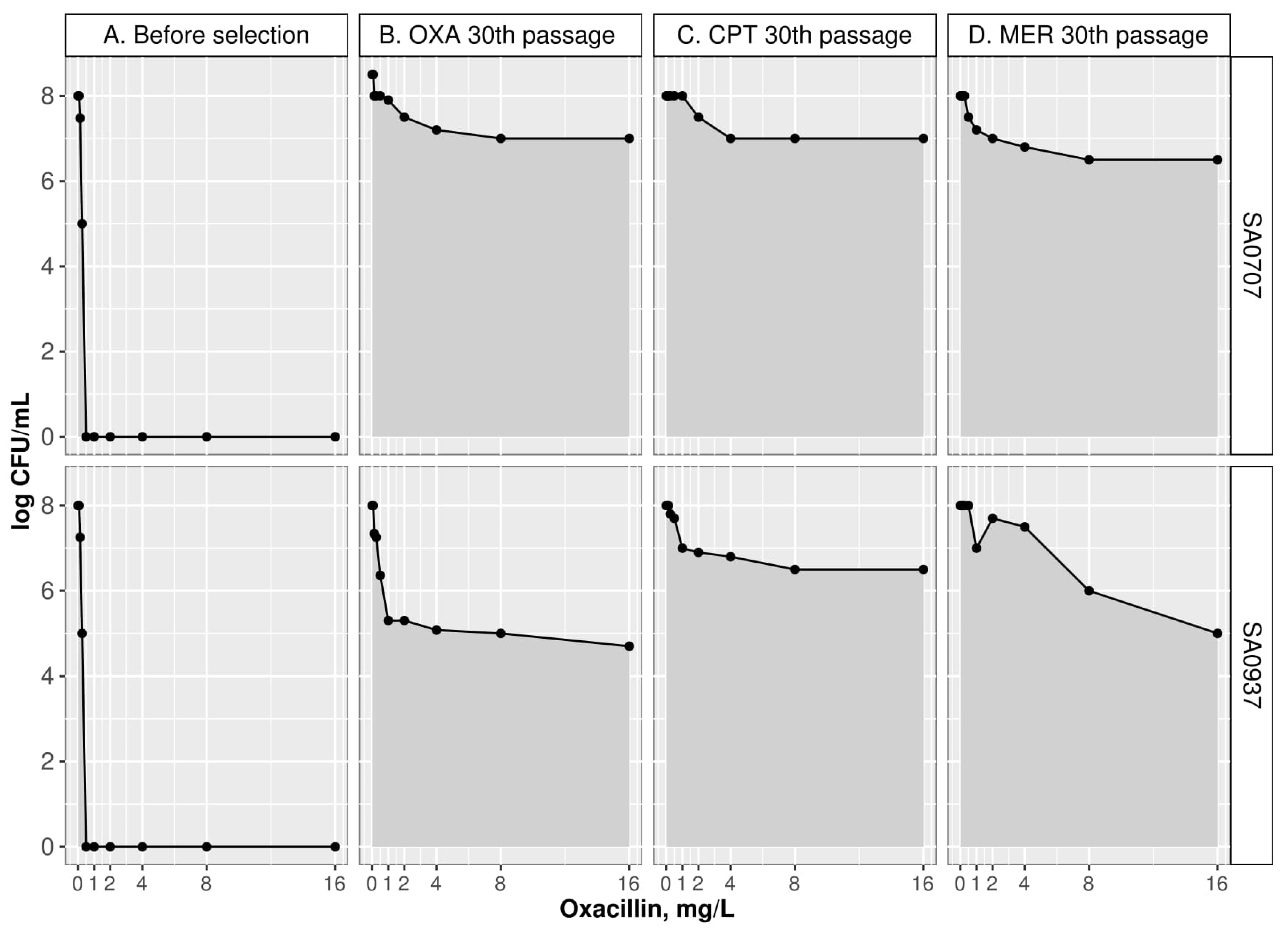
2.1. Phenotypic Changes during Selection with Beta-Lactams
2.2. Genetic Changes during Selection
2.2.1. Genetic Changes during Selection with Oxacillin
2.2.2. Genetic Changes during Selection with Ceftaroline
2.2.3. Genetic Changes during Selection with Meropenem
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Susceptibility Testing
4.2. Multistep Resistance Selection
4.3. PAP
4.4. Measurement of Growth Rates
4.5. Measurement of Induced Aautolytic Activity
4.6. Whole Genome Sequencing
4.7. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Senobar Tahaei, S.A.; Stájer, A.; Barrak, I.; Ostorházi, E.; Szabó, D.; Gajdács, M.G. Correlation Between Biofilm-Formation and the Antibiotic Resistant Phenotype in Staphylococcus aureus Isolates: A Laboratory-Based Study in Hungary and a Review of the Literature. Infect. Drug Resist. 2021, 14, 1155–1168. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178–182. [Google Scholar] [CrossRef]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Hiramatsu, K.; Tomasz, A.; de Lencastre, H.; Perreten, V.; Holden, M.T.G.; Coleman, D.C.; Goering, R.; Giffard, P.M.; Skov, R.L.; et al. Guidelines for Reporting Novel mecA Gene Homologues. Antimicrob. Agents Chemother. 2012, 56, 4997–4999. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and Zone Diameters. Version 11.0. 2021. Available online: http://www.eucast.org (accessed on 31 March 2021).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Annapolis Junction, MD, USA, 2021. [Google Scholar]
- Hryniewicz, M.M.; Garbacz, K. Borderline oxacillin-resistant Staphylococcus aureus (BORSA)—A more common problem than expected? J. Med. Microbiol. 2017, 66, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Argudin, M.A.; Roisin, S.; Nienhaus, L.; Dodemont, M.; de Mendonca, R.; Nonhoff, C.; Deplano, A.; Denis, O. Genetic Diversity among Staphylococcus aureus Isolates Showing Oxacillin and/or Cefoxitin Resistance Not Linked to the Presence of mec Genes. Antimicrob. Agents Chemother. 2018, 62, e00091-18. [Google Scholar] [CrossRef]
- Nomura, R.; Nakaminami, H.; Takasao, K.; Muramatsu, S.; Kato, Y.; Wajima, T.; Noguchi, N. A class A beta-lactamase produced by borderline oxacillin-resistant Staphylococcus aureus hydrolyses oxacillin. J. Glob. Antimicrob. Resist. 2020, 22, 244–247. [Google Scholar] [CrossRef]
- Giulieri, S.G.; Guerillot, R.; Kwong, J.C.; Monk, I.R.; Hayes, A.S.; Daniel, D.; Baines, S.; Sherry, N.L.; Holmes, N.E.; Ward, P.; et al. Comprehensive Genomic Investigation of Adaptive Mutations Driving the Low-Level Oxacillin Resistance Phenotype in Staphylococcus aureus. mBio 2020, 11. [Google Scholar] [CrossRef]
- Varela, M.C.; Roch, M.; Taglialegna, A.; Long, S.W.; Saavedra, M.O.; Rose, W.E.; Davis, J.J.; Hoffman, L.R.; Hernandez, R.E.; Rosato, R.R.; et al. Carbapenems drive the collateral resistance to ceftaroline in cystic fibrosis patients with MRSA. Commun. Biol. 2020, 3, 599. [Google Scholar] [CrossRef]
- Balslev, U.; Bremmelgaard, A.; Svejgaard, E.; Havstreym, J.; Westh, H. An outbreak of borderline oxacillin-resistant Staphylococcus aureus (BORSA) in a dermatological unit. Microb. Drug Resist. 2005, 11, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Speck, S.; Wenke, C.; Fessler, A.T.; Kacza, J.; Geber, F.; Scholtzek, A.D.; Hanke, D.; Eichhorn, I.; Schwarz, S.; Rosolowski, M.; et al. Borderline resistance to oxacillin in Staphylococcus aureus after treatment with sub-lethal sodium hypochlorite concentrations. Heliyon 2020, 6, e04070. [Google Scholar] [CrossRef]
- Gostev, V.; Sopova, J.; Kalinogorskaya, O.; Tsvetkova, I.; Lobzin, Y.; Klotchenko, S.; Sidorenko, S. In Vitro Ceftaroline Resistance Selection of Methicillin-Resistant Staphylococcus aureus Involves Different Genetic Pathways. Microb. Drug Resist. 2019, 25, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- da Costa, T.M.; de Oliveira, C.R.; Chambers, H.F.; Chatterjee, S.S. PBP4: A New Perspective on Staphylococcus aureus beta-Lactam Resistance. Microorganisms 2018, 6, 57. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Cai, X.; Ma, H.; Zhu, L.; Zhang, Y.; Chou, S.H.; Galperin, M.Y.; He, J. A decade of research on the second messenger c-di-AMP. FEMS Microb. Rev. 2020, 44, 701–724. [Google Scholar] [CrossRef]
- Chatterjee, A.; Poon, R.; Chatterjee, S.S. Stp1 Loss of Function Promotes beta-Lactam Resistance in Staphylococcus aureus That Is Independent of Classical Genes. Antimicrob. Agents Cchemother. 2020, 64, e02222-19. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Karinou, E.; Schuster, C.F.; Pazos, M.; Vollmer, W.; Grundling, A. Inactivation of the Monofunctional Peptidoglycan Glycosyltransferase SgtB Allows Staphylococcus aureus To Survive in the Absence of Lipoteichoic Acid. J. Bacteriol. 2019, 201, e00574-18. [Google Scholar] [CrossRef]
- Cafiso, V.; Bertuccio, T.; Purrello, S.; Campanile, F.; Mammina, C.; Sartor, A.; Raglio, A.; Stefani, S. dltA overexpression: A strain-independent keystone of daptomycin resistance in methicillin-resistant Staphylococcus aureus. Int. J. Antimicrob. Agents 2014, 43, 26–31. [Google Scholar] [CrossRef]
- Blake, K.L.; O’Neill, A.J.; Mengin-Lecreulx, D.; Henderson, P.J.; Bostock, J.M.; Dunsmore, C.J.; Simmons, K.J.; Fishwick, C.W.; Leeds, J.A.; Chopra, I. The nature of Staphylococcus aureus MurA and MurZ and approaches for detection of peptidoglycan biosynthesis inhibitors. Mol. Microbiol. 2009, 72, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.T.; Munita, J.M.; Arias, C.A. Mechanisms of drug resistance: Daptomycin resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 32–53. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Bening, S.C.; Manson, A.L.; Stokes, J.M.; Kohanski, M.A.; Badran, A.H.; Earl, A.M.; Cheney, N.J.; Yang, J.H.; Collins, J.J. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 2021, 371, eaba0862. [Google Scholar] [CrossRef]
- Barrick, J.E.; Yu, D.S.; Yoon, S.H.; Jeong, H.; Oh, T.K.; Schneider, D.; Lenski, R.E.; Kim, J.F. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 2009, 461, 1243–1247. [Google Scholar] [CrossRef]
- Skinner, S.; Murray, M.; Walus, T.; Karlowsky, J.A. Failure of Cloxacillin in Treatment of a Patient with Borderline Oxacillin-Resistant Staphylococcus aureus Endocarditis. J. Clin. Microbiol. 2009, 47, 859–861. [Google Scholar] [CrossRef] [PubMed]
- Moreillon, P. New and emerging treatment of Staphylococcus aureus infections in the hospital setting. Clin. Microbiol. Infect. 2008, 14 (Suppl. S3), 32–41. [Google Scholar] [CrossRef][Green Version]
- Gajdács, M. The Continuing Threat of Methicillin-Resistant Staphylococcus aureus. Antibiotics 2019, 8, 52. [Google Scholar] [CrossRef]
- Pfeltz, R.F.; Schmidt, J.L.; Wilkinson, B.J. A microdilution plating method for population analysis of antibiotic-resistant staphylococci. Microb. Drug Resist. 2001, 7, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef]
- Gustafson, J.E.; Berger-Bachi, B.; Strassle, A.; Wilkinson, B.J. Autolysis of methicillin-resistant and -susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 1992, 36, 566–572. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Barrick, J.E.; Colburn, G.; Deatherage, D.E.; Traverse, C.C.; Strand, M.D.; Borges, J.J.; Knoester, D.B.; Reba, A.; Meyer, A.G. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genom. 2014, 15, 1039. [Google Scholar] [CrossRef]
| Isolate | Ab Selection | Passage | OXA | CPT | MER | FOX | AMC | PEN | FOX DDM, mm | PAP (AUCderivative)/(AUCWT) |
|---|---|---|---|---|---|---|---|---|---|---|
| ECOFF * | ≤2 | ≤0.5 | ≤0.5 | ≤4 | - | ≤0.125 | 22≤ | - | ||
| SA0937 | WT | 0 | 0.125 | 0.25 | 0.06 | 4 | 0.25 | 0.06 | 30 | - |
| OXA | 5 | 16 | 4 | 0.25 | 8 | 16 | 8 | 32 | 2.24 | |
| OXA | 15 | ND ** | ND | ND | ND | ND | ND | 27 | 2.43 | |
| OXA | 30 | ND | ND | ND | ND | ND | ND | 22 | 2.31 | |
| Ab free | 40 | 2 | 1 | 1 | 8 | 1 | 0.25 | 23 | 1.93 | |
| CPT | 5 | 32 | 16 | 0.5 | 2 | 32 | 32 | 27 | 2.4 | |
| CPT | 15 | 32 | 16 | 1 | 4 | 16 | 32 | 28 | 2.62 | |
| CPT | 30 | 64 | 128 | 2 | 8 | 32 | 64 | 24 | 2.69 | |
| Ab free | 40 | 2 | 4 | 0.5 | 8 | 4 | 2 | 28 | 1.48 | |
| MER | 5 | 4 | 1 | 1 | 8 | 2 | 1 | 28 | 2.05 | |
| MER | 15 | 8 | 4 | 2 | 8 | 2 | 4 | 24 | 2.55 | |
| MER | 30 | 8 | 4 | 8 | 16 | 2 | 4 | 0 | 2.75 | |
| Ab free | 40 | 8 | 2 | 8 | 16 | 2 | 4 | 14 | 2.49 | |
| SA0707 | WT | 0 | 0.5 | 0.25 | 0.06 | 4 | 2 | 32 | 28 | - |
| OXA | 5 | 4 | 0.5 | 0.25 | 2 | 2 | 16 | 27 | 1.55 | |
| OXA | 15 | 32 | 2 | 1 | 2 | 32 | 64 | 29 | 2.73 | |
| OXA | 30 | 32 | 16 | 2 | 8 | 32 | 64 | 25 | 2.85 | |
| Ab free | 40 | 32 | 32 | 2 | 8 | 16 | 32 | 24 | 2.68 | |
| CPT | 5 | 1 | 1 | 0.5 | 4 | 4 | 128 | 22 | 1.08 | |
| CPT | 15 | 32 | 32 | 0.5 | 8 | 32 | 32 | 24 | 2.3 | |
| CPT | 30 | 128 | 64 | 2 | 16 | 64 | 128 | 22 | 2.82 | |
| Ab free | 40 | 128 | 64 | 2 | 16 | 64 | 128 | 22 | 2.73 | |
| MER | 5 | 2 | 1 | 1 | 8 | 4 | 16 | 28 | 1.9 | |
| MER | 15 | 8 | 2 | 4 | 4 | 4 | 16 | 14 | 2.37 | |
| MER | 30 | 8 | 2 | 4 | 8 | 4 | 4 | 0 | 2.71 | |
| Ab free | 40 | 8 | 2 | 4 | 8 | 4 | 4 | 14 | 2.52 |
| Figure | Strain | Wild Type | Antibiotics Used for Selection | ||
|---|---|---|---|---|---|
| OXA | CPT | MER | |||
| Growth Kinetics | |||||
| Dt, min | SA0937 | 27.25 (26.86–27.47) | 41.30 (39.0–42.6) | 33.53 (33.07–34.0) | 36.81 (34.2–39.5) |
| SA0707 | 25.42 (25.08–26.25) | 52.32 (51.01–53.68) | 38.31 (37.56–39.14) | 28.89 (28.02–31.23) | |
| r, min−1 | SA0937 | 0.025 (0.025–0.026) | 0.016 (0.016–0.017) | 0.02 (0.02–0.02) | 0.018 (0.017–0.02) |
| SA0707 | 0.027 (0.026–0.027) | 0.013 (0.012–0.014) | 0.018 (0.017–0.019) | 0.02 (0.019–0.021) | |
| Lag, min (Range) | SA0937 | 69–79 | 158–168 | 128–138 | 128–138 |
| SA0707 | 89–99 | 227–237 | 168–178 | 168–178 | |
| Induced Autolysis | |||||
| OD600/lysed cells, % | SA0937 | 55.8 (50.5–62.0) | 68.15 (62.4–75.3) | 35.0 (29.5–39.7) | 58.6 (53.6–64.1) |
| SA0707 | 14.0 (7.0–16.4) | 8.0 (2.0–13.0) | 14.4 (10.2–18.6) | 16.3 (10.0–19.4) | |
| SA0937 (blaZ-), Selection on: | SA0707 (blaZ+), Selection on: | ||||||
|---|---|---|---|---|---|---|---|
| Proteins | OXA | CPT | MER | OXA | CPT | MER | |
| CW | Pbp1 | - | H499R | G408V, W351L, W351R | - | - | A482V, P431L, H375D, W351K, L18F |
| Pbp2 | - | - | I19N, G587S, M559I, T552I, A416E, G142S | A450D | T552I, A416T | - | |
| Pbp3 | - | - | G286V, S634F | - | - | - | |
| Pbp4 | P ∆91 bp | P ∆1 bp, T201A, N138I | F241L, N141T | P ∆91 bp | P ∆91 bp | - | |
| GdpP | ∆1 bp | H621Y, R540Stop | E108Stop | Y475C | - | R289C | |
| VraS | T274K | ∆1 bp | C60Y | - | - | - | |
| VraT | P174Q | - | - | W119R | - | G226V | |
| MurAB | - | - | - | P ∆171 bp | P ∆171 bp | - | |
| GraR | - | - | - | G59E | ∆161 bp | - | |
| WTA | TagA | - | G171E | - | - | G171E | - |
| TagH | - | - | - | - | - | Q65Stop | |
| TagO | - | - | - | L323Stop | L41I | - | |
| DltA | - | - | - | - | A184V | ∆1 bp | |
| DltD | ∆236 bp | - | - | - | - | - | |
| SgtB | - | - | - | - | ∆1 bp | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gostev, V.; Kalinogorskaya, O.; Ivanova, K.; Kalisnikova, E.; Lazareva, I.; Starkova, P.; Sidorenko, S. In Vitro Selection of High-Level Beta-Lactam Resistance in Methicillin-Susceptible Staphylococcus aureus. Antibiotics 2021, 10, 637. https://doi.org/10.3390/antibiotics10060637
Gostev V, Kalinogorskaya O, Ivanova K, Kalisnikova E, Lazareva I, Starkova P, Sidorenko S. In Vitro Selection of High-Level Beta-Lactam Resistance in Methicillin-Susceptible Staphylococcus aureus. Antibiotics. 2021; 10(6):637. https://doi.org/10.3390/antibiotics10060637
Chicago/Turabian StyleGostev, Vladimir, Olga Kalinogorskaya, Ksenia Ivanova, Ekaterina Kalisnikova, Irina Lazareva, Polina Starkova, and Sergey Sidorenko. 2021. "In Vitro Selection of High-Level Beta-Lactam Resistance in Methicillin-Susceptible Staphylococcus aureus" Antibiotics 10, no. 6: 637. https://doi.org/10.3390/antibiotics10060637
APA StyleGostev, V., Kalinogorskaya, O., Ivanova, K., Kalisnikova, E., Lazareva, I., Starkova, P., & Sidorenko, S. (2021). In Vitro Selection of High-Level Beta-Lactam Resistance in Methicillin-Susceptible Staphylococcus aureus. Antibiotics, 10(6), 637. https://doi.org/10.3390/antibiotics10060637







