Colistin Treatment Affects Lipid Composition of Acinetobacter baumannii
Abstract
1. Introduction
2. Results
2.1. Colistin Susceptibility
2.2. Lipids Annotated in Acinetobacter baumannii 721164 by LC-HRMS2
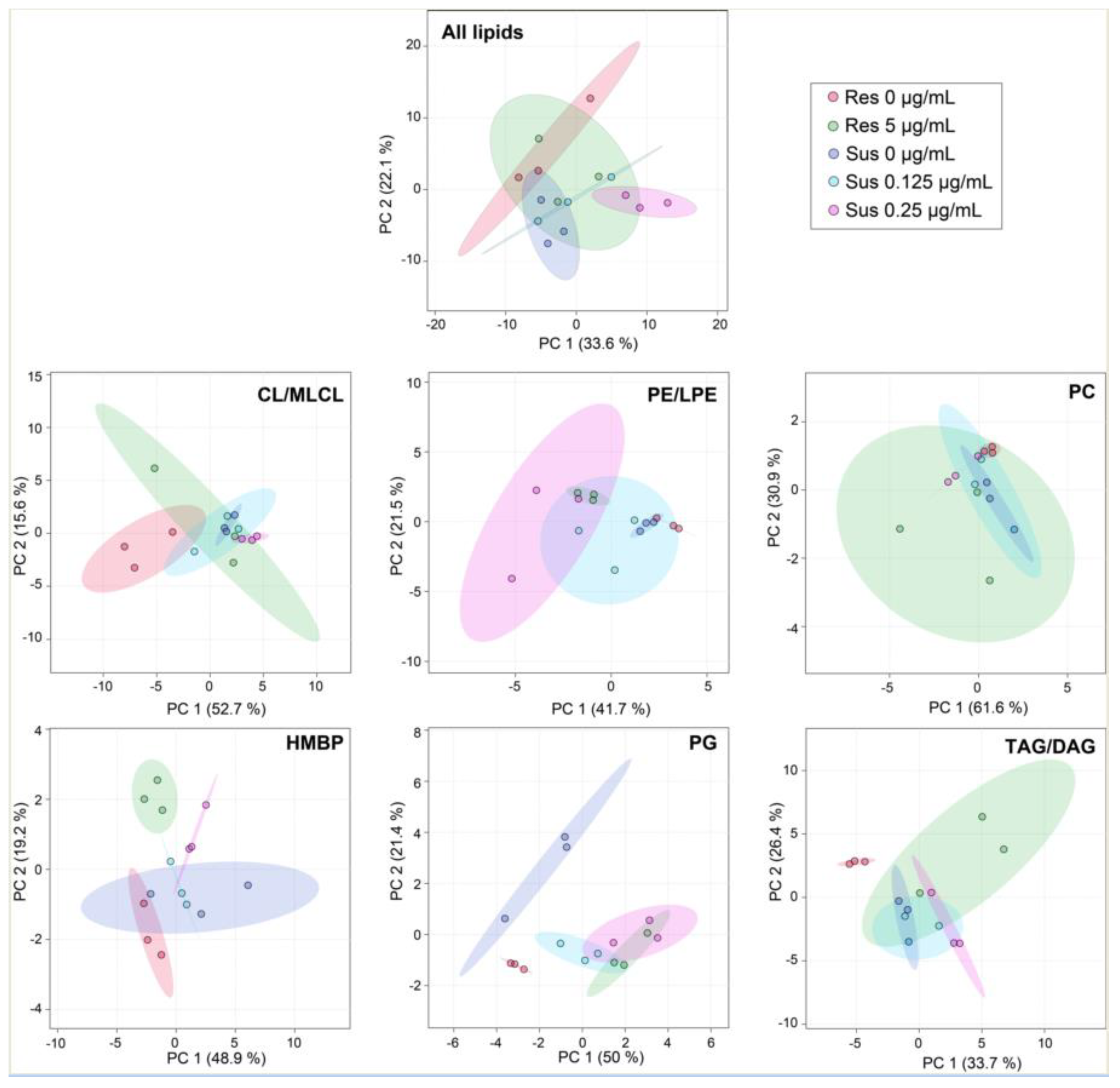
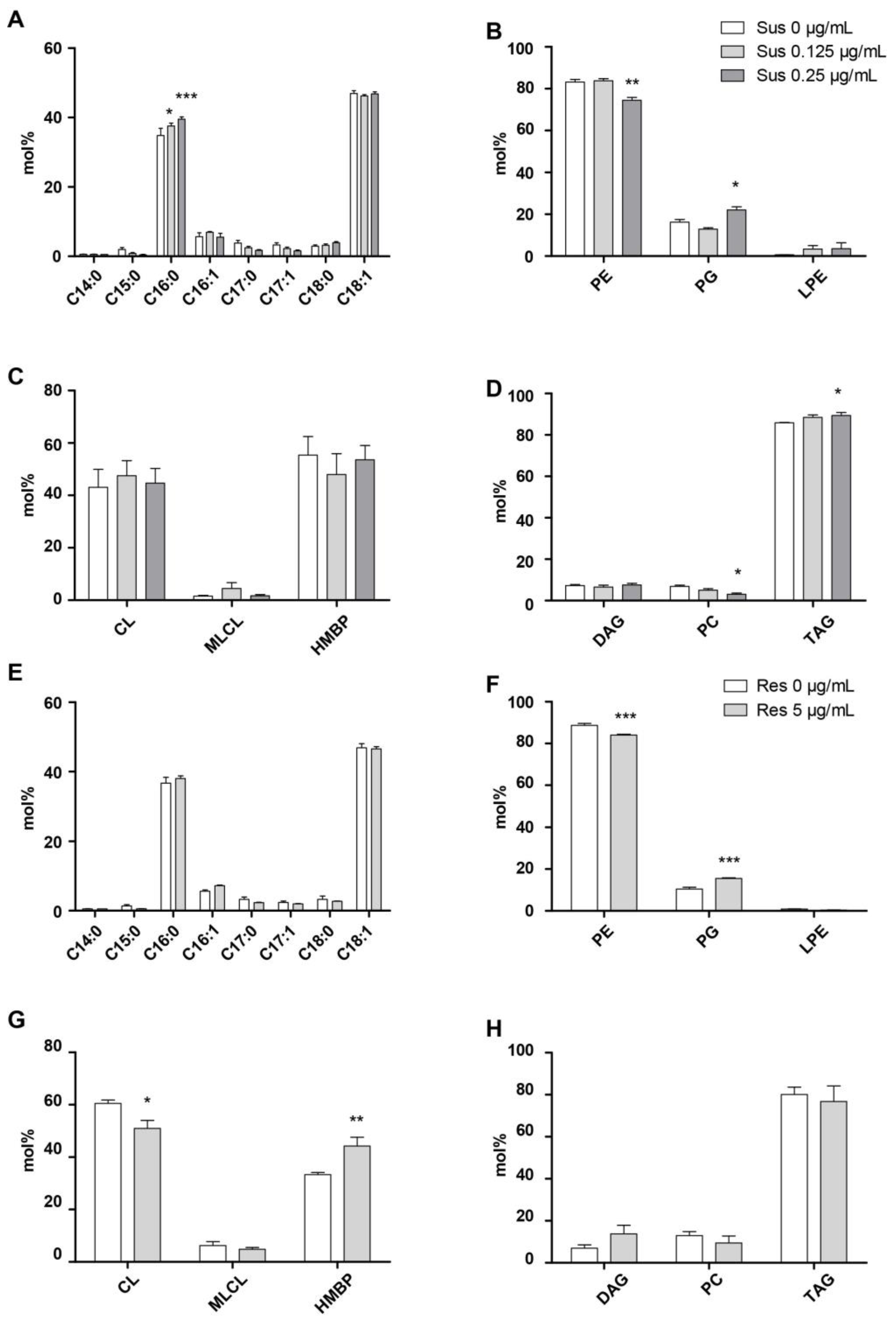
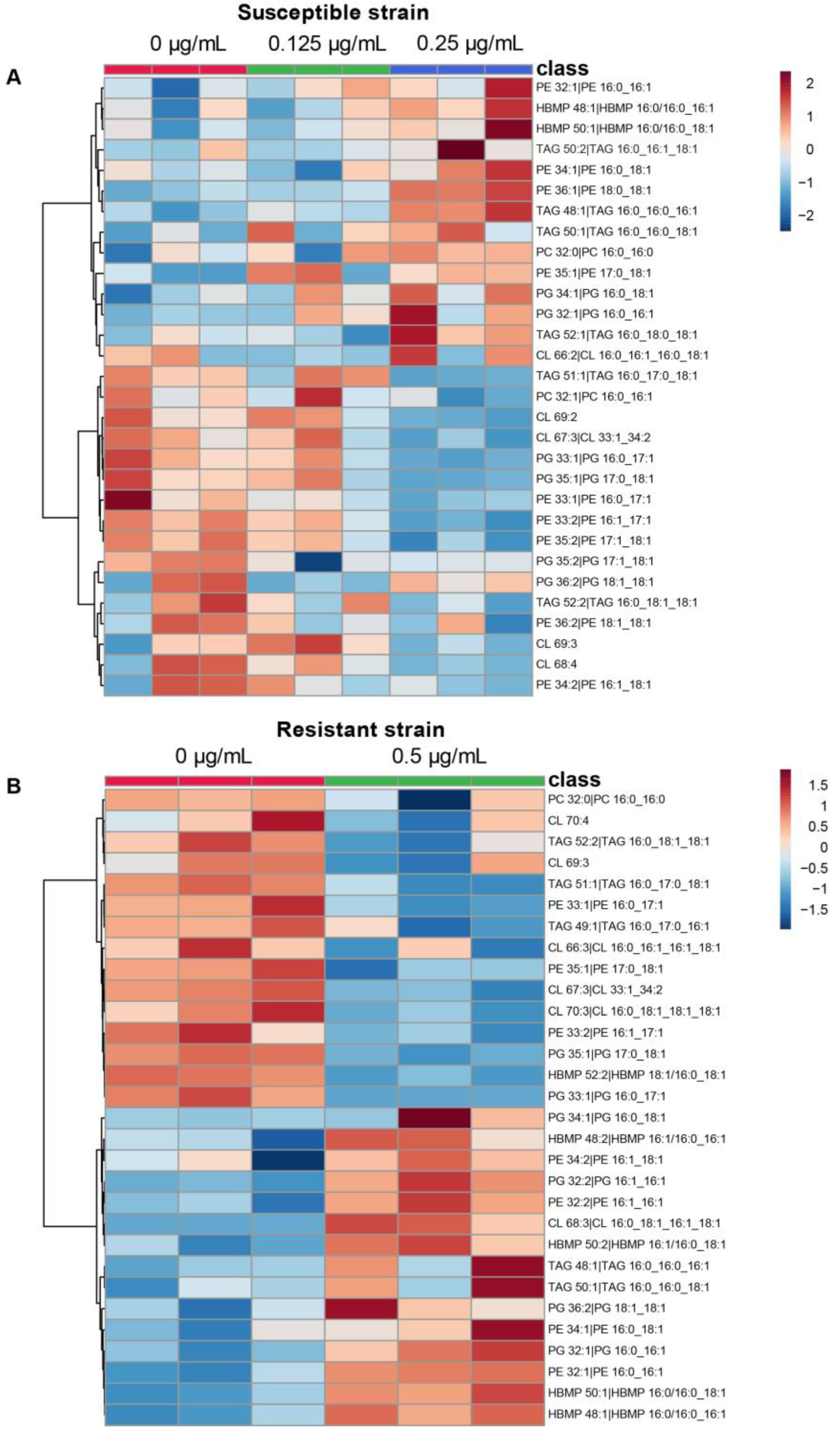
2.3. Colistin and Lipid Profiles of Acinetobacter baumannii by LC-HRMS2
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Growth Conditions
5.2. Susceptibility Testing to Colistin
5.3. Lipid Extraction
5.4. Fatty Acids Analysis by GC-FID
5.5. LC-HRMS2 Analyses
5.6. Data Processing and Annotation
5.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the infectious diseases society of america. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Willyard, C. Drug-resistant bacteria ranked. Nature 2017, 543, 15. [Google Scholar] [CrossRef]
- El-Sayed Ahmed, M.A.E.-G.; Zhong, L.-L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.-B. Colistin and its role in the era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, S.; Utsunomiya, C.; Ishikawa, S.; Sekiguchi, J. Purification and characterization of an autolysin of Bacillus polymyxa var. colistinus which is most active at acidic pH. J. Ferment. Bioeng. 1997, 83, 419–422. [Google Scholar] [CrossRef]
- Koch-Weser, J.; Sidel, V.; Federman, R.; Kanarek, M.; Finer, D.; Eaton, A. Adverse effects of sodium colistimethate. Ann. Intern. Med. 1970, 72, 857. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 2005, 40, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Schindler, M.; Osborn, M.J. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry 1979, 18, 4425–4430. [Google Scholar] [CrossRef] [PubMed]
- Koike, M.; Iida, K.; Matsuo, T. Electron microscopic studies on mode of action of polymyxin. J. Bacteriol. 1969, 97, 448–452. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef]
- Kaye, K.S.; Pogue, J.M.; Tran, T.B.; Nation, R.L.; Li, J. Agents of last resort: Polymyxin resistance. Infect. Dis. Clin. N. Am. 2016, 30, 391–414. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Needham, B.D.; Trent, M.S. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013, 11, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.F.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St. Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef]
- Soon, R.L.; Nation, R.L.; Cockram, S.; Moffatt, J.H.; Harper, M.; Adler, B.; Boyce, J.D.; Larson, I.; Li, J. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J. Antimicrob. Chemother. 2011, 66, 126–133. [Google Scholar] [CrossRef]
- Pelletier, M.R.; Casella, L.G.; Jones, J.W.; Adams, M.D.; Zurawski, D.V.; Hazlett, K.R.O.; Doi, Y.; Ernst, R.K. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013, 57, 4831–4840. [Google Scholar] [CrossRef]
- Adams, M.D.; Nickel, G.C.; Bajaksouzian, S.; Lavender, H.; Murthy, A.R.; Jacobs, M.R.; Bonomo, R.A. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 2009, 53, 3628–3634. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, Y.; Han, M.-L.; Li, M.; Song, J.; Velkov, T.; Li, C.; Li, J. Comparative analysis of phosphoethanolamine transferases involved in polymyxin resistance across 10 clinically relevant gram-negative bacteria. Int. J. Antimicrob. Agents 2018, 51, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Dößelmann, B.; Willmann, M.; Steglich, M.; Bunk, B.; Nübel, U.; Peter, S.; Neher, R.A. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob. Agents Chemother. 2017, 61. [Google Scholar] [CrossRef]
- Cafiso, V.; Stracquadanio, S.; Lo Verde, F.; Gabriele, G.; Mezzatesta, M.L.; Caio, C.; Pigola, G.; Ferro, A.; Stefani, S. Colistin resistant A. baumannii: Genomic and transcriptomic traits acquired under colistin therapy. Front. Microbiol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Kanipes, M.I.; Lin, S.; Cotter, R.J.; Raetz, C.R.H. Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. J. Biol. Chem. 2001, 276, 1156–1163. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Galindo-Lagunas, K.A.; Guan, Z.; Vinuesa, P.; Robinson, S.; Thomas-Oates, J.; Raetz, C.R.H.; Geiger, O. The lipid lysyl-phosphatidylglycerol is present in membranes of Rhizobium tropici CIAT899 and confers increased resistance to polymyxin B under acidic growth conditions. Mol. Plant-Microbe Interact. 2007, 20, 1421–1430. [Google Scholar] [CrossRef]
- Bisignano, C.; Ginestra, G.; Smeriglio, A.; La Camera, E.; Crisafi, G.; Franchina, F.; Tranchida, P.; Alibrandi, A.; Trombetta, D.; Mondello, L.; et al. Study of the lipid profile of ATCC and clinical strains of Staphylococcus aureus in relation to their antibiotic resistance. Molecules 2019, 24, 1276. [Google Scholar] [CrossRef]
- Sun, W.; Dunning, F.M.; Pfund, C.; Weingarten, R.; Bent, A.F. Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2-dependent defenses. Plant Cell 2006, 18, 764–779. [Google Scholar] [CrossRef]
- Schenk, E.R.; Nau, F.; Thompson, C.J.; Tse-Dinh, Y.-C.; Fernandez-Lima, F. Changes in lipid distribution in E. coli strains in response to norfloxacin. J. Mass Spectrom. 2015, 50, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Shireen, T.; Singh, M.; Dhawan, B.; Mukhopadhyay, K. Characterization of cell membrane parameters of clinical isolates of Staphylococcus aureus with varied susceptibility to alpha-melanocyte stimulating hormone. Peptides 2012, 37, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.M.; McElheny, C.L.; Gardner, F.M.; Chandler, C.E.; Bowler, S.L.; Mettus, R.T.; Spychala, C.N.; Fowler, E.L.; Opene, B.N.A.; Myers, R.A.; et al. A prospective study of Acinetobacter baumannii complex isolates and colistin susceptibility monitoring by mass spectrometry of microbial membrane glycolipids. J. Clin. Microbiol. 2018, 57. [Google Scholar] [CrossRef]
- Henry, R.; Crane, B.; Powell, D.; Deveson Lucas, D.; Li, Z.; Aranda, J.; Harrison, P.; Nation, R.L.; Adler, B.; Harper, M.; et al. The transcriptomic response of Acinetobacter baumannii to colistin and doripenem alone and in combination in an in vitro pharmacokinetics/pharmacodynamics model. J. Antimicrob. Chemother. 2015, 70, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-L.; Zhu, Y.; Creek, D.J.; Lin, Y.-W.; Anderson, D.; Shen, H.-H.; Tsuji, B.; Gutu, A.D.; Moskowitz, S.M.; Velkov, T.; et al. Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Mahamad Maifiah, M.H.; Cheah, S.-E.; Johnson, M.D.; Han, M.-L.; Boyce, J.D.; Thamlikitkul, V.; Forrest, A.; Kaye, K.S.; Hertzog, P.; Purcell, A.W.; et al. Global metabolic analyses identify key differences in metabolite levels between polymyxin-susceptible and polymyxin-resistant Acinetobacter baumannii. Sci. Rep. 2016, 6, 22287. [Google Scholar] [CrossRef] [PubMed]
- Jasim, R.; Han, M.-L.; Zhu, Y.; Hu, X.; Hussein, M.; Lin, Y.-W.; Zhou, Q. (Tony); Dong, C.; Li, J.; Velkov, T. Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and -resistant Klebsiella pneumoniae clinical isolates. Int. J. Mol. Sci. 2018, 19, 2356. [Google Scholar] [CrossRef]
- Hugh, R.; Reese, R. Designation of the type strain for Bacterium anitratum Schaub and Hauber 1948. Int. J. Syst. Bacteriol. 1967, 17, 245–254. [Google Scholar] [CrossRef]
- Lopalco, P.; Stahl, J.; Annese, C.; Averhoff, B.; Corcelli, A. Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Sci. Rep. 2017, 7, 2972. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; White, S.W.; Rock, C.O. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 2001, 40, 467–497. [Google Scholar] [CrossRef]
- White, S.W.; Zheng, J.; Zhang, Y.-M.; Rock, C.O. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Satlin, M.J.; Lewis, J.S.; Weinstein, M.P.; Patel, J.; Humphries, R.M.; Kahlmeter, G.; Giske, C.G.; Turnidge, J. Clinical and laboratory standards institute and european committee on antimicrobial susceptibility testing position statements on polymyxin B and colistin clinical breakpoints. Clin. Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Kamischke, C.; Fan, J.; Bergeron, J.; Kulasekara, H.D.; Dalebroux, Z.D.; Burrell, A.; Kollman, J.M.; Miller, S.I. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife 2019, 8. [Google Scholar] [CrossRef]
- Alvarez, H.; Steinbüchel, A. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Garrett, T.A.; Raetz, C.R.H.; Son, J.D.; Richardson, T.D.; Bartling, C.; Guan, Z. Non-enzymatically derived minor lipids found in Escherichia coli lipid extracts. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2011, 1811, 827–837. [Google Scholar] [CrossRef]
- Geiger, O.; López-Lara, I.M.; Sohlenkamp, C. Phosphatidylcholine biosynthesis and function in bacteria. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2013, 1831, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Vasilopoulos, G.; Moser, R.; Petersen, J.; Aktas, M.; Narberhaus, F. Promiscuous phospholipid biosynthesis enzymes in the plant pathogen Pseudomonas syringae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158926. [Google Scholar] [CrossRef]
- Boll, J.M.; Crofts, A.A.; Peters, K.; Cattoir, V.; Vollmer, W.; Davies, B.W.; Trent, M.S. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 2016, 113, E6228–E6237. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.D.; Wright, J.C.; Li, J.; Hood, D.W.; Moxon, E.R.; Richards, J.C. Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: Identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 2003, 185, 3270–3277. [Google Scholar] [CrossRef] [PubMed]
- Stahl, R.S.; Bisha, B.; Mahapatra, S.; Chandler, J.C. A model for the prediction of antimicrobial resistance in Escherichia coli based on a comparative evaluation of fatty acid profiles. Diagn. Microbiol. Infect. Dis. 2020, 96, 114966. [Google Scholar] [CrossRef] [PubMed]
- Mazzotta, A.S.; Montville, T.J. Characterization of fatty acid composition, spore germination, and thermal resistance in a Nisin-resistant mutant of Clostridium botulinum 169B and in the Wild-Type strain. Appl. Environ. Microbiol. 1999, 65, 659–664. [Google Scholar] [CrossRef]
- Dunnick, J.K.; O’Leary, W.M. Correlation of bacterial lipid composition with antibiotic resistance. J. Bacteriol. 1970, 101, 892–900. [Google Scholar] [CrossRef]
- Gonçalves, F.D.A.; de Carvalho, C.C.C.R. Phenotypic modifications in Staphylococcus aureus cells exposed to high concentrations of vancomycin and teicoplanin. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Boudjemaa, R.; Cabriel, C.; Dubois-Brissonnet, F.; Bourg, N.; Dupuis, G.; Gruss, A.; Lévêque-Fort, S.; Briandet, R.; Fontaine-Aupart, M.-P.; Steenkeste, K. Impact of bacterial membrane fatty acid composition on the failure of daptomycin to kill Staphylococcus aureus. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Dawaliby, R.; Trubbia, C.; Delporte, C.; Noyon, C.; Ruysschaert, J.-M.; Van Antwerpen, P.; Govaerts, C. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 2016, 291, 3658–3667. [Google Scholar] [CrossRef]
- Bogdanov, M.; Pyrshev, K.; Yesylevskyy, S.; Ryabichko, S.; Boiko, V.; Ivanchenko, P.; Kiyamova, R.; Guan, Z.; Ramseyer, C.; Dowhan, W. Phospholipid distribution in the cytoplasmic membrane of gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 2020, 6, eaaz6333. [Google Scholar] [CrossRef]
- Samantha, A.; Vrielink, A. Lipid A phosphoethanolamine transferase: Regulation, structure and immune response. J. Mol. Biol. 2020, 432, 5184–5196. [Google Scholar] [CrossRef]
- El-Halfawy, O.M.; Valvano, M.A. Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 2015, 28, 191–207. [Google Scholar] [CrossRef]
- Dafopoulou, K.; Zarkotou, O.; Dimitroulia, E.; Hadjichristodoulou, C.; Gennimata, V.; Pournaras, S.; Tsakris, A. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 2015, 59, 4625–4630. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. LC-MS-Based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS Library. In Lipidomics: Methods and Protocols; Bhattacharya, S.K., Ed.; Springer: New York, NY, USA, 2017; pp. 149–170. ISBN 978-1-4939-6996-8. [Google Scholar]
- Cazzola, H.; Lemaire, L.; Acket, S.; Prost, E.; Duma, L.; Erhardt, M.; Čechová, P.; Trouillas, P.; Mohareb, F.; Rossi, C.; et al. The impact of plasma membrane lipid composition on flagellum-mediated adhesion of enterohemorrhagic Escherichia coli. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Ikeda, K.; Takahashi, M.; Satoh, A.; Mori, Y.; Uchino, H.; Okahashi, N.; Yamada, Y.; Tada, I.; Bonini, P.; et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020, 38, 1159–1163. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboanalystR 3.0: Toward an optimized workflow for global metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
| Acinetobacter baumannii 721164 | Susceptible | Resistant |
|---|---|---|
| Colistin | 0.5 | >128 |
| Lipids | m/z | Retention Time | Assignment |
|---|---|---|---|
| Cardiolipin (CL) | 1293.89026 | 9.502 | CL 60:1 |
| 1291.87561 | 9.099 | CL 60:2 | |
| 1289.85425 | 8.62 | CL 60:3 | |
| 1305.89014 | 9.326 | CL 61:2 | |
| 1303.8728 | 8.866 | CL 61:3 | |
| 1321.9187 | 9.991 | CL 62:1 | |
| 1319.90637 | 9.495 | CL 62:2 | |
| 1317.89062 | 9.138 | CL 62:3 | |
| 1335.93335 | 10.148 | CL 63:1 | |
| 1333.92212 | 9.805 | CL 63:2 | |
| 1331.90564 | 9.362 | CL 63:3 | |
| 1347.93762 | 10.034 | CL 64:2|CL 16:0_16:1_16:0_16:1 | |
| 1345.9209 | 9.483 | CL 64:3|CL 16:0_16:1_16:1_16:1 | |
| 1361.9541 | 10.21 | CL 65:2 | |
| 1359.93982 | 9.837 | CL 65:3 | |
| 1357.92249 | 9.389 | CL 65:4 | |
| 1375.96924 | 10.205 | CL 66:2|CL 16:0_16:1_16:0_18:1 | |
| 1373.953 | 10.067 | CL 66:3|CL 16:0_16:1_16:1_18:1 | |
| 1371.93896 | 9.656 | CL 66:4 | |
| 1389.98438 | 10.276 | CL 67:2 | |
| 1387.96814 | 10.219 | CL 67:3|CL 33:1_34:2 | |
| 1385.95239 | 9.863 | CL 67:4 | |
| 1403.99841 | 10.299 | CL 68:2|CL 16:0_18:1_16:0_18:1 | |
| 1401.98572 | 10.219 | CL 68:3|CL 16:0_18:1_16:1_18:1 | |
| 1399.96814 | 10.092 | CL 68:4 | |
| 1418.01538 | 10.313 | CL 69:2 | |
| 1415.99963 | 10.279 | CL 69:3 | |
| 1413.98303 | 10.223 | CL 69:4 | |
| 1430.01562 | 10.303 | CL 70:3|CL 16:0_18:1_18:1_18:1 | |
| 1427.99902 | 10.266 | CL 70:4 | |
| 1444.02808 | 10.317 | CL 71:3 | |
| 1456.02783 | 10.303 | CL 72:4 | |
| Monolysocardiolipin (MLCL) | 1107.68982 | 6.044 | MLCL 48:0 |
| 1121.70483 | 6.354 | MLCL 49:0 | |
| 1117.67041 | 8.435 | MLCL 49:2 | |
| 1135.72205 | 6.712 | MLCL 50:0 | |
| 1137.74084 | 7.17 | MLCL 50:2|MLCL 16:0_34:2 | |
| 1149.73999 | 7.012 | MLCL 51:0 | |
| 1163.75537 | 7.218 | MLCL 52:0 | |
| 1165.77185 | 7.785 | MLCL 52:2|MLCL 18:0_34:2 | |
| 1157.70374 | 6.694 | MLCL 52:3 | |
| 1191.78455 | 7.806 | MLCL 53:0 | |
| 1187.75085 | 7.762 | MLCL 53:2 | |
| 1185.7345 | 7.264 | MLCL 53:3 | |
| Hemibismonoacylglycerophosphate (HBMP) | 901.65656 | 7.196 | HBMP 44:1|HBMP 16:1/12:0_16:0 |
| 929.68842 | 7.81 | HBMP 46:1|HBMP 16:0/14:0_16:1 | |
| 927.67273 | 7.244 | HBMP 46:2|HBMP 14:0/14:1_18:1 | |
| 957.72144 | 8.389 | HBMP 48:1|HBMP 16:0/16:0_16:1 | |
| 955.70532 | 7.872 | HBMP 48:2|HBMP 16:1/16:0_16:1 | |
| 953.69006 | 7.32 | HBMP 48:3|HBMP 16:1/16:1_16:1 | |
| 970.72339 | 8.17 | HBMP 49:3 | |
| 985.75183 | 8.934 | HBMP 50:1|HBMP 16:0/16:0_18:1 | |
| 983.73761 | 8.36 | HBMP 50:2|HBMP 16:1/16:0_18:1 | |
| 981.72137 | 7.924 | HBMP 50:3|HBMP 16:1/16:1_18:1 | |
| 1000.76843 | 9.183 | HBMP 51:2 | |
| 1011.7688 | 8.998 | HBMP 52:2|HBMP 18:1/16:0_18:1 | |
| Phosphatidylcholine (PC) | 734.5694 | 6.325 | PC 32:0|PC 16:0_16:0 |
| 732.55377 | 6.661 | PC 32:1|PC 16:0_16:1 | |
| 730.53809 | 4.832 | PC 32:2|PC 16:1_16:1 | |
| 760.58508 | 6.182 | PC 34:1|PC 16:0_18:1 | |
| 758.5694 | 5.525 | PC 34:2|PC 16:1_18:1 | |
| Lysophosphatidylethaniolamine (LPE) | 450.26407 | 1.067 | LPE 16:1 |
| 464.28006 | 1.321 | LPE 17:1 | |
| 478.29681 | 1.648 | LPE 18:1 | |
| Phosphatidylethanolamine (PE) | 662.479 | 5.642 | PE 30:0|PE 14:0_16:0 |
| 660.46484 | 4.993 | PE 30:1|PE 14:0_16:1 | |
| 674.48035 | 5.368 | PE 31:1|PE 15:0_16:1 | |
| 690.5108 | 6.406 | PE 32:0|PE 16:0_16:0 | |
| 688.49481 | 5.736 | PE 32:1|PE 16:0_16:1 | |
| 686.47961 | 5.093 | PE 32:2|PE 16:1_16:1 | |
| 702.51129 | 6.114 | PE 33:1|PE 16:0_17:1 | |
| 700.49573 | 5.446 | PE 33:2|PE 16:1_17:1 | |
| 716.52686 | 6.519 | PE 34:1|PE 16:0_18:1 | |
| 714.51105 | 5.83 | PE 34:2|PE 16:1_18:1 | |
| 730.54224 | 8.1 | PE 35:1|PE 17:0_18:1 | |
| 730.54279 | 6.858 | PE 35:1|PE 17:0_18:1 | |
| 728.52765 | 6.2 | PE 35:2|PE 17:1_18:1 | |
| 744.55786 | 7.261 | PE 36:1|PE 18:0_18:1 | |
| 742.54224 | 6.598 | PE 36:2|PE 18:1_18:1 | |
| Phosphatidylglycerol (PG) | 693.47375 | 4.525 | PG 30:0|PG 14:0_16:0 |
| 691.45935 | 4.051 | PG 30:1|PG 14:0_16:1 | |
| 705.47412 | 4.327 | PG 31:1|PG 15:0_16:1 | |
| 719.49042 | 4.616 | PG 32:1|PG 16:0_16:1 | |
| 717.47479 | 4.155 | PG 32:2|PG 16:1_16:1 | |
| 733.50592 | 4.935 | PG 33:1|PG 16:0_17:1 | |
| 731.49005 | 4.407 | PG 33:2|PG 16:1_17:1 | |
| 747.52142 | 5.227 | PG 34:1|PG 16:0_18:1 | |
| 745.50616 | 4.751 | PG 34:2|PG 16:1_18:1 | |
| 761.53699 | 5.61 | PG 35:1|PG 17:0_18:1 | |
| 759.52203 | 5.016 | PG 35:2|PG 17:1_18:1 | |
| 775.55298 | 5.948 | PG 36:1|PG 18:0_18:1 | |
| 773.53888 | 5.322 | PG 36:2|PG 18:1_18:1 | |
| Diacylglycerol (DAG) | 586.54303 | 7.577 | DAG 32:0|DAG 16:0_16:0 |
| 584.53033 | 6.908 | DAG 32:1|DAG 16:0_16:1 | |
| 582.51422 | 6.23 | DAG 32:2|DAG 16:1_16:1 | |
| 598.54559 | 7.278 | DAG 33:1|DAG 16:0_17:1 | |
| 614.57684 | 8.293 | DAG 34:0|DAG 16:0_18:0 | |
| 612.55981 | 7.641 | DAG 34:1|DAG 16:0_18:1 | |
| 610.54663 | 7.159 | DAG 34:2|DAG 16:1_18:1 | |
| 626.57391 | 7.997 | DAG 35:1|DAG 17:0_18:1 | |
| 642.60565 | 8.969 | DAG 36:0|DAG 18:0_18:0 | |
| 640.59149 | 8.353 | DAG 36:1|DAG 18:0_18:1 | |
| 638.57568 | 7.692 | DAG 36:2|DAG 18:1_18:1 | |
| Triacylglycerol (TAG) | 768.71069 | 10.794 | TAG 44:0|TAG 14:0_14:0_16:0 |
| 766.69482 | 10.333 | TAG 44:1|TAG 12:0_16:0_16:1 | |
| 796.74341 | 11.257 | TAG 46:0|TAG 14:0_16:0_16:0 | |
| 794.72626 | 10.814 | TAG 46:1|TAG 14:0_16:0_16:1 | |
| 792.7085 | 10.435 | TAG 46:2|TAG 14:0_16:1_16:1 | |
| 810.75525 | 11.472 | TAG 47:0|TAG 15:0_16:0_16:0 | |
| 808.74298 | 11.046 | TAG 47:1|TAG 15:0_16:0_16:1 | |
| 824.77271 | 11.622 | TAG 48:0|TAG 16:0_16:0_16:0 | |
| 822.75842 | 11.276 | TAG 48:1|TAG 16:0_16:0_16:1 | |
| 820.74347 | 10.845 | TAG 48:2|TAG 16:0_16:1_16:1 | |
| 818.72485 | 10.461 | TAG 48:3|TAG 14:1_16:1_18:1 | |
| 838.7934 | 11.838 | TAG 49:0|TAG 16:0_16:0_17:0 | |
| 836.77765 | 11.492 | TAG 49:1|TAG 16:0_17:0_16:1 | |
| 834.75635 | 11.105 | TAG 49:2|TAG 16:0_16:1_17:1 | |
| 852.79919 | 11.729 | TAG 50:0|TAG 16:0_16:0_18:0 | |
| 850.78961 | 11.7 | TAG 50:1|TAG 16:0_16:0_18:1 | |
| 848.77393 | 11.296 | TAG 50:2|TAG 16:0_16:1_18:1 | |
| 846.75836 | 10.867 | TAG 50:3|TAG 16:1_16:1_18:1 | |
| 866.8172 | 11.865 | TAG 51:0|TAG 16:0_17:0_18:0 | |
| 864.80768 | 11.853 | TAG 51:1|TAG 16:0_17:0_18:1 | |
| 862.7915 | 11.503 | TAG 51:2|TAG 16:0_17:1_18:1 | |
| 878.82141 | 11.957 | TAG 52:1|TAG 16:0_18:0_18:1 | |
| 876.80676 | 11.706 | TAG 52:2|TAG 16:0_18:1_18:1 | |
| 874.78894 | 11.313 | TAG 52:3|TAG 16:1_18:1_18:1 | |
| 892.83417 | 12.027 | TAG 54:1|TAG 18:0_18:0_18:1 | |
| 890.8208 | 11.855 | TAG 54:2|TAG 18:0_18:1_18:1 | |
| 906.85333 | 12.085 | TAG 54:3|TAG 18:1_18:1_18:1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Y.; Acket, S.; Beaumont, E.; Galez, H.; Duma, L.; Rossez, Y. Colistin Treatment Affects Lipid Composition of Acinetobacter baumannii. Antibiotics 2021, 10, 528. https://doi.org/10.3390/antibiotics10050528
Tao Y, Acket S, Beaumont E, Galez H, Duma L, Rossez Y. Colistin Treatment Affects Lipid Composition of Acinetobacter baumannii. Antibiotics. 2021; 10(5):528. https://doi.org/10.3390/antibiotics10050528
Chicago/Turabian StyleTao, Ye, Sébastien Acket, Emma Beaumont, Henri Galez, Luminita Duma, and Yannick Rossez. 2021. "Colistin Treatment Affects Lipid Composition of Acinetobacter baumannii" Antibiotics 10, no. 5: 528. https://doi.org/10.3390/antibiotics10050528
APA StyleTao, Y., Acket, S., Beaumont, E., Galez, H., Duma, L., & Rossez, Y. (2021). Colistin Treatment Affects Lipid Composition of Acinetobacter baumannii. Antibiotics, 10(5), 528. https://doi.org/10.3390/antibiotics10050528






