Abstract
Bacterial infections have been treated effectively by antibiotics since the discovery of penicillin in 1928. A worldwide increase in the use of antibiotics led to the emergence of antibiotic resistant strains in almost all bacterial pathogens, which complicates the treatment of infectious diseases. Antibiotic-resistant bacteria play an important role in increasing the risk associated with the usage of surface waters (e.g., irrigation, recreation) and the spread of the resistance genes. Many studies show that important pathogenic antibiotic-resistant bacteria can enter the environment by the discharge of sewage treatment plants and combined sewage overflow events. Mussels have successfully been used as bio-indicators of heavy metals, chemicals and parasites; they may also be efficient bio-indicators for viruses and bacteria. In this study an influence of the discharge of a sewage treatment plant could be shown in regard to the presence of E. coli in higher concentrations in the mussels downstream the treatment plant. Antibiotic-resistant bacteria, resistant against one or two classes of antibiotics and relevance for human health could be detected in the mussels at different sampling sites of the river Rhine. No multidrug-resistant bacteria could be isolated from the mussels, although they were found in samples of the surrounding water body.
1. Introduction
1.1. Antibiotic-Resistant Bacteria in Water Systems
Numerous bacterial species occur naturally in surface water bodies, but pathogenic antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) can also be found [1,2,3,4]. Faecal pathogenic bacteria, including ARB and ARGs, enter surface water bodies through various pathways, i.e., point sources such as effluents of sewage treatment plants (STPs) and combined sewer overflows [5,6,7], as well as diffuse surface or subsurface run-off from non-sealed landscapes [8]. Wastewater from health care facilities, such as hospitals or nursing homes, and the agro-food industry are further known sources of human-pathogenic and/or (multi-)resistant bacteria and various types of resistances [9,10]. Antibiotics excreted into wastewater (WW) are an additional reason for the spread of antibiotic resistance [11].
Acquired resistance genes are often located on plasmids and can be passed onto other bacteria via horizontal gene transfer. In this process, WW and STPs are of particular importance due to the high density of bacteria [12]. The WHO classified ARB as a serious threat to modern medicine [13] and categorized specific organisms based on their clinical significance and resistance capabilities into three priority classes. Carbapenem-resistant Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales were thereby grouped as the first priority (“critical”) whereas vancomycin-resistant Enterococcus faecium (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) are in the second priority group (“high”) [14].
1.2. Mussel Species in the River Rhine
The river Rhine is a large, well-monitored, cross-border-river that receives substantial amounts of treated wastewater. It originates in the Swiss canton of Grisons and flows through Germany and the Netherlands into the North Sea. Besides a large and diverse flora, the river Rhine also contains a variety of fish and small animal species such as insects, snails and mussels. Endemic mussels include the duck mussel (Anodonta anatina), the painter’s mussel (Unio pictorum) and the common river mussel (Unio crassus) [15]. The river Rhine is also populated by invasive mussel species, such as Corbicula spp. [16] and Dreissena spp. [17]. Both species are so numerous that they account for the majority of current shellfish in the river Rhine.
Corbicula spp. occur in large populations and are a popular food source [18]. They were first detected in European surface waters in 1980 [19] and spread throughout connected water and river systems. Mussels of the genus Corbicula occurring in the river Rhine are C. fluminea and C. fluminalis.
Of the genus Dreissena, the species D. polymorpha and D. rostriformis bugensis are of interest in the river Rhine. They vary widely in shell shape, colour and stripe pattern on the shell [20]. Dreissena spp. have byssus threads, which they use to attach themselves to stones, wood, other large mussels, metal or other solid surfaces [21,22]. They are sessile, but can dissolve the byssus threads by enzymes. Both mentioned species occur in rivers, lakes and harbours, but are most common in canals [22]. D. polymorpha was discovered for the first time in the Upper Rhine attached to a raft in the Mannheim harbour in 1840 [17]. The species D. rostriformis bugensis was first detected much later in an estuary of the river Rhine in the Netherlands (Hollands Diep) in 2006 [23]. From there, it migrated upstream and was found in 2008 in the Lower Rhine region between Dormagen and Bimmen [24].
Mussels use suspension filtration to acquire nutrients. Water is taken up into the interior of the shell through the siphon and phytoplankton, microorganisms and organic material are filtered out [25,26,27]. Mussels are established bio-indicators for heavy metals, chemicals and parasites but are also considered for viruses and bacteria [28,29]. They can accumulate large quantities of microorganisms from their surrounding waters, including opportunistic bacteria (Aeromonas, Vibrio, Pseudomonas), protozoan parasites (Cryptosporidium, Giardia), viruses (adenoviruses, hepatoviruses) and pathogenic bacteria (Escherichia coli, Salmonella) [30]. They may therefore jeopardize human health, particularly when consumed as seafood [31]. Both mussel genera investigated in this study are edible [18], albeit the ingestion of D. polymorpha is not recommended due to the proven transmission of the parasites Toxoplasma gondii, Giardia spp. and Cryptosporidium spp. [32,33].
Mussels living in surface waters that contain pathogenic bacteria and ARB may represent a reservoir of those microorganisms, and thus can be used as indicators for their presence in a water environment.
2. Results
In total, 22 mussels were obtained from three different sampling sites (Table 1). Nine mussels belonging to Corbicula spp. were harvested upstream the municipal STP of Bonn and eight mussels belonging to the same species were selected downstream of the municipal STP. Five mussels of Dreissena spp. could be harvested from the sampling location in the river in Cologne. The Corbicula spp. sampled upstream of the municipal STP had an average tissue weight of 1.4 g, whereas mussels found downstream the municipal STP showed an average weight of 0.4 g. The Dreissena spp. soft mussel tissue weighted 0.2 g each. Water temperatures over the study period ranged between 22.0 °C and 28.3 °C.

Table 1.
Individual mussel soft tissue weight measured without shells and sampled at the three different sampling sites.
2.1. Bacterial Flora in the Mussel Tissue
The abundance of potentially clinically relevant pathogens in the mussels was investigated by using different agar plates. The total bacterial counts (TBCs) (Columbia blood agar), Gram-negative bacteria (MacConkey agar) and coliform bacteria (chromocult coliform (CC) agar), as well as E. coli (CC agar) were determined to evaluate the bacterial flora of the two mussel species and a potential influence of WW and the STP. The bacterial abundance was calculated in colony forming units per 10 g of mussel soft tissue to enable comparison of the samples (Figure 1).
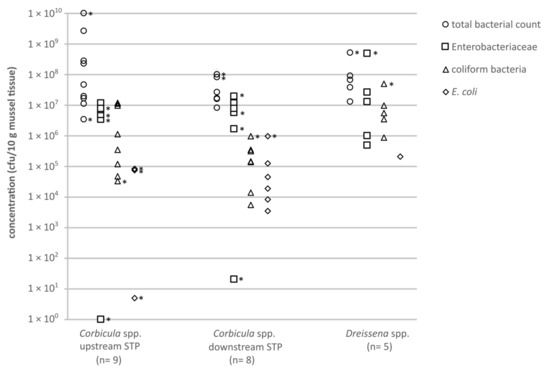
Figure 1.
Concentrations (cfu/10 g mussel soft tissue) of clinically relevant bacteria from mussels, cultivated on different agars: circles = total bacteria count (TBC; Columbia blood agar); squares = Enterobacteriaceae (MacConkey agar); triangles = coliform bacteria (CC agar); diamonds = E. coli (CC agar). Asterisks mark category values of ≥x cfu/10 g mussel tissue, the smallest possible value is shown.
The TBCs of bacteria in the mussel soft tissue of Corbicula spp. were diverse. Mussels sampled upstream of the municipal STP in Bonn showed results between 3.4 × 106 and 1.0 × 1010 cfu/10 g (Figure 1). In mussels sampled downstream of the municipal STP, the TBCs ranged between 8.0 × 106 and 1.0 × 108 cfu/10 g. The TBCs on Columbia blood agar detected for the genus Corbicula were slightly higher upstream compared to downstream of the municipal STP.
The concentrations of Gram-negative bacteria, in the mussels Corbicula spp. ranged from 1 up to >2.0 × 107 cfu/10 g upstream the STP and 20 up to >2.0 × 107 cfu/10 g downstream the STP. The bacterial concentrations were below the individual limit of detection (LODi) for two mussels samples (<43 and <48 cfu/10 g). Varying detection limits resulted from the different weights of the sampled mussel soft tissue (Table 1).
The concentrations of coliform bacteria in Corbicula spp. ranged between 3.4 × 104 and 1.2 × 107 cfu/10 g upstream the STP. Downstream the STP, concentrations of 5.5 × 103 up to >1.0 × 106 cfu/10 g were detected. In five out of nine samples upstream the STP, the number of E. coli in Corbicula spp. soft tissue was below the LODi. The maximum concentration was >8.3 × 104 cfu/ 10 g. The concentration of E. coli in the mussels Corbicula spp. found downstream the local STP was below the LODi, with a maximum of >1 × 106 cfu/10 g. Results <LODi are not shown in the figure.
For mussel species of the genus Dreissena (D1–D5) the concentration of bacteria grown on Columbia blood agar ranged between 1.3 × 107 and >5.0 × 108 cfu/10 g. The concentrations for the Gram-negative bacteria and coliform bacteria varied between 5.0 × 105 and >5.0 × 106 cfu/10 g and 9.0 × 105 and >5.0 × 107 cfu/10 g, respectively. In four out of five samples, E. coli could not be detected.
2.2. Distribution of Antibiotic-Resistant Bacteria
Methicillin-resistant S. aureus, vancomycin-resistant E. faecium and E. faecalis, extended spectrum beta-lactamase (ESBL)-producing Klebsiella spp., Enterobacter spp. and Citrobacter spp. (grouped as KEC) and ESBL-producing Proteus spp. could not be detected in neither the sampled Corbicula spp. nor the Dreissena spp. mussels. The numbers of colonies of these bacterial species were below the individual detection limits, which varied between <36 and <1000 cfu/10 g depending on the weight of the mussel tissue. Only one isolate of an ESBL-producing E. coli was obtained from a mussel of the Corbicula spp. upstream the STP. Pseudomonas spp., resistant against third-generation cephalosporins (3GCR), were detected frequently in all mussel species and sampling sites; none of these were P. aeruginosa 3GCR (Figure 2). All resistant Acinetobacter spp. detected belonged to the A. calcoaceticus-baumannii complex. Results <LODi are not shown in the figure. Each isolate was confirmed by matrix-assisted laser desorption/ionisation time of flight mass spectrometry (MALDI-TOF-MS) and tested for its susceptibility against the antibiotic substances shown in Table 2.
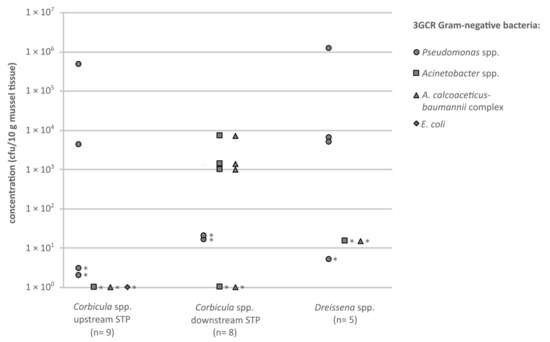
Figure 2.
Concentrations (cfu/10 g mussel soft tissue) of antibiotic-resistant (3GCR) Gram-negative bacteria, cultivated on CHROMagar ESBL (circles = Pseudomonas spp.; squares = Acinetobacter spp.; triangles = A. calcoaceticus-baumannii complex; diamonds = E. coli), originating from the tissue of the investigated mussels. Asterisks mark category values of ≥x cfu/10 g mussel tissue, the smallest possible value is shown.

Table 2.
Antibiotic-resistant isolates of the bacteria (isolated on the CHROMagar ESBL plates) from different sampling sites and matrices, detection of carbapenemase genes within the isolates and classification of German (3/4MRGN = multi-resistant Gram-negatives with resistance against three or four antibiotic groups tested) and international (XDR = extensively drug-resistant; showing resistance against all but one or two reserve antibiotic classes) multidrug resistance levels.
MRSA, VRE and ESBL-producing Proteus spp. could not be detected in any water sample consisting of 100 mL river water from the mussel sampling sites. ESBL-producing E. coli and KEC were detected in low concentrations from 1 up to 6 cfu/100 mL and ≥2 cfu/100 mL in one sample, respectively (Figure 3). These two KEC isolates were identified by MALDI-TOF-MS as K. pneumoniae. Both isolates were detected in the sampled water downstream of the local STP. Pseudomonas spp. 3GCR, could be detected in all samples at different concentrations (≥4–30 cfu/100 mL); but only one isolate of P. aeruginosa 3GCR could be obtained from a water sample downstream of the municipal STP. Acinetobacter spp. 3GCR were present in all water samples ranging from 5 to >100 cfu/100 mL), all belonging to the A. calcoaceticus-baumannii complex.
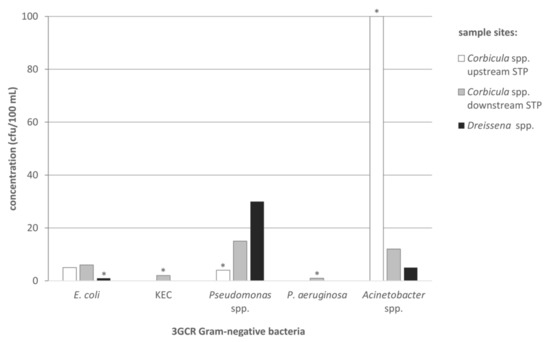
Figure 3.
Concentrations (cfu/100 mL) of antibiotic-resistant Gram-negative bacteria (E. coli, KEC, Pseudomonas spp., P. aeruginosa, Acinetobacter spp.), cultivated on ESBL CHROMagar, originated from the water of the investigated sampling sites (white bars = Corbicula spp. upstream of the STP; grey bars = Corbicula spp. downstream of the STP; black bars = Dreissena spp.) during the time of mussel harvesting. Asterisks mark category values of ≥x cfu/100 mL river water, the smallest possible value is shown.
The isolates were also investigated for their individual resistance profiles. All bacterial isolates from CHROMagar ESBL found in mussel soft tissue and water showed resistance to the broad-spectrum cephalosporin cefotaxime and 28.6% additionally against ceftazidime (Table 2). Also, 14.3% of the isolates showed phenotypic resistance against ciprofloxacin.
Isolates with a multidrug resistance status originated from river water. One isolate belonging to the A. calcoaceticus-baumannii complex and one E. coli isolate had a 3MRGN status and one P. aeruginosa (3GCR) isolate was classified as 4MRGN. Further investigation showed that this P. aeruginosa strain is a member of ST316 and harboured the carbapenemase gene blaVIM. Most ARB detected in the mussels belong to the A. calcoaceticus-baumannii complex and one isolate could be identified as ESBL-producing E. coli. The latter was a single isolate gained from a Corbicula spp. mussel sampled upstream the municipal STP (Cu). This isolate showed phenotypic resistance against the cephalosporins cefotaxime and ceftazidime as well as against the fluoroquinolone ciprofloxacin. However, it was still susceptible to the ureidopenicillin piperacillin in combination with tazobactam, as well as the carbapenems imipenem and meropenem. No extensively drug resistant (XRD) strains could be isolated from the mussel tissue or the river water.
3. Discussion
Ensuring that the examined mussels were vital, the environmental factor temperature was evaluated. According to a previous study, mussels of the species Corbicula spp. and Dreissena spp. can tolerate water temperatures of 2 to 36 °C [34]. The average water temperature of the river Rhine during the sampling period varied from 22.0 to 28.3 °C. These temperatures are in the normal range of the living conditions for both genera of mussels.
3.1. Influence of Treated Wastewater on the Bacterial Flora in River Mussels
The concentrations of potential clinically relevant bacteria were determined in the mussel soft tissue of each mussel species at each sampling site to analyse the influence of various sampling parameters. Cultivated isolates on Colombia blood agar, Gram-negative bacteria cultivated on MacConkey agar, as well as coliform bacteria and E. coli cultivated on CC agar were included in the investigation. This study presents an orientational investigation on the occurrence of antibiotic resistant bacteria in mussels. Although it is based on a limited number of samples, some valuable insights can be gained.
Mussels are known to be able to accumulate heavy metals in their tissue and shell, showing a high tolerance to the effects of these metals [35]. As suspension filter feeders, they have high filtration rates [25,26,27]. Bighiu et al. (2019) [36] demonstrated in lab scale experiments that mussels also accumulate bacteria. The concentration of the indicator bacteria E. coli and enterococci were 132 times higher compared to the respective water sample, when sewage effluent had been used to feed the mussels (mussel tissue and shell were analysed). They also showed that bacteria can persist up to 48 h in the mussels before digestion and that peak exposures with faecal bacteria by water contamination can be monitored by investigating mussels [36].
The bacterial flora within the mussel tissue of Corbicula spp. in this study was diverse and relatively constant across sampling sites, except for the more frequent detection of E. coli downstream of the municipal STP. The presence of the faecal indicator E. coli can be linked to the influence of WW at the sampling site. Even though the individual LODi for the mussels upstream the STP was lower due to their higher weight, E. coli could only be detected in four out of nine samples. Araujo et al. (1993) described a high variety in the sizes of adults of Corbicula spp. Small mussels seem to be more affected by the composition of their habitat, as they use amino acids as an energy source; larger mussels primarily draw on their larger storage reserves of carbohydrates [37]. The effect by chemical components in the water on the growth rate of Corbicula spp. could already be shown by Belanger et al. (1990) [38]. Whether the size variations of the mussels stemmed from a natural variation or were due to the water quality at the sampling sites could not be finally evaluated by this study setting. Nevertheless, all sampled mussels downstream the STP were smaller, pointing to an effect of water quality on mussel growth.
3.2. Antibiotic-Resistant Bacteria in the Mussel Tissue
Bighiu et al. (2019) [36] found multidrug resistant bacteria at 2–5 times higher concentrations in the mussels (shell and tissue) than in the water sample. The water samples analysed in this study show relatively low concentrations of ARB except for bacteria from the A. calcoaceticus-baumannii complex, which could be found in concentrations >100 cfu/100 mL river water (sample upstream the STP). The selected ARB were not detected in six of seventeen Corbicula spp. mussels examined, of which four out of nine were collected upstream of the STP and two out of eight collected downstream of the STP. Four out of five mussels of the genus Dreissena were tested positive for at least one of the investigated ARB.
In most of the mussel tissue samples (73%) of Corbicula spp. and Dreissena spp., ESBL-producing E. coli, bacteria belonging to the A. calcoaceticus-baumannii complex or Pseudomonas spp. resistant against third-generation cephalosporins were detected. These resistant bacteria seem to be relatively persistent within mussels. However, ESBL-producing E. coli in mussels was a single isolate gained from a sample of a Corbicula spp. upstream of the STP, and VRE were not detected within the mussel tissue. This is in accordance with the limited persistence of faecal bacteria in aquatic environments. In water samples, VRE were not detected whereas ESBL-producing E. coli could be extracted in low concentrations. The 4MRGN P. aeruginosa strain, isolated from the river water downstream the STP, harboured the carbapenemase gene blaVIM. As a member of ST316, it does not belong to known high risk strains and has been mainly reported in Asia [39,40,41]. In case of a positive result for ARB in the mussel tissue, the concentrations varied within 3 log10 cfu/10 g. None of the isolates belonged to the XDR group confirming results published by Müller et al. (2018) [42].
In addition to studies which used mussels as bio-indicators for faecal contamination, various studies investigated the ability of C. fluminea and D. polymorpha to remove E. coli from water [25,43]. It was demonstrated that D. polymorpha can digest and metabolise 100% of the bacteria and thus remove them irreversibly from the water body [43]. In C. fluminea, the removal rate was up to 98%, most likely because of the slightly lower filtration rate of 33 mL × g−1 × h−1 [44] compared to 5–400 mL × g−1 × h−1 in D. polymorpha [25]. These results suggest that both mussel species can filter and digest bacteria from water, so that overall, a smaller diversity of bacterial species was present as shown in the present study.
The detected ARB probably originated from the mucus of the mussels, clung to the surface of the mussel tissue or had not yet been digested. Although the water quality and ecology of the river Rhine have been improved over the last decades, e.g., through optimization and modernization of STPs supported by international programmes like “Rhine 2020—Program on the sustainable development of the Rhine” [45], the wastewater discharges into the river Rhine still emit substantial amounts of resistant ESBL-producing Gram-negative bacteria [9]. Our study demonstrates the persistence of resistant human pathogens in the river water and in mussels. Following the water course, such bacteria may spread unnoticed, posing risks to human health through various pathways. Multiple studies have shown that the environment, including water bodies like rivers, is affected by the spread of antibiotic resistance [46]. More multidrug resistant isolates were detected in water than in mussels. However, when present, the concentrations of detected ARB were higher in the mussels than in the corresponding water sample.
4. Materials and Methods
4.1. Sampling Sites
Corbicula spp. was sampled at two sampling sites located on the right bank of the river Rhine near the city of Bonn, Germany. The study was designed to include sampling sites with and without the influence of treated WW. The first site was upstream of the municipal STP and not influenced by its discharges, as there is no further STP for at least 7.5 km upstream of the sampling site (Corbicula spp. upstream STP). The second site was located on the same river bank but approx. 500 m downstream of the outlet of the STP (which is located on the left river side and connected to various hospitals and one maximum care hospital) (Corbicula spp. downstream STP). Thus, this site was directly influenced by treated WW. All sites used for harvesting of Corbicula spp. were reachable from the banks of the river Rhine and no more than 2 m away from the shore; the water level varied between approximately 0.5 and 1.0 m.
Mussels of the genus Dreissena were sampled at one sampling site located on the left bank of the river Rhine in Cologne, Germany. The mussels were collected from stones below the grid of the Ecological Rhine station of the University of Cologne. Mussels of the genus Dreissena were collected 1 to 4 m from the shore at a water depth of no more than 0.3 m. A STP is located 3.4 km upstream of this sampling site, which processes WW from urban areas with no connection to hospitals.
4.2. Sampling Procedure
The mussels were taken from their natural habitat, resulting in different sampling schemes. Corbicula spp. which were buried in the sand could be found at the water edge. They were harvested with a shovel or a sample beaker (tied to a telescopic pole) and dug out of the sand. Mussels of the genus Dreissena were picked from stones, detached Dreissena spp. were not examined. To loosen mussels of Dreissena spp. from the substrate, the byssus threads were severed with a knife or carefully detached by hand.
Only living and healthy mussels were analysed. Vitality was tested by gently trying to pull both halves of the shell apart. If this was not successful, it could be assumed that the mussels were in a vital condition. The mussels were separated according to type and location and transferred in sterile bottles that were filled with the water from the respective sampling location to keep the mussels alive until examination in the laboratory. In addition to the analysis of the mussels, water samples were taken for each sampling site and sampling campaign. For this, 100 mL of water were taken five times at 2-min intervals to generate mixed samples according to the German standard method for the examination of water, wastewater and sludge [47].
To further ensure that the examined mussels were vital and thus undergoing suspension filtration, the temperature was evaluated. All samples were cooled during the transport to the laboratory and kept at 2–8 °C until being processed within the next 24 h.
4.3. Preparation of Mussel Samples
All following steps were carried out under sterile conditions: The mussels were individually transferred into a petri dish each and measured in width and length. The byssus threads of Dreissena spp. were removed using a scalpel and discarded. To open the shells, the scalpel was inserted into the lateral inflow or outflow channels and passed along the shell until both halves of the shell could be pulled apart. The anterior and posterior shell adductors and retractors were detached with the scalpel and the mantle was removed from the hypostracum, using tweezers. The soft tissue was weighed, transferred to a sterile 50 mL sampling container that contained 10 mL of 0.9% (w/v) NaCl and crushed until a homogeneous suspension was obtained.
4.4. Determination of the Relevant Bacterial Flora in Mussels
Enabling the inclusion of different bacterial species, the mussel tissue samples were cultivated using different nutrient media. The TBC was performed using Columbia blood agar (Oxoid, Wesel, Germany) after an incubation at 37 °C for 24 h. E. coli and coliform bacteria were analysed by using CC agar (Merck, Darmstadt, Germany) supplemented with a selective supplement (Merck; 2.5 mg vancomycin and 2.5 mg cefsulodine per 500 mL media) to inhibit the growth of Pseudomonas spp., Aeromonas spp. and Gram-positive bacteria; incubation was performed at 37 °C for 24 h. Gram-negative bacteria were determined using MacConkey agar (Oxoid, Wesel, Germany) after an incubation at 37 °C for 24 h. The homogenised mussel sample was diluted and spread onto the culture media. For each sample, if possible, two to three dilution steps were tested to obtain at least one evaluable dilution. After incubation on blood agar and MacConkey agar plates, the TBC was determined by counting all colonies, independent of appearance. For the CC agar, the colonies were counted separately by colour, according to the manufacturer’s instructions. While E. coli grows in dark blue to violet colonies, Enterobacter aerogenes, Citrobacter freundii and other coliforms form pink to red colonies. The resulting bacterial concentrations were calculated according to ISO 8199 [48].
4.5. Detection of Antibiotic-Resistant Bacteria
The water samples and the mussel cell suspensions were tested for the presence of antibiotic-resistant bacteria using chromogenic agar plates (CHROMagar ESBL, CHROMagar MRSA and CHROMagar VRE; MAST Diagnostica, Reinfeld, Germany), which are normally used for the isolation and differentiation of ARB from human material. A detailed description with performance characteristics for environmental samples was recently published by Schreiber et al. (2021) [49]. Depending on the expected target bacteria and background flora, 1 mL and/or a dilution of the sample was spread directly on agar plates. Volumes > 1 mL up to 100 mL were filtered through a cellulose nitrate filter (mixed cellulose ester, diameter 50 mm, pore size 45 µm). The plates were then incubated for 24 h at 42 °C for CHROMagar MRSA and CHROMagar ESBL and for 48 h at 42 °C for CHROMagar VRE. Of those media, nine different bacterial species were selected as target organisms, considering the priority list of ARB to guide research, discovery and development of new antibiotics [14]. Microbiological parameters included clinically relevant Gram-negative bacteria and resistant phenotypes: ESBL-producing Klebsiella spp., Enterobacter spp., Citrobacter spp. (grouped as “KEC”, due to morphological similarity on the CHROMagar ESBL plates), E. coli, Proteus mirabilis as well as P. aeruginosa and species belonging to the Acinetobacter calcoaceticus-baumannii complex showing resistance to 3rd-generation cephalosporins (3GCR). As Gram-positive clinically relevant resistant bacteria the following species were selected: methicillin-resistant S. aureus (MRSA) and the vancomycin-resistant enterococci (VRE) E. faecium and E. faecalis.
Confirmation and identification of bacterial species were performed using matrix-assisted laser desorption/ionisation time of flight mass spectrometry (MALDI-TOF MS) and with a VITEK® MS mass spectrometer (bioMérieux, Marcy l’Etoile, France), employing the MylaTM software.
For identification of the resistance profiles, bacteria belonging to the A. calcoaceticus-baumannii complex, Enterobacterales and P. aeruginosa were tested for antibiotic resistance using the microdilution assay Micronaut-S MDR MRGN-Screening 3 system (MERLIN, Bornheim-Hersel, Germany). The interpretation of the susceptibility status was performed according to the European Committee on Antimicrobial Susceptibility Testing criteria (EUCAST, Version 9.0, 2019).
With regard to multidrug resistance, all Gram-negative isolates were tested for their susceptibility against three of four clinically relevant antibiotic classes (piperacillin/tazobactam, fluoroquinolones (ciprofloxacin), third-generation cephalosporins (cefotaxime and/or ceftazidime) and carbapenems (meropenem and/or imipenem)) were classified as 3MRGN (multidrug resistant Gram-negative bacteria). All isolates that showed resistance to all four antibiotic groups mentioned above were classified as 4MRGN. The use of piperacillin/tazobactam is a modification of the test standards defined by the German Commission for Hospital Hygiene and Infection Prevention (KRINKO), which includes solely piperacillin. However, the latter is not used as a single drug in clinical therapy in Germany [50,51]. Therefore, we tested the commonly used combination with tazobactam. In case of the detection of a carbapenemase gene, the isolate was deemed 4MRGN independently of the phenotypical resistance against the tested antibiotics.
4.6. Molecular Typing of Resistant Isolates
For multilocus sequence typing (MLST) of multidrug resistant isolates (4MRGN), two to three colonies from fresh Columbia agar plates were incubated for 5 min at 95 °C in 80 μL of ultra purewater. The suspension was spun down at 14,000× g and the PCR was conducted according to previously described DLST standard procedures for P. aeruginosa [39,40]. The PCR products were purified from 1% agarose gels using the GeneJET Gel Extraction Kit (Fisher Scientific GmbH, Schwerte, Germany) and sequenced by GATC Biotech AG (Konstanz, Germany). Sequence types were determined via the specific MLST website for P. aeruginosa (https://pubmlst.org/organisms/pseudomonas-aeruginosa/) (accessed on 15 March 2021) [52].
4.7. Determination of Individual Limit of Detection
An individual limit of detection (LODi) occurred depending on the weight of the mussel and the initial expected bacteria concentration and background flora. The general LODi can be calculated by using the volume (10 mL) divided by soft tissue weight. For the very small mussels (0.1 g soft tissue) the LODi was 100 cfu/10 g or even 1000 cfu/10 g, in those cases that only 10 mL of the soft tissue suspension could be analysed and was found negative for the target species.
4.8. Graphical Representation of the Results
The data are presented as individual data points (Figure 1 and Figure 2) and as bars in Figure 3 (single values are shown). The limitation of the microbiological analysis, caused majorly by accompanying background flora, leads to results that indicate a minimum value and no exact value. In the case of results that indicate values ≥x cfu/10 g for the mussel tissue or ≥x cfu/100 mL for the water samples, the smallest possible value is entered in the graph. Such a value is marked with an asterisk in the respective figures.
5. Conclusions
The presence of faecal bacteria in the mussels points to an impact of treated wastewater on the individual mussel. Antibiotic-resistant isolates could be detected in the soft tissue of mussels of the genera Dreissena and Corbicula from different sampling sites of the river Rhine. No multidrug-resistant bacteria could be isolated from the mussel tissue, but were found in samples of the corresponding river water. In accordance with other work, a higher number of antibiotic-resistant bacteria (not multidrug-resistant) could be detected in the mussels as compared to the water. The diversity of antibiotic-resistant bacteria was higher in the water samples, reflected by different persistence. The cleaning performance of mussels naturally inhabiting river systems on the microbiological water quality cannot be estimated by these results and needs to be examined in further studies. The presence of antibiotic-resistant Gram-negative bacteria in the river Rhine and within mussels indicates that some resistance genes are widespread or even ubiquitous in water courses.
Author Contributions
N.Z.: Conceptualization, Data curation, Investigation, Methodology, Writing-original draft, Writing—review & editing, supervision; I.L.: Conceptualization, Data curation, Investigation, Writing—original draft, Methodology; S.M.E.: Investigation, Writing—original draft; E.S.: Methodology, Writing—review & editing; G.B.: Writing—review & editing; T.K.: Writing—review & editing; C.S.: Project administration, Writing—review & editing, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This study constitutes part of the collaborative research project “HyReKA” (Biological and hygienic–medical relevance and control of antibiotic-resistant pathogens in clinical, agricultural and municipal wastewater and their relevance in raw water). The authors thank the Federal Ministry of Education and Research of Germany (=BMBF) for funding the “HyReKA” joint project (funding ID: 02WRS1377).
Institutional Review Board Statement
The authors declare no conflict of interest. This study complies with the guidelines of the Declaration of Helsinki (1964) by the World Medical Association (No. 160/120 HyReKA Ethikantrag).
Data Availability Statement
All data supporting the results are shown in this study.
Acknowledgments
We also thank the staff of the Ecological Rhine station from the University of Cologne for their valuable advice on mussel habitats. Additional thanks to our dear colleague Dennis Schmiege for his for his support in improving the level of language and comprehensibility. This publication made use of the Pseudomonas aeruginosa MLST website (https://pubmlst.org/organisms/pseudomonas-aeruginosa/) (accessed on 15 March 2021) situated at the University of Oxford.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding Human Health Risks Caused by Antibiotic Resistant Bacteria (ARB) and Antibiotic Resistance Genes (ARG) in Water Environments: Current Knowledge and Questions to Be Answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.-L.; Cantón, R. Antibiotics and Antibiotic Resistance in Water Environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Nappier, S.P.; Liguori, K.; Ichida, A.M.; Stewart, J.R.; Jones, K.R. Antibiotic Resistance in Recreational Waters: State of the Science. Int. J. Environ. Res. Public. Health 2020, 17, 8034. [Google Scholar] [CrossRef]
- Schreiber, C.; Kistemann, T. Antibiotic Resistance among Autochthonous Aquatic Environmental Bacteria. Water Sci. Technol. 2013, 67, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic Resistance Genes in Treated Wastewater and in the Receiving Water Bodies: A Pan-European Survey of Urban Settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef]
- Gasse, J. Quantifizierung Der Emissionen Aus Abwasseranlagen Und Deren Auswirkungen Auf Die Hygienische Qualität von Flie\s Sgewässern; Oldenbourg Industrieverl: Munich, Germany, 2009. [Google Scholar]
- Schreiber, C.; Heinkel, S.-B.; Zacharias, N.; Mertens, F.-M.; Christoffels, E.; Gayer, U.; Koch, C.; Kistemann, T. Infectious Rain? Evaluation of Human Pathogen Concentrations in Stormwater in Separate Sewer Systems. Water Sci. Technol. 2019, 80, 1022–1030. [Google Scholar] [CrossRef]
- Schreiber, C.; Rechenburg, A.; Rind, E.; Kistemann, T. The Impact of Land Use on Microbial Surface Water Pollution. Int. J. Hyg. Environ. Health 2015, 218, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Voigt, A.M.; Zacharias, N.; Timm, C.; Wasser, F.; Sib, E.; Skutlarek, D.; Parcina, M.; Schmithausen, R.M.; Schwartz, T.; Hembach, N.; et al. Association between Antibiotic Residues, Antibiotic Resistant Bacteria and Antibiotic Resistance Genes in Anthropogenic Wastewater—An Evaluation of Clinical Influences. Chemosphere 2020, 241, 125032. [Google Scholar] [CrossRef]
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in Understanding Antimicrobial Resistance across Domesticated Animal, Human, and Environmental Systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef]
- Kümmerer, K. Antibiotics in the Aquatic Environment—A Review. Chemosphere 2009, 75, 417–441. [Google Scholar] [CrossRef] [PubMed]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The Global Threat of Antimicrobial Resistance: Science for Intervention. N. Microbes N. Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-156474-8. [Google Scholar]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Schöll, F.; Becker, C.; Tittizer, T. Das Makrozoobenthos Des Schiffbaren Rheins von Basel Bis Emmerich 1986-1995. Lauterbornia 1995, 21, 115–137. [Google Scholar]
- Bernauer, D.; Jansen, W. Recent Invasions of Alien Macroinvertebrates and Loss of Native Species in the Upper Rhine River, Germany. Aquat. Invasions 2006, 1, 55–71. [Google Scholar] [CrossRef]
- Kinzelbach, R. The Main Features of the Phylogeny and Dispersal of the Zebra Mussel Dreissena polymorpha. Zebra Mussel Dreissena Polymorpha 1992, 4, 5–17. [Google Scholar]
- Chijimatsu, T.; Tatsuguchi, I.; Abe, K.; Oda, H.; Mochizuki, S. A Freshwater Clam (Corbicula fluminea) Extract Improves Cholesterol Metabolism in Rats Fed on a High-Cholesterol Diet. Biosci. Biotechnol. Biochem. 2008, 0809081039. [Google Scholar] [CrossRef]
- Mouthon, J. Sur La Présence En France et Au Portugal de Corbicula (Bivalvia, Corbiculidae) Originaire d’Asie. Basteria 1981, 45, 109–116. [Google Scholar]
- Pathy, D.A.; Mackie, G.L. Comparative Shell Morphology of Dreissena Polymorpha, Mytilopsis leucophaeata, and the “Quagga” Mussel (Bivalvia: Dreissenidae) in North America. Can. J. Zool. 1993, 71, 1012–1023. [Google Scholar] [CrossRef]
- Bonner, T.P.; Rockhill, R.L. Ultrastructure of the Byssus of the Zebra Mussel (Dreissena polymorpha, Mollusca: Bivalvia). Trans. Am. Microsc. Soc. 1994, 302–315. [Google Scholar] [CrossRef]
- Matthews, J.; Van der Velde, G.; De Vaate, A.B.; Collas, F.P.; Koopman, K.R.; Leuven, R.S. Rapid Range Expansion of the Invasive Quagga Mussel in Relation to Zebra Mussel Presence in The Netherlands and Western Europe. Biol. Invasions 2014, 16, 23–42. [Google Scholar] [CrossRef]
- Molloy, D.P.; bij de Vaate, A.; Wilke, T.; Giamberini, L. Discovery of Dreissena rostriformi bugensiss (Andrusov 1897) in Western Europe. Biol. Invasions 2007, 9, 871–874. [Google Scholar] [CrossRef]
- Haybach, A.; Christmann, K.H. Erster Nachweis Der Quaggamuschel Dreissena Rostriformis Bugensis (Andrusov, 1897)(Bivalvia: Dreissenidae) Im Niederrhein von Nordrhein-Westfalen. Lauterbornia 2009, 67, 69–72. [Google Scholar]
- Gomes, J.F.; Lopes, A.; Gonçalves, D.; Luxo, C.; Gmurek, M.; Costa, R.; Quinta-Ferreira, R.M.; Martins, R.C.; Matos, A. Biofiltration Using C. Fluminea for E. Coli Removal from Water: Comparison with Ozonation and Photocatalytic Oxidation. Chemosphere 2018, 208, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, C.C.; Hakenkamp, C.C. The Functional Role of Burrowing Bivalves in Freshwater Ecosystems. Freshw. Biol. 2001, 46, 1431–1446. [Google Scholar] [CrossRef]
- Winters, A.D.; Marsh, T.L.; Faisal, M. Heterogeneity of Bacterial Communities within the Zebra Mussel (Dreissena polymorpha) in the Laurentian Great Lakes Basin. J. Gt. Lakes Res. 2011, 37, 318–324. [Google Scholar] [CrossRef]
- Boufafa, M.; Kadri, S.; Redder, P.; Bensouilah, M. Occurrence and Distribution of Fecal Indicators and Pathogenic Bacteria in Seawater and Perna Perna Mussel in the Gulf of Annaba (Southern Mediterranean). bioRxiv 2020, 2020.10.04.325167. [Google Scholar] [CrossRef]
- Ilic, N.; Velebit, B.; Teodorovic, V.; Djordjevic, V.; Karabasil, N.; Vasilev, D.; Djuric, S.; Adzic, B.; Dimitrijevic, M. Influence of Environmental Conditions on Norovirus Presence in Mussels Harvested in Montenegro. Food Environ. Virol. 2017, 9, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Ghozzi, K.; Marangi, M.; Papini, R.; Lahmar, I.; Challouf, R.; Houas, N.; Dhiab, R.B.; Normanno, G.; Babba, H.; Giangaspero, A. First Report of Tunisian Coastal Water Contamination by Protozoan Parasites Using Mollusk Bivalves as Biological Indicators. Mar. Pollut. Bull. 2017, 117, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Zannella, C.; Mosca, F.; Mariani, F.; Franci, G.; Folliero, V.; Galdiero, M.; Tiscar, P.G.; Galdiero, M. Microbial Diseases of Bivalve Mollusks: Infections, Immunology and Antimicrobial Defense. Mar. Drugs 2017, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Graczyk, T.K.; Marcogliese, D.J.; de Lafontaine, Y.; Da Silva, A.J.; Mhangami-Ruwende, B.; Pieniazek, N.J. Cryptosporidium Parvum Oocysts in Zebra Mussels (Dreissena polymorpha): Evidence from the St. Lawrence River. Parasitol. Res. 2001, 87, 231–234. [Google Scholar] [CrossRef]
- Ladeiro, M.P.; Aubert, D.; Villena, I.; Geffard, A.; Bigot, A. Bioaccumulation of Human Waterborne Protozoa by Zebra Mussel (Dreissena polymorpha): Interest for Water Biomonitoring. Water Res. 2014, 48, 148–155. [Google Scholar] [CrossRef]
- Karatayev, A.Y.; Burlakova, L.E.; Padilla, D.K. Contrasting Distribution and Impacts of Two Freshwater Exotic Suspension Feeders, Dreissena polymorpha and Corbicula fluminea. In The Comparative Roles of Suspension-Feeders in Ecosystems; Dame, R.F., Olenin, S., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 239–262. [Google Scholar]
- Andrès, S.; Baudrimont, M.; Lapaquellerie, Y.; Ribeyre, F.; Maillet, N.; Latouche, C.; Boudou, A. Field Transplantation of the Freshwater Bivalve Corbicula fluminea along a Polymetallic Contamination Gradient (River Lot, France): I. Geochemical Characteristics of the Sampling Sites and Cadmium and Zinc Bioaccumulation Kinetics. Environ. Toxicol. Chem. 1999, 18, 2462–2471. [Google Scholar] [CrossRef]
- Bighiu, M.A.; Haldén, A.N.; Goedkoop, W.; Ottoson, J. Assessing Microbial Contamination and Antibiotic Resistant Bacteria Using Zebra Mussels (Dreissena Polymorpha). Sci. Total Environ. 2019, 650, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.; Moreno, D.; Ramos, M.A. The Asiatic Clam Corbicula fluminea (Muller, 1774) (Bivalvia, Corbiculidae) in Europe. Am. Malacol. Bull. 1993, 10, 39–49. [Google Scholar]
- Belanger, S.E.; Farris, J.L.; Cherry, D.S.; Cairns, J., Jr. Validation of Corbicula fluminea Growth Reductions Induced by Copper in Artificial Streams and River Systems. Can. J. Fish. Aquat. Sci. 1990, 47, 904–914. [Google Scholar] [CrossRef]
- Feng, W.; Sun, F.; Wang, Q.; Xiong, W.; Qiu, X.; Dai, X.; Xia, P. Epidemiology and Resistance Characteristics of Pseudomonas Aeruginosa Isolates from the Respiratory Department of a Hospital in China. J. Glob. Antimicrob. Resist. 2017, 8, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Wang, J.; Zhou, Z.; Wang, H.; Jiang, Y.; Yu, Y. Multilocus Sequence Typing Reveals Genetic Diversity of Carbapenem- or Ceftazidime-Nonsusceptible Pseudomonas Aeruginosa in China. Antimicrob. Agents Chemother. 2013, 57, 5697–5700. [Google Scholar] [CrossRef] [PubMed]
- Musila, L.; Kyany’a, C.; Maybank, R.; Stam, J.; Oundo, V.; Sang, W. Detection of Diverse Carbapenem and Multidrug Resistance Genes and High-Risk Strain Types among Carbapenem Non-Susceptible Clinical Isolates of Target Gram-Negative Bacteria in Kenya. PLoS ONE 2021, 16, e0246937. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of Multi-Resistant Gram-Negative Bacteria into German Wastewater and Surface Waters. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Silverman, H.; Achberger, E.C.; Lynn, J.W.; Dietz, T.H. Filtration and Utilization of Laboratory-Cultured Bacteria by Dreissena polymorpha, Corbicula fluminea, and Carunculina texasensis. Biol. Bull. 1995, 189, 308–319. [Google Scholar] [CrossRef]
- Cohen, R.R.; Dresler, P.V.; Phillips, E.J.; Cory, R.L. The Effect of the Asiatic Clam, Corbicula fluminea, on Phytoplankton of the Potomac River, Maryland. Limnol. Oceanogr. 1984, 29, 170–180. [Google Scholar] [CrossRef]
- International Commission for the Protection of the Rhine (ICPR). Assessment “Rhine 2020”. Available online: https://www.iksr.org/fileadmin/user_upload/DKDM/Dokumente/Broschueren/EN/bro_En_Assessment_%E2%80%9CRhine_2020%E2%80%9D.pdf (accessed on 9 March 2021).
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef] [PubMed]
- DIN 38402-11:2009-2. In German Standard Methods for the Examination of Water, Waste Water and Sludge—General Information (Group A)—Part 11: Sampling of Waste Water (A 11); Beuth Verlag GmbH: Berlin, Germany, 1995.
- ISO 8199:2018-10. In Water Quality—General Requirements and Guidance for Microbiological Examinations by Culture; ISO: Geneva, Switzerland, 2018.
- Schreiber, C.; Zacharias, N.; Essert, S.M.; Wasser, F.; Müller, H.; Sib, E.; Precht, T.; Parcina, M.; Bierbaum, G.; Schmithausen, R.M.; et al. Clinically Relevant Antibiotic-Resistant Bacteria in Aquatic Environments—An Optimized Culture-Based Approach. Sci. Total Environ. 2021, 750, 142265. [Google Scholar] [CrossRef] [PubMed]
- KRINKO. Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen. Bundesgesundheitsblatt—Gesundheitsforsch.—Gesundh. 2012, 55, 1311–1354. [Google Scholar] [CrossRef]
- KRINKO. Ergänzung zur Empfehlung der KRINKO „Hygienemaßnahmen bei Infektionen oder Besiedlung mit multiresistenten gramnegativen Stäbchen“ (2012) im Zusammenhang mit der von EUCAST neu definierten Kategorie „I“ bei der Antibiotikaresistenzbestimmung: Konsequenzen für die Definition von MRGN. Epidemiol. Bull. 2019, 82–83. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
