Abstract
The emergence of multidrug-resistant pathogens presents a global challenge for treating and preventing disease spread through zoonotic transmission. The water and foodborne Enterohaemorrhagic Escherichia coli (EHEC) are capable of causing intestinal and systemic diseases. The root cause of the emergence of these strains is their metabolic adaptation to environmental stressors, especially acidic pH. Acid treatment is desired to kill pathogens, but the protective mechanisms employed by EHECs cross-protect against antimicrobial peptides and thus facilitate opportunities for survival and pathogenesis. In this review, we have discussed the correlation between acid tolerance and antibiotic resistance, highlighting the identification of novel targets for potential production of antimicrobial therapeutics. We have also summarized the molecular mechanisms used by acid-adapted EHECs, such as the two-component response systems mediating structural modifications, competitive inhibition, and efflux activation that facilitate cross-protection against antimicrobial compounds. Moving beyond the descriptive studies, this review highlights low pH stress as an emerging player in the development of cross-protection against antimicrobial agents. We have also described potential gene targets for innovative therapeutic approaches to overcome the risk of multidrug-resistant diseases in healthcare and industry.
1. Introduction
Low, acid pH kills bacteria. Humans have evolved acid conditions to protect from food-borne pathogens, and pathogens have evolved ways to bypass those protections. Acid tolerance, the ability to survive acid conditions, is a clinically important phenotype of foodborne pathogens and an overwhelming issue for public health [1,2]. Most microorganisms, including pathogenic bacteria, prefer to grow at pH 6–7 [3]. Globally, the use of acid-based antimicrobial agents is widely practiced because it kills microbes at workplaces, in hospitals, on medical equipment, as food preservatives, in soil niches and during wastewater treatment [2,4]. Commercially, wastewater is treated with nitrous acid (disinfectant) while diluted acetic acid, hypochlorous acid, chlorhexidine, ethanol, acetate, hydrogen peroxide and boric acid are commonly used to treat wounds and infections in hospitals [5,6,7]. Acidified-chlorinated water (a blend of hydrochloric acid and mild organic acids) is often sprayed on meat and lettuce leaves to attain a pH of 2.5 [8,9]. This helps kill certain Enterohaemorrhagic Escherichia coli (EHEC), which causes severe poultry, bovine and human extra-intestinal diseases at a very low infectious dose (10–500 cells) [10,11].
EHECs colonize farm animals and are easily transmitted to humans, especially through poorly cooked beef and raw milk. The uncontrolled use of inappropriate veterinary antibiotics and growth promoters during animal husbandry and farming is a major reason for the spread of EHEC strains [12]. Besides, excessive administration of disinfectants, inadequate hygienic practices and contaminated meat and/or dairy products also facilitate the emergence of acid-tolerant strains. These EHECs can survive in diverse acidic environments (pH 2–3) including soil, farm water, apple cider, meat and even in the human gastrointestinal tract [2,13]. The ability to survive extremely acidic gastric fluid increases the risk of foodborne diseases in humans caused by EHECs [14,15]. This adaptation not only provides survival opportunities but also facilitates several cross-protective benefits, including enhanced antimicrobial resistance, biofilm formation, pathogenic adhesion, and colonization [16,17].
More than 400 serotypes of EHECs are known to cause several life-threatening diseases, such as hemorrhagic colitis, intussusception, bloody diarrhea, inflammatory bowel disease, and systemic hemolytic uremic syndrome (HUS). HUS is a multi-symptomatic syndrome caused by the highly prevalent serotype O157:H7. Infected patients suffer from thrombocytopenia, acute renal failure, and hemolytic anemia leading to death [18]. In severe circumstances, multi-organ failure has been reported [18]. EHECs epitomize overwhelming health concerns, especially in newborns, children, and immunocompromised patients; these patients suffer high rates of morbidity and mortality [19]. Globally, the well-reported virulent serotypes of EHECs are O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, O157:H7, and O157:H. Among these, EHEC O157:H7 strains are highly virulent and have been found responsible for several foodborne outbreaks across the globe having a mortality rate of 5% [20,21,22,23]. More importantly, newly emerging EHECs, especially EHEC O157:H7 and EHEC O80:H2 serotype strains, are multidrug-resistant (MDR-EHECs); no effective antibiotic has been reported for treating diseases caused by these strains [18,21,24,25]. EDL933, a well-known EHEC O157:H7 strain, first reported in 1983, affected 47 people in Oregon and Michigan [26]. So far, this deadly strain is reported to cause 73,000 illnesses, 2200 hospitalizations, and 60 deaths annually in the United States [27]. In addition, the EHEC O80:H2 strain was reported in France where it caused severe symptoms of HUS in association with bacteremia; all antibiotics were ineffective [25]. Similarly, another EHEC serotype, O104:H4, was also found resistant to almost all known antibiotics in Europe; it resulted in 3800 disease cases and 53 deaths [25,28]. These strains possess multidrug-resistance-encoding regions that provide enhanced resistance to several known antibiotic determinants, including chloramphenicol, aminopenicillin, cefotaxime, neomycin, aminoglycoside, sulfamethoxazole, nalidixic acid, beta-lactams, cotrimoxazole, amoxicillin-clavulanic acid, imipenem, norfloxacin, tetracycline, phenicols, streptomycin, trimethoprim, ciprofloxacin, kanamycin and carbapenems [18,19,29,30,31,32,33,34].
Antibiotic resistance genes can be transmitted from animals to humans in several ways; for EHECs, the foodborne route is probably the most important. In EHECs, the multidrug-resistance genes are encoded by plasmids that can easily be transmitted from one organism to another through horizontal gene transfer or bacteriophages, resulting in the emergence of zoonotic infections in humans [18,19,30]. Using antibiotics for the treatment of EHEC infections has long been controversial as several studies correlate the use of antibiotics with death and an increased rate of cerebellar hemorrhage [25,35]. It has been suggested that antibiotics targeting DNA synthesis (quinolones and ciprofloxacin) should not be used during acute EHEC infections, as treatment increases several systemic complications [36,37]. For example, treatment with fluoroquinolones induces Shiga toxin secretions, resulting in even higher mortality rates [25,36,38,39]. These complications are highly dependent on several factors including, type of antibiotic used, dose of antibiotic, time of administration, route of antibiotic administration, type of EHEC strain and severity of the infection [35].
EHECs are widely distributed in domestic ruminants (sheep, goats, pigs, turkeys and cattle) and use food as a vector to infect humans. Designing effective intervention technologies and risk-management options are required to overcome antimicrobial resistance in the food chain. These foodborne pathogens suffer from various physical and chemical stresses during cooking, such as heating, freezing, acid, and salt treatments [40]. These treatments can efficiently kill certain pathogens; however, pathogens that survive these treatments become genetically and physiologically strongly adapted. Fecal contamination of water and ingestion of EHEC-contaminated food products (meat, milk, raw vegetables) create a greater risk of transmission of resistant genes from pathogenic bacteria to commensal gut flora [41,42]. However, the extremely acidic pH of the mammalian gastrointestinal tract can kill almost all types of microbes, except acid-tolerant microbes. They can then transfer resistance genes to microbial flora in the gut [43]. EHECs are well known for their adaptation to the acidic environment; their tolerance level is comparable to that of acidophiles. Furthermore, foodborne, acid-adapted strains also confer cross-protection to antibiotics, which plays a vital role in the spread of multidrug-resistant pathotypes [16,44,45,46,47,48]. The human gastrointestinal tract is reported to provide the best environment for the emergence, transmission, and spread of antibiotic-resistance genes in bacterial populations. Several factors assist this transmission of genes from one bacterium to another including high cell density, antibiotic exposure and innate ability of gene transfer [43]. The human body appears to serve as an “antibiotic resistance gene bank” to generate resistant pathotypes, which may emerge as a great public health challenge [49,50].
All EHECs are highly acid-tolerant, especially, EHEC O157:H7 strains, which are known to be the best-adapted strains. The resistance potential of all EHEC O157:H7 strains against almost all marketed antibiotics is posing a challenge to treat life-threatening diseases caused by these strains. These strains employ sophisticated antibiotic inactivation, structural modifications, target replacement and antibiotic efflux activation mechanisms [19]. There are several ways through which different antimicrobial agents target intracellular processes by blocking the binding proteins, affecting cell division, modifying ribosomal proteins, and causing competitive inhibition [51]. In penetrating the cell, these antimicrobial agents first need to breach the outer membrane of the bacterial cell [52].
Several antimicrobial agents tend to kill pathogens by affecting the integrity of the outer membrane. The outer membrane of Gram-negative bacteria is composed of a negatively charged hydrophobic lipid bilayer (lipopolysaccharides) and pore-forming proteins. The negatively charged lipopolysaccharides can easily be disrupted by positively charged antimicrobial peptides (cationic antimicrobial peptides). Therefore, lipopolysaccharide modification provides a way to protect against outer-membrane disruption [52].
The involvement of various two-component systems (TCS) and global regulators also facilitate the expression of various efflux pumps and competitor proteins. Microorganisms use efflux pumps to regulate their internal environment by eliminating harmful compounds such as metabolites and antimicrobial agents [53]. In acid-adapted EHECs, the activation of multiple efflux pumps is one of the major strategies for developing antimicrobial resistance [54,55,56,57]. Understanding the relationship between acid tolerance and genetic adaptability demands detailed insight into cellular responses to changing environments [54,58]. This review examines the expression of genes that are induced by acidic pH and how acid tolerance facilitates antibiotic resistance.
2. Acid Tolerance Potential of EHECs
Extremely virulent serotypes of EHECs (O157:H7 strains) have regulatory aspects of acid tolerance that are not found in other E. coli strains [59]. Although EHECs can acquire antibiotic resistance by horizontal gene transfer, they can also develop de novo resistance during exposure to various environmental stresses, especially low pH. The mechanisms that initially allow the bacteria to survive stress subsequently result in resistance to even higher antibiotic concentrations measured by minimum inhibitory concentration (MIC) [60]. The set of genes involved in protecting against acid stress are also responsible for the acquisition of antibiotic resistance [51]. Thus, it is important to understand the regulation of several transcriptional regulators (GadE, H-NS) and two-component signal transduction kinases (EvgAS, PhoPQ, RcsB) that are activated in response to low pH. At the molecular level, key players involved in mediating acid tolerance include enzymatic cascades of specific decarboxylases and families of two-component signal-transduction kinases.
The signal-transduction kinases consist of a sensor kinase and a response regulator that controls acid-tolerance, pathogenicity, and antibiotic resistance [61,62,63,64]. These two-component kinases work in coordination with multiple global regulators, transcription factors, and several other local regulatory proteins [1,14,65,66]. This regulation further involves several regulatory proteins, chaperons, and periplasmic proteins that protect the EHECs against DNA damage and protein coagulation.
Four main systems regulate acid tolerance: oxidative, glutamate-dependent, arginine-dependent and lysine-dependent acid resistance systems. They work in tandem to protect cells from acid stress (Figure 1; Table 1) [67,68,69,70,71,72]. These mechanisms exchange intracellular protons for an amino acid (glutamate, arginine, or lysine) and expel amines into the extracellular media in exchange for the corresponding amino acid [73].
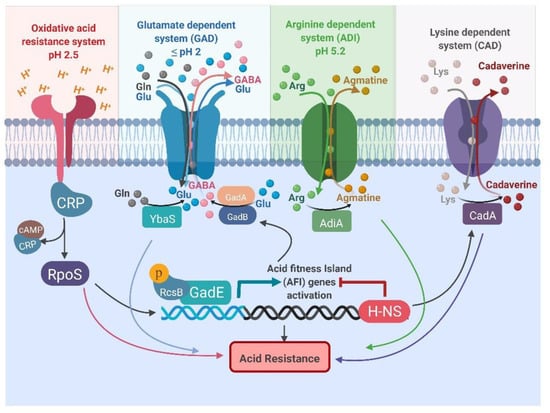
Figure 1.
Representation of the oxidative, glutamate-dependent, arginine-dependent, and lysine-dependent acid resistance systems in Escherichia coli. All abbreviations are listed at the end of the manuscript.
The oxidative system does not involve an externally-derived amino acid; it is regulated by sigma factor RpoS and catabolite repressor protein (CRP). It provides the lowest level of protection at pH 2.5 at the expense of energy, as shown in Figure 1 [74]. The glutamate-dependent system is adapted to protect under extremely acidic conditions and is highly efficient. The arginine-dependent system is only induced under anaerobic conditions. It requires arginine decarboxylase (AdiA) and an arginine:agmatne antiporter (AdiC) to provide a modest level of protection under mild acidic conditions (Table 1) [75]. The lysine-dependent system also works under slightly acidic environments; the efficiency of resistance is lower than seen with other systems [68,76]. This system involves a lysine decarboxylase (CadA) and a bifunctional lysine:cadaverine antiporter (CadB) (Table 1). The pH inside certain compartments of the mammalian gastrointestinal tract drops below 2; which renders all acid resistance systems inactive except the glutamate-dependent acid one.

Table 1.
Overview of the genes involved in acid resistance regulatory systems in E. coli.
Table 1.
Overview of the genes involved in acid resistance regulatory systems in E. coli.
| Protection Mechanism | Main Substrate | Decarboxylases | Antiporter | Final Product | Regulators | Level of Protection | pH | Reference |
|---|---|---|---|---|---|---|---|---|
| Oxidative system | Glucose | - | - | - | RpoS | Least | 2.5 | [77,78,79] |
| Glutamate dependent system (GAD) | l-Glutamate | GadA, GadB | GadC | GABA | GadE, GadX, GadW | Highest | ≤2 | [67,75,79,80,81,82] |
| Arginine dependent system (ADI) | l-Arginine | AdiA | AdiC | Agmatine | - | Modest | 5.2 | [75,79] |
| Lysine dependent system (CAD) | Lysine | CadA | CadB | Cadaverine | CadC | Quite ineffective | NA | [75,79] |
Note: All abbreviations are defined at the end of the manuscript.
2.1. Glutamate-Dependent Acid Resistance System
The glutamate-dependent system provides the highest level of protection under extremely acidic conditions. This system involves two glutamate decarboxylases, GadA and GadB, that work in coordination with the gamma-aminobutyric acid (GABA) antiporter (GadC) and a set of these three genes known as GAD [83]. Extracellular glutamate is exchanged with intracellular GABA through the GABA antiporter GadC and subsequently decarboxylated by GAD [84]. During the decarboxylation of L-glutamate, the α-carboxylic group is released as carbon dioxide and a proton is incorporated into the GABA molecule, which is exported across the inner membrane in exchange for more glutamate through GadC [67]. In addition, the antiporter increases the availability of glutamate to the GAD enzymes, thereby, enhancing the efficiency of the system by acidifying the cytoplasm [85].
The functional side chain of glutamate imported by GadC has a pKa of 4.1. Before entering the cytoplasm, during acid stress (pH 2.5), this side chain gets more than 50% protonated and these protons dissociate to acidify the cytoplasm. Therefore, the cytoplasmic pH drops to 3.6, which is an optimal pH for glutamate decarboxylase while rendering arginine and lysine decarboxylases inactive, as their optimal pH is 5.25 and 5.5, respectively [86,87]. This defense strategy works by reversing the membrane potential to maintain more protons inside as compared to the external environment [76]. The inner membrane potential remains more positive and gradually slows the flow of protons into the cell, thereby maintaining homeostasis.
2.2. Control of Glutamate-Dependent System
The gap between an environmental stimulus and gene regulation is bridged by sensors and regulators of two-component systems. A two-component system typically consists of a sensory kinase that monitors the environmental conditions and modulates phosphorylation of the respective response regulator. The response regulator then regulates gene expression, which changes the behavior of the bacterial cell. To cope with acid stress in the gastrointestinal tract, several two-component systems play specific roles in maintaining homeostasis and cell integrity. The selection of resistance mechanism depends upon the energy source and extracellular environmental conditions.
The protection conferred by the glutamate-dependent system is significantly higher than the other systems, allowing up to 80% survival. As a consequence, the glutamate-dependent system is considered a key player in acid regulation. This system comprises a complex network of two-component regulators with a wide array of interactions to cope with mild to extreme acid stress. The key interacting regulators of this network are GadE, EvgAS, PhoPQ, YdeO, GadW, RcsB, and GadX, which regulate gene expression spatially and temporally (Figure 2) [44,67,87,88,89,90,91,92,93,94,95,96,97]. This system is activated either by mild acidic pH during the exponential growth phase or by entry into stationary phase. Two-component systems are regulated by the induction of mild acid shock, while the rpoS-gadX-gadY-gadW circuit is activated during stationary phase. Once triggered, this system activates a cascade of regulatory genes that then activate the central regulators GadE and YdeO (Figure 2) [87,95,98,99]. The increased expression of central regulators results in the activation of several acid-resistance genes at different loci. This activation involves more than 20 proteins (CRP, Dps, EvgA/S, GadE, GadX, GadW, H-NS, Lon, PhoP/Q, RNaseE, sigma factor 70, sigma factor RpoS, SspA, TrmE, TopA, TorS/R and YdeO) and several non-coding RNAs (DsrA, GadY, and GcvB) (Figure 2) [91,99,100,101,102,103,104,105,106,107,108].
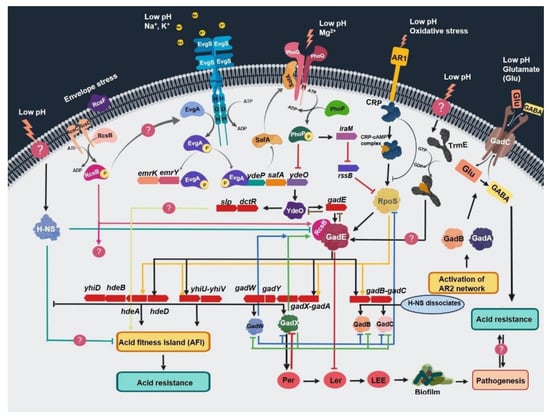
Figure 2.
Schematic representation of acid stress regulation by different two-component signal transduction systems, acid-resistance networks, and their interconnecting assemblies. All abbreviations are defined at the end of the manuscript.
The sensor kinase EvgS detects the low pH signal and activates response regulator EvgA, which then starts a gene transcription cascade leading to activation of the ydeP–safA–ydeO circuit. The activated YdeO increases the expression of GadE and other genes involved in the regulation of acid-fitness-island (AFI) genes, namely, slp–dctR–yhiD–hdeB–hdeA–hdeD–yhiU–yhiV–gadW–gadY–gadX–gadA. It also activates glutamate-dependent acid-resistance genes, namely, gadA, gadB, and gadC (Figure 2). Activation of these genes and gadE requires the heterodimerization of RcsB with GadE. Activation of GadX also stimulates the LEE (locus of enterocyte effacement) to mediate acid-induced regulation of pathogenic traits including biofilm formation, multidrug resistance, and enhanced colonization, as shown in Figure 2.
2.2.1. EvgAS: An Acid-Resistance Regulator
EvgAS is indispensable for protecting against low pH through a range of interacting mechanisms that depend upon the conditions (stress in exponential phase or entry into stationary phase) [90]. Under acid stress, the sensory kinase EvgS phosphorylates the response regulator EvgA. The activated EvgA then phosphorylates a transcriptional regulator YdeO (the AraC/XylS super-family transcriptional regulator). This phosphorylation depends on a small membrane protein SafA (sensor-associating factor A) from the ydeO-safA operon. The phosphorylated YdeO ultimately activates GadE, which regulates various decarboxylases and provides resistance against acid stress. It also regulates several other cellular processes as shown in Figure 2 [109]. EvgA strongly binds to the promoter regions of several genes involved in the regulation of acid resistance, such as ydeP, safA, yfdX, frc, yegR, and gadE. The role of YdeP, YfdX and YegR needs to be investigated in the context of acid resistance.
2.2.2. PhoPQ: Role in Acid Regulation
PhoPQ is a two-component signaling system that responds to multiple environmental stimuli, including low pH, osmotic shock, low concentration of divalent cations and antimicrobial peptides (AMPs) [103,110,111]. It consists of a histidine kinase PhoQ that interacts with SafA and starts a phosphorylation cascade resulting in activation of the response-regulator PhoP. During the exponential phase, transcription factor PhoP activates IraM, which then interacts with RpoS [112]. RpoS is a central regulator of the stress that encodes sigma factor-38 and allows the cell to survive environmental challenges [113]. Due to this interaction, the level of RpoS increases and subsequently recruits RNA polymerase (RNAP) to RpoS-regulated promoters, including the gadE promoter [114,115]. Mg2+ stress concurrently activates PhoPQ and other regulatory proteins, thereby enhancing pathogenesis by increasing pathogen survival [94].
2.2.3. RcsB: An Essential Activator/Repressor
RcsB is a response regulator that functions both as an activator and a repressor. It works in coordination with GadE to form a heterodimer on the GAD box that activates transcription of acid-resistance genes (Figure 2). It is an essential element for gadA and gadB promoter activity [82,83,84,85]. All acid-resistance promoters activated by GadE are also dependent on RcsB for their activation; the regulation mechanism of RcsB is still unknown.
3. Cross-Protection in EHECs
Cross-protection is the defensive adaptability of a strain when exposed to certain environmental stresses, including acid stress. Cross-protection mechanisms are either non-specific for the choice of substrate (multidrug efflux pumps), share a few common regulatory sets of genes (glutamate-dependent pathway genes), or undergo structural modifications (lipopolysaccharide chain modification) [116,117]. Foodborne EHECs encounter several acidic treatments from farm to gut and gradually adapt. This exposure to acidic conditions helps them develop cross-protection against other environmental stresses, including antimicrobial agents [118]. In addition to EHECs, acid-adapted pathogens other than EHEC pathotypes’ have also been reported in several major outbreaks all over the world, as shown in Table 2. This cross-protection poses serious concerns to the consumers (humans) and could lead to the emergence of new pathotypes.

Table 2.
Acid-adapted pathogens other than EHEC pathotypes’ and their acquired antibiotic resistance.
4. Metabolic Adaptations
Once ingested, EHECs experience severe environmental challenges including extreme pH fluctuation and nitrosative stress (nitric acid) from volatile organic acids formed as a result of anaerobic fermentation in the gastrointestinal tract [30]. Mostly, EHECs favor pH 6–8 for their growth; to survive low pH stress they develop a transmembrane gradient to maintain homeostasis [2,126,127]. When grown at acidic pH, genes involved in metabolism, energy production and class I heat shock proteins are down-regulated to lower metabolic cost [128]. These strains consume intracellular protons through amino acid decarboxylation during acid stress, which highly acidifies the cytoplasm, resulting in increased acid tolerance [44,76]. During oxidative respiration, electron transport of membrane-bound systems, including the atp operon, is down-regulated to inhibit the import of protons [129]. While in the intestine, the enhanced expression of the Long Polar Fimbriae gene (lpf-2) mediates bacterial colonization in response to anaerobic nitrosative stress [126]. Genes involved in motility, type III secretion system (T3SS), bacterial chemotaxis, biofilm formation, adhesion, iron uptake and oxidative resistance are upregulated [1,127]. Cellular adhesion capacity (the intimin gene eae) of EHEC O157:H7 is enhanced by the histone-like, nucleoid-associated H-NS protein that regulates bacterial fitness and uncontrolled virulence [1,30]. In addition, the expression of fur, which is involved in iron uptake, is also up-regulated. In particular, low pH helps bacteria survive acid stress by enhancing motility, adhesion, and iron utilization, thereby assisting the pathogen in enhancing apoptosis of epithelial cells and become more virulent [2]. This mechanistic regulation helps the pathogen achieve homeostatic balance by modifying metabolic pathways at the cost of energy generated from redox or ATP-driven reactions.
5. Acid-Adaptive Antibiotic Resistance Strategies
Generally, growth-inhibiting stresses, such as low pH, high temperature, or nutritional deficiency, induce several metabolic rearrangements at the cellular and metabolic levels that influence differential regulation of more than 500 genes to ensure tight homeostasis. In response to extracellular acid stress, pathogens undergo several cellular and global transcriptional changes that alter their responsiveness to a wide array of antibiotics. These regulatory changes help the organism adapt to extreme environmental stresses and subsequently enable cross-protection consistent with the survival of the organism [44,60].
While passing through the gastrointestinal tract, EHECs experience anaerobic conditions and nitrosative stress that trigger enhanced expression of the multidrug efflux genes (mdtEF) and several two-component signaling systems including EvgAS, PhoPQ, RcsB, PmrAB, ArcAB, BaeSR, KdpA, and CpxAR [44,130,131,132]. These two-component systems combat cell envelope disruption caused by proton imbalance and antibiotic-induced accumulation of mistranslated peptides that can cause cell damage by disturbing homeostasis [133]. Multidrug-resistance efflux pumps are essential for withstanding antibiotic challenges and other environmental toxins. During anaerobic conditions, the global transcription factor ArcA increases the expression of MdtEF (more than 20 fold), which dramatically enhances efflux activity leading to antibiotic resistance [130]. Incubation at low pH also aids the development of antibiotic resistance, which persists even after environmental conditions shift [16,60]. Overall, these stress-induced genetic alterations confer genetic plasticity that results in enhanced population diversity, strengthening of the envelope and resistance to a wide array of antibiotics.
5.1. Acid-Adaptive Structural Modifications
Gram-negative bacteria have a highly asymmetric outer membrane with phosphatidylethanolamine lipids at the inner side, while the external side is enriched with lipopolysaccharides [134,135,136]. The lipopolysaccharide membrane is composed of three vital parts, including a gel-like hydrophobic anchor (lipid A), branched oligosaccharides (core region), and polymer of repeating saccharide subunits (O-antigen) (Figure 3) [135,136]. The structure of this membrane is enriched with many phosphoryl and carboxyl groups bridged with divalent cations that facilitate low permeability and antibiotic resistance [134,136].
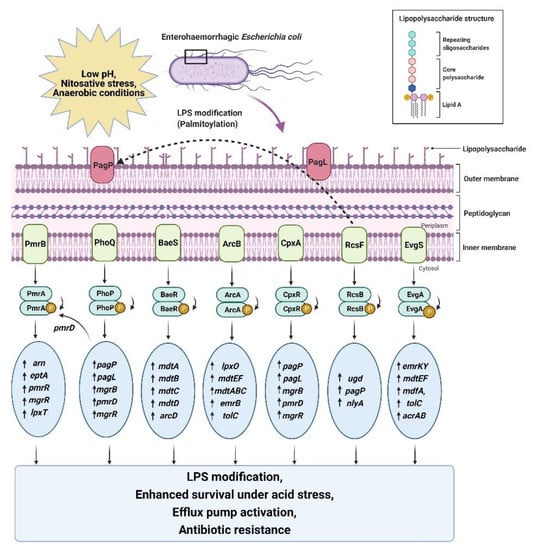
Figure 3.
Schematic representation of the acid-induced activation of a specific set of genes by two-component systems leading to LPS modification, efflux pump activation, enhanced survival, and antibiotic resistance in EHECs. All abbreviations are defined at the end of the manuscript.
5.1.1. Acid-Induced LPS Modification
PhoPQ upregulates the transcription of acid-resistance genes under acidic stress; the same set of genes also mediate LPS modifications in EHECs [103,137]. Several environmental changes, such as acidic pH, osmotic stress, low concentration of divalent cations (Mg2+), and the presence of antimicrobial peptides (AMP), trigger this pathway [103,134]. EHECs have evolved this defensive strategy to remodel the outer membrane by adding a palmitoyl chain, a hydroxyl group, and a positively charged aminoarabinose sugar moiety to the lipid A anchor, acetylation of the O-antigen and hydroxylation of fatty acids through the PhoPQ two-component regulatory system (Figure 3) [111,137]. These induced modifications help EHECs become more virulent by increasing cationic antimicrobial peptide resistance and suppressing TLR4 immune responses. They also increase permeability to large lipophilic agents [138]. EHEC serotype O157:H7 is reported to develop increased resistance to cationic antimicrobial peptides, especially Polymyxin B, in response to acid stress, bile salts, and ferric ions in the human gut (Table 3) [103,137].
5.1.2. Acid-Induced Antimicrobial Resistance by RcsB
The bacterial cell envelope comprises outer and inner membranes that act as a protective barrier. The outer membrane is an asymmetric bilayer of phospholipid and lipopolysaccharides (LPS). There is a thin peptidoglycan layer lying in the periplasmic space between the outer and inner membrane of the cell. EHECs are at high risk of losing cell envelope integrity and improper protein folding under extremely acidic conditions due to excessive osmotic pressure [139,140,141]. Penicillin-binding proteins (PBPs) keep adding new subunits to the outer membrane during cell growth and repair. Genetic profiling of EHECs confirmed that pH stress inhibits penicillin-binding proteins, which in turn activates the rcs phosphorelay to retain envelope integrity and develop resistance to amdinocillin (mecillinam) and cefsulodin (a member of the beta-lactam group of antibiotics) [141]. When acid stress is encountered, the expression of ugd is increased and incorporates a 4-amino acid modification to the lipid A anchor (Figure 3) [142]. The expression of PagP is also increased, which regulates lipid A palmitoylation, thereby limiting bacterial recognition by the host immune response. Activated PagP also triggers the expression of RcsB-GadE-regulated genes cpsB, rprA, gadA and gadB [96,142]. The Rcs phosphorelay cascade is widely distributed among EHECs; knockout mutants of rcsB are hyper-susceptible to beta-lactams, which suggests that RcsB is a global regulator of cell envelope integrity [139,140,141].
5.1.3. CpxAR-Mediated Peptidoglycan Cross-Linking
The Cpx-TCS (conjugative pilus expression) is a well-studied TCS that counters cell envelope perturbations. Most induction stimuli for Cpx include misfolded proteins, alkaline pH, salt, changes in lipid composition and attachment to abiotic surfaces. Acid-induced activation of Cpx regulates proton influx and cell wall stability by influencing membrane porins and cross-linking between lipopolysaccharide and peptidoglycan in the outer membrane [143]. Under low pH stress, the activated CpxRA upregulates the expression of several proteins including CydAB, GadAC, CadA, and HdeABD [143]. Most of the activated genes are controlled by the glutamate-dependent acid resistance system that extends the function of Cpx acid-induced tolerance.
During acid stress, several Cpx-regulated proteases, multidrug efflux genes and peptidoglycan amidase genes also reduce susceptibility to cationic antimicrobial peptides (polymyxin B), aminoglycosides (kanamycin), novobiocin and beta-lactams [141,144,145]. Knockout mutants of the Cpx two-component system reduce susceptibility towards antimicrobial agents, as shown in Table 3 [141,145].
5.2. Target Replacement
PhoPQ- and PmrAB-Mediated Competitive Inhibition
During stress, PhoP regulates the transcription of several stress-responsive and virulence pathway genes, including pagL, pagP, and pmrD, to induce modifications in the lipid A anchor (Figure 3). PmrD is a small regulatory RNA that triggers PhoPQ-mediated activation of PmrAB [111,146,147]. Under acid stress, histidine and glutamate residues of PmrB sense low pH and phosphorylate PmrA. Activated PmrA triggers the arn operon and eptA, which then modify the aminoarabinose and phosphoethanolamine residues in lipid A, respectively [111,148]. In parallel, PmrR blocks the regulatory domain of lipid A phosphotransferase LpxT (a competitive inhibitor of EptA) and facilitates EptA-mediated phosphoethanolamine modification. These PmrA-dependent modifications confer resistance to cationic antimicrobial peptides, including polymyxin. WD101 (a pmrA mutant strain) showed 40-fold lower susceptibility to polymyxin compared with its isogenic parent, W3110 [111,149].
DNA microarray studies confirmed that coupling of the above mentioned two-component systems results in significantly reduced susceptibility for polymyxin B and colistin (both are membrane-disrupting CAPs) [51,137]. Polymyxin B and colistin are last-resort antibiotics for multidrug-resistant EHECs [51]. The lipopolysaccharide membrane serves as the first site for interacting with, as well as combating against, cationic antimicrobial peptides. Thus, the two-component systems trigger genes responsible for structural modification that prevents binding of antimicrobial peptides.
5.3. Acid-Adaptive Activation of Drug Efflux Pumps
In Gram-negative bacteria, multidrug-resistant efflux pumps play an indispensable role in exporting toxins or harmful metabolites and antimicrobials of different families across the inner and outer membranes. Thus, efflux pumps decrease intracellular drug concentration.
5.3.1. Activation of EvgAS-Regulated Drug Efflux Genes
EvgAS regulates multiple regulatory mechanisms including acid tolerance, drug efflux transporters and bacterial drug-resistance pathways. Extracellular acid stress leads to cytoplasmic acidification that permits EvgA to activate emrKY, mdtEF, mdfA, tolC and acrAB drug efflux (TolC-dependent pumps) genes [150,151]. Low pH induces the expression of emrAB and emrKY multidrug-resistance efflux genes, which confer a growth advantage and facilitate multidrug resistance against extended-spectrum β-lactamases (ESBLs) (Table 3) [150,151,152,153]. In addition to the development of resistance to ESBLs, the evgA-ydeO-gadE regulatory cascade also facilitates antimicrobial tolerance to other drugs, including gallium nitrate (GaNt) [91]. Gallium nitrate is an FDA-approved drug widely used for the treatment of carcinogenic hypercalcemia and is effective against several clinically significant MDR bacteria. Advanced genomic techniques confirmed the gain-of-function mutation in the EvgSA two-component system in these tolerant strains [91]. Deletion mutants of evgS and evgA failed to confer GaNt tolerance. The regulation of GaNt tolerance by EvgS substitutional mutant (E701G) depends on phosphor-transfer from EvgS to EvgA. The phosphorylated EvgA up-regulates the transcription of safA, ydeO, and gadE. Deletion of gadE in the E701G mutant failed to confer GaNt tolerance, while deletion mutants of ydeO and safA showed partial reversal of tolerance. Thus, GadE acts as a key regulator of EvgS mediated GaNt tolerance and is the central regulator of glutamate-dependent acid resistance system [91].
5.3.2. Activation of the KdpA Proton Pump
Potassium ions are needed for a variety of cellular functions, including intracellular pH regulation and cross-membrane potential. KdpA is a part of the KdpFABC ion channel involved in the ATP-driven transport of potassium ions across the cytoplasm [154]. Recent comparative studies on acid-adapted and non-adapted EHEC strains revealed activation of the KdpA proton pump in response to low pH [16]. During acid stress, this system blocks the flow of protons across the cell to increase the survival rate by more than 100 hrs [16]. Transcriptomic studies revealed upregulation of KdpA, BhsA (outer membrane protein), and ArnA in acid-adapted E. coli O157:H7 strain [16,155]. The enhanced expression of ArnA confers resistance to polymyxin B and colistin in growth cultures (Table 3). BhsA renders the outer membrane hydrophobic by modifying the lipopolysaccharides in a way that renders the cell surface more hydrophobic than hydrophilic [155]. This outer-membrane modification helps by-pass disruptive damage from cationic antimicrobial peptides and also increases cell aggregation [155]. These findings confirm that the KdpFABC ion channel regulates the development of antibiotic resistance in acid-adapted EHEC strains.

Table 3.
Role of different two-component systems involved in mediating antibiotic resistance in response to acid stress. All abbreviations are listed at the end of the manuscript.
Table 3.
Role of different two-component systems involved in mediating antibiotic resistance in response to acid stress. All abbreviations are listed at the end of the manuscript.
| Treatment under Acid Stress | Two-Component Systems Involved | Acquired Antibiotic Resistance/Tolerance | Phenotypic Expression 1 | Reference |
|---|---|---|---|---|
| ΔtatC, over-expressed nlpE | CpxRA | Cationic antimicrobial peptides (CAPs) | Increased tolerance | [145] |
| ΔrcsF, ΔrcsB, ΔcpxR | RcsCB, CpxRA | Mecillinam and cefsulodin | Increased tolerance | [139] |
| ΔcpxR | CpxRA | Cephalexin | Increased tolerance | [156] |
| W3110 tolC732::kan, W3110 acrB747::kan, W3110 mdtB774::kan, W3110 mdtF769::kan, W3110 emrY776::kan, W3110 emrB767::kan, W3110 marR751::kan | MarRAB, AcrAB, EmrKY, MdtABC and TolC | Extended-spectrum β-lactamases (ESBLs) | Increased tolerance | [152] |
| Δmar | MarRAB, AcrAB and TolC | Beta-lactamase, rifampicin, spectinomycin, streptomycin, tetracycline, nalidixic acid | Increased resistance | [157] |
| ΔrcsF and ΔrcsB | RcsBC | Cefsulodin | Increased tolerance | [139] |
| RcsBC, CpxRA, BaeSR | Mecillinam and cefsulodin | |||
| baeR cloned pTrc99A plasmid | BaeRS, MdtABC, ArcAB | Ceftriaxone, | 8 fold increased resistance | [158,159] |
| novobiocin, | ||||
| deoxycholate | ||||
| pH stress only | ArcAB, MarRAB | Ceftriaxone, | Presence ofhyper-resistantcolonies | [60] |
| amikacin, | ||||
| nalidixic acid | ||||
| ArcAB, MarRAB, MdtABC | Multidrugresistance | [144,160] | ||
| RcsCB | Cationic antimicrobial peptides (CAPs) | Intrinsic resistance | [161] | |
| Aztreonam | [162] | |||
| Beta-lactams | [161] | |||
| Daptomycin | [96,163,164] | |||
| ΔdpiA, ΔcpxR | RcsBC, CpxRA | Ampicillin | Increased tolerance | [165] |
| ΔpmrA, ΔpmrB | PmrAB, arn operon | PolymyxinB | Increased tolerance | [137] |
| ΔacrB | BaeSR, RcsBC, CpxRA, EvgAS, ArcAB | Multidrug resistance | 16- to 32-fold increased resistance | [166] |
| ΔmarR | MarRAB | Norfloxacin | Increased tolerance | [44]. |
| ΔacrBΔevgAS, ΔacrBΔemrKY, ΔacrBΔyhiUVΔemrKY, ΔacrBΔyhiUVΔemrKY/pUCevgA | ArcAB, EvgAS, EmrKY | Multidrug resistance | 4 fold increased resistance | [167,168] |
| Overexpression of baeR, evgA, rcsB | BaeSR, RcsBC, CpxRA, EvgAS, ArcAB | Multidrug resistance | 16- to 32-fold increased resistance | [166] |
1 Resistance is an increase in MIC above the breakpoint; tolerance is the loss of killing with no change in MIC.
5.3.3. Activation of TolC-Dependent Efflux Pumps
Nitrosative stress is a type of acid stress that is induced by the high concentration of nitric acid in the gastric fluid. This stress affects the transcription of several regulatory proteins [169]. EHECs activate several multidrug-resistance efflux pumps that contribute to both intrinsic and acquired antibiotic resistance [169,170].
AcrAB-TolC Regulation under Anaerobic Conditions
AcrAB-TolC is a resistance nodulation division (RND-type) efflux pump that contains an outer membrane channel (TolC), an inner membrane channel (AcrB), and a periplasmic protein (AcrA). As a housekeeping TCS, it is constitutively expressed to provide intrinsic resistance towards various toxins [130]. Under anaerobic conditions, AcrA (response regulator) triggers the upregulation of acid-induced efflux genes (gadE-mdtEF operon) by more than 20-fold (the activation of the gadE-mdtEF operon under aerobic conditions is controlled by EvgSA [130]). TolC is also reported to enhance GAD-EvgA acid tolerance, while bile salts and fatty acids present in the stomach trigger AcrAB-mediated activation of another global regulator rob [152,170]. This enhancing regulation of efflux genes results in increased drug resistance and survival of EHECs under nitrosative anaerobic environmental conditions in the human gut. Significantly reduced survival rate has been reported in knock-out mutant strains of MdtEF and MdtABC (BaeSR regulated efflux pump) [130,152]. Further studies confirmed that AcrAB-TolC deletion mutants showed attenuated colonization in mice and chickens [170]. In contrast, under extremely acidic conditions, knockout mutants of tolC, emrB, mdtC, and mdtB showed extremely low survival rates (Table 3) [152]. These findings suggest a role for efflux pumps in the development of multidrug resistance and enhanced survival rate for EHECs while passing through the stomach.
Activation of Multiple Antibiotic Resistance Operon
Members of the enterobacteriaceae family have a locus called the multiple antibiotic resistance (marRAB operon), which can confer cross-resistance to several antibiotics including tetracycline, ampicillin, norfloxacin, chloramphenicol, nalidixic acid, and β-lactams [171]. The MarR transcriptional regulator belongs to the AraC/XylS regulatory family that is responsible for inducing adaptive changes in response to environmental stress. Antibiotic resistance induced by the Mar operon is influenced by low pH-mediated acidification of the cytoplasm [44]. Experimental studies show that norfloxacin-sensitive, wild-type EHECs display a significantly enhanced norfloxacin-resistant phenotype when subjected to low pH (Table 4) [44]. Acid-triggered mar regulation also upregulates the transcription of inaA, whereas deletion mutants of this gene showed increased chloramphenicol and nalidixic acid resistance. Several studies also show that inaA is located within the mar locus [73,172,173,174,175,176].
BaeSR: Multidrug-Resistance Efflux Pump Regulator
BaeSR is one of the stress-triggered systems involved in the regulation of TolC-dependent multidrug efflux pumps (MdtABCD) and Spy (periplasmic chaperone) (Figure 3) [177,178,179]. As mentioned earlier, knockout mutants of mdtABC showed a significantly reduced survival rate under extremely acidic conditions (Table 3) [152]. As expected, Overexpression of the response regulator BaeR results in enhanced expression of MdtA and the AcrD efflux pump that mediates beta-lactam, cephalosporin and novobiocin resistance in large mammals (calves, pigs, and chickens) (Table 3 and Table 4) [151,179].
5.3.4. Prophage-Encoded AraC-Like Transcriptional Regulators
EHEC serotype O157:H7 strains possess a locus of enterocyte effacement pathogenicity island (LEE-PAI), which regulates virulence genes of T3SS, intimin and Tir (translocation receptor) [59,61]. These genes are required to colonize, adhere to, produce intestinal lesions, and destroy intestinal microvilli. When EHECs occupy favorable environmental niches in the host intestine, LEE-PAI causes many virulence factors to be expressed [59]. Expression of LEE-PAI genes is tightly regulated by a set of transcriptional regulators (GadE, QseA, H-NS, IHF (integration host factor), Ler, and GrlA). Further studies showed that prophage-encoded loci of EHEC O157:H7 strains (specifically the EDL933 strain) carry a set of AraC-like transcriptional regulators PatE, PsrA, and PsrB. Mutational studies of these genes suggest that PatE and PsrB act as positive regulators of glutamate-dependent acid-resistance genes and trigger several key virulence determinants in acidic environments [59].
6. Acquired Antibiotic Resistance among EHEC Serotypes
In addition to acid tolerance, the glutamate-dependent acid resistance pathway performs several extended cross-protective functions, such as strengthening the cell envelope, enhanced attachment, colonization, biofilm formation, multidrug resistance, and bacterial pathogenicity [180,181]. Recent studies on multidrug-resistant EHECs confirm that under low pH stress, different serotypes respond variably toward acid resistance. This variation is attributed to the expression of glutamate-dependent regulation of RpoS [182]. RpoS deletion mutants of EDL933 and other O157:H7 strains show down-regulation of GadA and acid fitness island genes [183,184]. In comparison to other serotypes, the EHEC O157:H7 and EHEC O26:H11 strains show enhanced resistance and improved survival in the mammalian gut [185]. Transcriptomic profiles of virulent EHEC serotypes confirm that the expression level of gadA, gadB, and gadE genes is significantly upregulated when exposed to low pH [185]. As expected, an O157:H7 knockout mutant of the central regulator GadE resulted in 40-fold decreased expression of GadA and enhanced susceptibility towards acid stress. Likewise, knockout mutagenesis of EHEC strains confirmed a non-colonizing phenotype for rcsB, arcA, cpxR, excluding evgS in mouse models [126,127]. Surprisingly, ArcA induced expression of GadE-MdtEF is highly dependent on the anaerobic environment provided by the human stomach [131]. These findings confirm that EHECs cannot survive in the human gut in the absence of the acid-adapted regulatory changes [65,98,186,187].
Interestingly, these adaptations not only help the pathogens survive but also facilitate high virulence and enhanced colonization in animal models. Although mutant studies provide information about the adaptation, colonization, and infection pattern of EHECs, appropriate animal models still need to be developed [188]. For example, mouse models have a gastric pH is less acidic than that of humans [189,190]. Nevertheless, several statements can be made from other systems. For example, GadC deletion mutants of serotype O157:H7, when grown in a calf model, show reduced survival [191]. Moreover, low pH-pretreated gadE and dctR transposon mutants of O157:H7 show strong adherence to human epithelial type 2 and human colorectal adenocarcinoma cell lines, increasing apoptosis [15]. Other studies confirm an acid-induced YadK adhesin that allows EHEC to adhere strongly to epithelial cells and facilitate bacterial-host attachment, resulting in increased colonization and pathogenesis [192]. Multiple studies with EHEC O157:H7 and other serotypes have also reported the importance of acid-induced development of other phenotypes that enhance survival and improve virulence [44,87,90,91,94,129,132,185,188,193,194]. EHECs use low pH stress to adapt and regulate a wide array of genes that enhance survival and increase pathogenicity.

Table 4.
Effect of low pH-mediated cross-protection against antibiotics and minimum inhibitory concentrations (MIC) of different EHEC serotypes.
Table 4.
Effect of low pH-mediated cross-protection against antibiotics and minimum inhibitory concentrations (MIC) of different EHEC serotypes.
| Acid-Adapted Strains | pH | Acquired Resistance | MIC | Reference |
|---|---|---|---|---|
| EHEC O157:H7 ATCC 43889 | 2.75 | Polymixin B, Colistin | Increased | [16] |
| E. coli ATCC25922 | Acidic | Colistin | Increased | [195] |
| E. coli (EHEC) ATCC 43889E. coli ATCC 10536 | 2 | Tetracycline | Increased | [41] |
| Foodborne EHEC strain | 4 | Nalidixic acid, amikacin, ceftriaxone | 5 fold increase | [60] |
| E. coli K-12 | 2 | Multidrug resistance | Increased | [45] |
| E. coli O157:H7 strain | 4.8 | Trimethoprim, ampicillin, and ofloxacin | Increased | [196] |
| EHEC Gut flora | 2.5–4 | Multidrug resistance | Increased | [43,197,198,199,200,201,202] |
| Tetracycline | [48] | |||
| Rifampicin resistant E. coli | 2.5–4 | Sulphonamide, gentamicin and ampicillin | Increased | [203] |
| E. coli O157:H7 | 3.7 | Streptomycin | Increased | [204] |
| 29A and 29B EHEC strains | 2.5–4 | Ampicillin | Increased | [205] |
| E. coli IID 5208 | 3.2 | Chitosan | Increased | [206] |
| Foodborne E. coli | Acidic | Aminoglycosides, cephalosporins, and quinolones | Increased | [207] |
| E. coli ATCC 12806 | Acidic | Ampicillin-sulbactam, amoxicillin-clavulanic acid, cefotaxime, trimethoprim-sulphamethoxazole, tetracycline, ciprofloxacin, nitrofurantoin | Not evaluated | [208] |
| E. coli O157:H7 | Acidic | Amoxicillin, tetracycline, ciprofloxacin, chloramphenicol, streptomycin, erythromycin, and gentamicin | Increased | [209] |
| E. coli BW25113 | 3 | Trimethoprim | Increased | [46] |
| E. coli O157:H7, E. coli O26:H7 | 4.2–4.4 | Ampicillin, kanamycin, streptomycin, trimethoprim, nalidixic acid, rifampicin, sulphonamides, chloramphenicol, chloramphenicol, tetracycline, minocycline, doxycycline | Increased | [210] |
| E. coli O157:H7 | 1.5 | Trimethoprim, ampicillin, ofloxacin | Increased | [211] |
| E. coli | Acidic | Ampicillin | Increased | [212] |
| EHEC W3110 | Acidic | Chloramphenicol | Increased | [47] |
| EHEC EV18 strain | Acidic | Norfloxacin | Increased | [44]. |
| E. coli K12 | Acidic | Cephalosporins, ceftiofur, cefotaxime | 2-fold increased | [151] |
Note: MIC above the breakpoint indicates that the organism is resistant.
7. Effect on Pathogenicity and Biofilm Formation
The molecular mechanisms underlying the pathogenic regulation of infectious E. coli indicate that biofilm formation correlates significantly with pathogenicity. Approximately 42 genes are regulated within a biofilm matrix in response to acid stress [213,214,215,216], including differential expression of rpoS [217] gadAB, gadC, hdeABD and yjiD (anti-adapter protein iraD, which inhibits rpoS). Knockout mutants of the genes mentioned above, when grown in glutamate-rich medium, increased biofilm formation [218]. Transcriptomic analysis of another gene cluster, ymgABC, revealed a significant role in regulating acid stress, with the ymgB gene product being downregulated in biofilm-forming cells [219]. To confirm a role in acid regulation, ten isogenic mutants of E. coli strain K-12 (ΔymgB, ΔymgA, ΔymgC, ΔycgZ, and ΔgadB, ΔgadA, ΔgadE, ΔhdeB, ΔhdeA, and ΔhdeD) were grown in glutamate enriched medium resulting in enhanced biofilm formation. These results highlight the importance of acid-resistance genes in biofilm formation [219].
Additionally, activation of several TCS response-regulators stimulates the expression of acid-fitness-island genes under acid stress that play an important role in pathogenesis regulation. In EHECs, NtrC, RcsB, and GadX are involved in the upregulation of the LEE (locus of enterocyte effacement) pathogenicity island, which indicates that nitrogen metabolism and glutamate-dependent-system genes play important roles in pathogenesis regulation [104,218,220]. Biofilm formation by another E. coli strain (MG1655) significantly increased at pH 5.5, while at lower pH the expression of flagellar synthesis genes and several virulence factors was strongly induced [1,221]. These studies highlight the biological relevance of acid stress in the regulation of pathogenesis in pathogenic E. coli [1,222,223].
8. Risk of Acquired Resistance in Non-Pathogenic Bacteria
Under respiratory stress, expression of the GAD operon is equally essential for pathogenic and non-pathogenic E. coli [82,224,225]. In an acidic environment, GadC consumes protons to promote GABA production that generates a proton motive force along with ATP production. Specifically, commensal bacteria and lactic acid bacteria (LAB) harbor GAD to produce GABA and act as probiotics in the GIT [224,226]. GABA plays an important role in bacteria that helps in the fermentation of protein-rich foods, such as cheese, rice germ, kimchi, yogurt, green tea, and sourdough [82,225,227]. Recent studies have found that fermentation of grapes by Lactobacillus plantarum DSM 19463 results in the production of GAD-derived GABA, which plays an important role in inducing the expression of β-defensin-2, hyaluronan synthase, and filaggrin genes responsible for skin protection in humans [228]. These remarkable findings lead to novel cosmetic formulations to treat antimicrobial problems related to skin.
Non-pathogenic bacteria maintain long-term commensalism with the host by stimulating the host immune system and inhibiting the colonization of gut pathogens [229,230,231]. To survive pH fluctuations in different compartments of the gut, the commensal bacteria also undergo the same extent of outer-membrane lipopolysaccharide modifications that contribute to ampicillin resistance. These changes result in modification of lipid A by LpxF phosphatase in commensal isolates of Bacteroidetes thetaiotaomicron that show significantly high polymyxin B resistance and enhanced colonization [232]. These adaptations in gut microbiota occur in response to environmental change. Clinical studies also report acquired tetracycline resistance in 22–33% EHECs in the gastric fluid by horizontal gene transfer, indicating an alarming health concern [41]. In some cases, commensal bacteria are reported to cause diseases, such as Crohn’s disease (CD), inflammatory bowel disease (IBD), and ulcerative colitis (UC) [233,234,235,236]. These adaptive pathogenic changes in gut microbiota occur in response to environmental factors, biodiversity, and genetic adaptability [230]. These findings indicate that modification of the lipid A anchor and other processes can promote a long-term commensal relationship between host and bacteria. On the other hand, horizontal gene transfer provides an open passage for the evolution of opportunistic pathogens having reduced antimicrobial susceptibility.
9. Conclusions and Future Perspective
EHECs have adapted to survive pre- and post-ingestion acid stress, thereby contributing to enhanced pathogenesis. Low pH positively regulates several metabolic pathways, such as motility, biofilm, chemotaxis, periplasmic secretory systems, and multidrug resistance that collectively regulate virulence. We highlighted the role of several signal-transduction cascades that enhance acid tolerance that results in the acquisition of antibiotic resistance. At present, almost all reported drugs are ineffective at controlling the spread of EHECs. Globally, the increasing resistance towards various classes of antibiotics, specifically cationic antimicrobial peptides and extended-spectrum β-lactamases, has become an overwhelming problem, making EHEC infections untreatable. EHECs have established complex regulatory mechanisms involving structural modification and efflux activation that provide an alarming condition for the emergence of new multidrug-resistant pathogens having improved colonization and infection capabilities.
Several factors influence the organism’s choice of the resistance mechanism. Cytoplasmic acidification offers a baseline level of defense that can act in tandem with modifications/mutations to reduce antibiotic susceptibility. Bacterial exposure to low pH is associated with acquired antimicrobial resistance to various therapeutic antibiotics. Active efflux and structural modifications of the bacterial membrane are the best-documented mechanisms responsible for bacterial cross-protection to antibiotics. The judicious and rational use of acidic treatments is crucial to reduce the risk of selecting antimicrobial-resistant bacteria. Antibiotic resistance acquisition strategies are extremely diverse; knowledge of this phenomenon at the molecular level provides an understanding of the details and appreciation to scale this important health problem. An even deeper understanding of the defensive responses deployed by the pathogens may reveal novel targets for agents that will help overcome the spread of foodborne diseases.
We emphasize that rigorous hygiene measures must be followed and all available antimicrobial agents should be used wisely to control the spread of multidrug-resistant strains. The risk of acid-adapted cross-protection by subsequent antimicrobial inactivation necessitates the identification of novel determinants that can influence the future epidemiology and health impact of multidrug-resistant infections. More efforts should be placed to develop novel non-antibiotic approaches such as vaccines, immuno-stimulants, phage therapies, prebiotics, and probiotics to treat EHEC infections.
Author Contributions
S.W.S. and T.X. conceived the original screening and manuscript plans. S.W.S. and A.A. (Ahmad Ali) wrote the manuscript. S.W.S. and A.A. (Asma Ahsan) drew all the figures in the manuscript. S.S. drew all the tables in the manuscript. T.X. and F.S. reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (grants 31672571).
Acknowledgments
The authors are grateful to Saad Sarfraz (Station de Neucfchateau, CIRAD, Sainte-Marie, Capesterre Belle Eau, Guadeloupe, France) and Muhammad Arslan (University of Alberta, Canada) for providing technical support.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations were used in the manuscript:
| Glu | l-glutamate |
| Gln | Glutamine |
| GABA | Gamma-aminobutyric acid |
| Arg | l-arginine |
| AdiA | Arginine decarboxylase |
| Lys | Lysine |
| CRP | Global regulatory cyclic AMP receptor protein |
| NA | Not applicable |
| EHEC | Enterohaemorrhagic Escherichia coli |
| HUS | Hemolytic uremic syndrome |
| TCS | Two-component system |
| LEE | Locus of enterocyte effacement |
| AFI | Acid fitness island |
| AR2 | Glutamate-dependent acid resistance system |
| AR1 | Oxidative system |
| RNAP | RNA polymerase |
| T3SS | Type-three secretion system |
| AMP | Antimicrobial peptides |
| CAPs | Cationic antimicrobial peptides |
| ESBLs | Extended-spectrum β-lactamases |
| LPS | Lipopolysaccharides |
| PBPs | Penicillin-binding proteins |
| IHF | Integration host factor |
| RND | Resistance nodulation division |
| LEE-PAI | Locus of enterocyte effacement pathogenicity island |
| LAB | Lactic acid bacteria |
| CD | Crohn’s disease |
| IBD | Inflammatory bowel disease |
| UC | Ulcerative colitis |
| GAD | Glutamate-dependent system |
| ADI | Arginine-dependent system |
| CAD | Lysine-dependent system |
| MIC | Minimum inhibitory concentration |
| GaNt | Gallium nitrate |
References
- Maurer, L.M.; Yohannes, E.; Bondurant, S.S.; Radmacher, M.; Slonczewski, J.L. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 2005, 187, 304–319. [Google Scholar] [CrossRef]
- Foster, J.W. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Microbiol. 2004, 2, 898–907. [Google Scholar] [CrossRef]
- Gullian-Klanian, M.; Sánchez-Solis, M.J. Growth kinetics of Escherichia coli O157:H7 on the epicarp of fresh vegetables and fruits. Braz. J. Microbiol. 2018, 49, 104–111. [Google Scholar] [CrossRef]
- Wang, L.; Bassiri, M.; Najafi, R.; Najafi, K.; Yang, J.; Khosrovi, B.; Hwong, W.; Barati, E.; Belisle, B.; Celeri, C.; et al. Hypochlorous acid as a potential wound care agent: Part I. Stabilized hypochlorous acid: A component of the inorganic armamentarium of innate immunity. J. Burns Wounds 2007, 6, e5. [Google Scholar]
- Pijuan, M.; Wang, Q.; Ye, L.; Yuan, Z. Improving secondary sludge biodegradability using free nitrous acid treatment. Bioresour. Technol. 2012, 116, 92–98. [Google Scholar] [CrossRef]
- Drosou, A.; Falabella, A.; Kirsner, R.S. Antiseptics on Wounds: An Area of Controversy. Wounds 2003, 15, 149–166. [Google Scholar]
- Nagoba, B.S.; Selkar, S.P.; Wadher, B.J.; Gandhi, R.C. Acetic acid treatment of pseudomonal wound infections—A review. J. Infect. Public Health 2013, 6, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.L.; Kalchayanand, N.; Bosilevac, J.M. Pre- and post-harvest interventions to reduce pathogen contamination in the U.S. beef industry. Meat Sci. 2014, 98, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-M.; Hung, Y.-C.; Doyle, M.P.; Ezeike, G.O.I.; Kim, C. Pathogen Reduction and Quality of Lettuce Treated with Electrolyzed Oxidizing and Acidified Chlorinated Water. J. Food Sci. 2001, 66, 1368–1372. [Google Scholar] [CrossRef]
- Karmali, M.A.; Gannon, V.; Sargeant, J.M. Verocytotoxin-producing Escherichia coli (VTEC). Vet. Microbiol. 2010, 140, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Scheiring, J.; Andreoli, S.P.; Zimmerhackl, L.B. Treatment and outcome of Shiga-toxin-associated hemolytic uremic syndrome (HUS). Pediatr. Nephrol. 2008, 23, 1749–1760. [Google Scholar] [CrossRef]
- Iramiot, J.S.; Kajumbula, H.; Bazira, J.; Kansiime, C.; Asiimwe, B.B. Antimicrobial resistance at the human–animal interface in the Pastoralist Communities of Kasese District, South Western Uganda. Sci. Rep. 2020, 10, 14737. [Google Scholar] [CrossRef]
- Jakobsen, L.; Spangholm, D.J.; Pedersen, K.; Jensen, L.B.; Emborg, H.D.; Agerso, Y.; Aarestrup, F.M.; Hammerum, A.M.; Frimodt-Moller, N. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int. J. Food Microbiol. 2010, 142, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Kanjee, U.; Houry, W.A. Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 2013, 67, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Ferens, W.A.; Hovde, C.J. Escherichia coli O157:H7: Animal reservoir and sources of human infection. Foodborne Pathog. Dis. 2011, 8, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.; Kim, S.M.; Kim, H.J. Transcriptome changes and polymyxin resistance of acid-adapted Escherichia coli O157:H7 ATCC 43889. Gut Pathog. 2020, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Yousef, A.E.; Juneja, V.K. Microbial Stress Adaptation and Food Safety, 1st ed.; CRC Press: Boca Raton, FL, USA, 2002; p. 384. [Google Scholar]
- Cointe, A.; Birgy, A.; Mariani-Kurkdjian, P.; Liguori, S.; Courroux, C.; Blanco, J.; Delannoy, S.; Fach, P.; Loukiadis, E.; Bidet, P.; et al. Emerging Multidrug-Resistant Hybrid Pathotype Shiga Toxin-Producing Escherichia coli O80 and Related Strains of Clonal Complex 165, Europe. Emerg. Infect. Dis. 2018, 24, 2262–2269. [Google Scholar] [CrossRef]
- Hassan, R.; Tantawy, M.; Gouda, N.A.; Elzayat, M.G.; Gabra, S.; Nabih, A.; Diab, A.A.; El-Hadidi, M.; Bakry, U.; Shoeb, M.R.; et al. Genotypic characterization of multiple drug resistant Escherichia coli isolates from a pediatric cancer hospital in Egypt. Sci. Rep. 2020, 10, 4165. [Google Scholar] [CrossRef] [PubMed]
- Wi, S.M.; Yoon, J.W. Acid resistance mechanisms in enterohemorrhagic Escherichia coli O157:H7. J. Prev. Vet. Med. 2018, 42, 124–132. [Google Scholar] [CrossRef]
- Delannoy, S.; Beutin, L.; Fach, P. Towards a molecular definition of enterohemorrhagic Escherichia coli (EHEC): Detection of genes located on O island 57 as markers to distinguish EHEC from closely related enteropathogenic E. coli strains. J. Clin. Microbiol. 2013, 51, 1083–1088. [Google Scholar] [CrossRef]
- Segura, A.; Bertoni, M.; Auffret, P.; Klopp, C.; Bouchez, O.; Genthon, C.; Durand, A.; Bertin, Y.; Forano, E. Transcriptomic analysis reveals specific metabolic pathways of enterohemorrhagic Escherichia coli O157:H7 in bovine digestive contents. BMC Genom. 2018, 19, 766. [Google Scholar] [CrossRef]
- Saile, N.; Voigt, A.; Kessler, S.; Stressler, T.; Klumpp, J.; Fischer, L.; Schmidt, H. Escherichia coli O157:H7 Strain EDL933 Harbors Multiple Functional Prophage-Associated Genes Necessary for the Utilization of 5-N-Acetyl-9-O-Acetyl Neuraminic Acid as a Growth Substrate. Appl. Environ. Microbiol. 2016, 82, 5940–5950. [Google Scholar] [CrossRef] [PubMed]
- Panos, G.Z.; Betsi, G.I.; Falagas, M.E. Systematic review: Are antibiotics detrimental or beneficial for the treatment of patients with Escherichia coli O157:H7 infection? Aliment. Pharmacol. Ther. 2006, 24, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Soysal, N.; Mariani-Kurkdjian, P.; Smail, Y.; Liguori, S.; Gouali, M.; Loukiadis, E.; Fach, P.; Bruyand, M.; Blanco, J.; Bidet, P.; et al. Enterohemorrhagic Escherichia coli Hybrid Pathotype O80:H2 as a New Therapeutic Challenge. Emerg. Infect. Dis. 2016, 22, 1604–1612. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef]
- Lim, J.Y.; Yoon, J.; Hovde, C.J. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5–14. [Google Scholar] [CrossRef]
- Menne, J.; Nitschke, M.; Stingele, R.; Abu-Tair, M.; Beneke, J.; Bramstedt, J.; Bremer, J.P.; Brunkhorst, R.; Busch, V.; Dengler, R.; et al. Validation of treatment strategies for enterohaemorrhagic Escherichia coli O104:H4 induced haemolytic uraemic syndrome: Case-control study. BMJ 2012, 345, e4565. [Google Scholar] [CrossRef]
- Venturini, C.; Beatson, S.A.; Djordjevic, S.P.; Walker, M.J. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2010, 24, 1160–1166. [Google Scholar] [CrossRef]
- Um, M.M.; Brugère, H.; Kérourédan, M.; Oswald, E.; Bibbal, D. Antimicrobial Resistance Profiles of Enterohemorrhagic and Enteropathogenic Escherichia coli of Serotypes O157:H7, O26:H11, O103:H2, O111:H8, O145:H28 Compared to Escherichia coli Isolated from the Same Adult Cattle. Microb. Drug Resist. Larchmt. N. Y. 2018, 24, 852–859. [Google Scholar] [CrossRef]
- Medina, A.; Horcajo, P.; Jurado, S.; De La Fuente, R.; Ruiz-Santa-Quiteria, J.A.; Domínguez-Bernal, G.; Orden, J.A. Phenotypic and Genotypic Characterization of Antimicrobial Resistance in Enterohemorrhagic Escherichia coli and Atypical Enteropathogenic E. coli Strains from Ruminants. J. Vet. Diagn. Investig. 2011, 23, 91–95. [Google Scholar] [CrossRef]
- Guiral, E.; Gonçalves Quiles, M.; Muñoz, L.; Moreno-Morales, J.; Alejo-Cancho, I.; Salvador, P.; Alvarez-Martinez, M.J.; Marco, F.; Vila, J. Emergence of Resistance to Quinolones and β-Lactam Antibiotics in Enteroaggregative and Enterotoxigenic Escherichia coli Causing Traveler’s Diarrhea. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef] [PubMed]
- Hussein, S.H.; Samir, R.; Aziz, R.K.; Toama, M.A. Two putative MmpL homologs contribute to antimicrobial resistance and nephropathy of enterohemorrhagic E. coli O157:H7. Gut Pathog. 2019, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Mühlen, S.; Dersch, P. Treatment Strategies for Infections With Shiga Toxin-Producing Escherichia coli. Front. Cell. Infect. Microbiol. 2020, 10, 169. [Google Scholar] [CrossRef]
- Zhang, Q.; Donohue-Rolfe, A.; Krautz-Peterson, G.; Sevo, M.; Parry, N.; Abeijon, C.; Tzipori, S. Gnotobiotic piglet infection model for evaluating the safe use of antibiotics against Escherichia coli O157:H7 infection. J. Infect. Dis. 2009, 199, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Agger, M.; Scheutz, F.; Villumsen, S.; Mølbak, K.; Petersen, A.M. Antibiotic treatment of verocytotoxin-producing Escherichia coli (VTEC) infection: A systematic review and a proposal. J. Antimicrob. Chemother. 2015, 70, 2440–2446. [Google Scholar] [CrossRef]
- Corogeanu, D.; Willmes, R.; Wolke, M.; Plum, G.; Utermöhlen, O.; Krönke, M. Therapeutic concentrations of antibiotics inhibit Shiga toxin release from enterohemorrhagic E. coli O104:H4 from the 2011 German outbreak. BMC Microbiol. 2012, 12, 160. [Google Scholar] [CrossRef]
- Pedersen, M.G.; Hansen, C.; Riise, E.; Persson, S.; Olsen, K.E. Subtype-specific suppression of Shiga toxin 2 released from Escherichia coli upon exposure to protein synthesis inhibitors. J. Clin. Microbiol. 2008, 46, 2987–2991. [Google Scholar] [CrossRef]
- Wesche, A.M.; Gurtler, J.B.; Marks, B.P.; Ryser, E.T. Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J. Food Prot. 2009, 72, 1121–1138. [Google Scholar] [CrossRef]
- Hwang, D.; Kim, S.M.; Kim, H.J. Modelling of tetracycline resistance gene transfer by commensal Escherichia coli food isolates that survived in gastric fluid conditions. Int. J. Antimicrob. Agents 2017, 49, 81–87. [Google Scholar] [CrossRef]
- Kobayashi, M.; Sasaki, T.; Agui, N. Possible food contamination with the excreta of housefly with enterohemorrhagic Escherichia coli O157:H7. Med Entomol. Zool. 2002, 53, 83–87. [Google Scholar] [CrossRef]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Fernández, E.Z.; Schuldiner, S. Acidification of Cytoplasm in Escherichia coli Provides a Strategy to Cope with Stress and Facilitates Development of Antibiotic Resistance. Sci. Rep. 2020, 10, 9954. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, S.H.; Lee, A.V.; Pham, M.T.N.; Kassaye, B.B.; Li, H.; Tallada, S.; Lis, C.; Lang, M.; Liu, Y.; Ahmed, N.; et al. Salicylate, Bile Acids and Extreme Acid Cause Fitness Tradeoffs for Multidrug Pumps in Escherichia coli K-12. bioRxiv 2020. [Google Scholar] [CrossRef]
- Mitosch, K.; Rieckh, G.; Bollenbach, T. Noisy Response to Antibiotic Stress Predicts Subsequent Single-Cell Survival in an Acidic Environment. Cell Syst. 2017, 4, 393–403.e395. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.P.; Li, H.; Engmann, M.L.; Bischof, K.M.; Kunka, K.S.; Harris, M.E.; Tancredi, A.C.; Ditmars, F.S.; Basting, P.J.; George, N.S.; et al. Inverted Regulation of Multidrug Efflux Pumps, Acid Resistance and Porins in Benzoate-Evolved Escherichia coli K-12. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef]
- Liao, X.; Ma, Y.; Daliri, E.B.-M.; Koseki, S.; Wei, S.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Interplay of antibiotic resistance and food-associated stress tolerance in foodborne pathogens. Trends Food Sci. Technol. 2020, 95, 97–106. [Google Scholar] [CrossRef]
- Cabello, F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Bhagirath, A.Y.; Li, Y.; Patidar, R.; Yerex, K.; Ma, X.; Kumar, A.; Duan, K. Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. Int. J. Mol. Sci. 2019, 20, 1781. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta BBA Proteins Proteom. 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Soto, S.M. Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 2013, 4, 223–229. [Google Scholar] [CrossRef]
- Martins, A.; Spengler, G.; Rodrigues, L.; Viveiros, M.; Ramos, J.; Martins, M.; Couto, I.; Fanning, S.; Pagès, J.-M.; Bolla, J.M.; et al. pH Modulation of efflux pump activity of multi-drug resistant Escherichia coli: Protection during its passage and eventual colonization of the colon. PLoS ONE 2009, 4, e6656. [Google Scholar] [CrossRef] [PubMed]
- Reygaert, W.C. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Hancock, R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Nové, M.; Kincses, A.; Molnár, J.; Amaral, L.; Spengler, G. The Role of Efflux Pumps and Environmental pH in Bacterial Multidrug Resistance. In Vivo Athens Greece 2020, 34, 65–71. [Google Scholar] [CrossRef]
- Cheng, H.-Y.; Yu, R.-C.; Chou, C.-C. Increased acid tolerance of Escherichia coli O157:H7 as affected by acid adaptation time and conditions of acid challenge. Food Res. Int. 2003, 36, 49–56. [Google Scholar] [CrossRef]
- Yang, J.; Russell, T.W.; Hocking, D.M.; Bender, J.K.; Srikhanta, Y.N.; Tauschek, M.; Robins-Browne, R.M. Control of Acid Resistance Pathways of Enterohemorrhagic Escherichia coli Strain EDL933 by PsrB, a Prophage-Encoded AraC-Like Regulator. Infect. Immun. 2015, 83, 346–353. [Google Scholar] [CrossRef]
- McMahon, M.A.S.; Xu, J.; Moore, J.E.; Blair, I.S.; McDowell, D.A. Environmental stress and antibiotic resistance in food-related pathogens. Appl. Environ. Microbiol. 2007, 73, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Houry, W.A. Acid stress response in enteropathogenic gammaproteobacteria: An aptitude for survival. Biochem. Cell Biol. 2010, 88, 301–314. [Google Scholar] [CrossRef]
- Ortega, A.D.; Quereda, J.J.; Pucciarelli, M.G.; García-del Portillo, F. Non-coding RNA regulation in pathogenic bacteria located inside eukaryotic cells. Front. Cell. Infect. Microbiol. 2014, 4, 162. [Google Scholar] [CrossRef]
- Castillo-Juárez, I.; Maeda, T.; Mandujano-Tinoco, E.A.; Tomás, M.; Pérez-Eretza, B.; García-Contreras, S.J.; Wood, T.K.; García-Contreras, R. Role of quorum sensing in bacterial infections. World J. Clin. Cases. 2015, 3, 575–598. [Google Scholar] [CrossRef]
- Pitman, S.; Cho, K.H. The Mechanisms of Virulence Regulation by Small Noncoding RNAs in Low GC Gram-Positive Pathogens. Int. J. Mol. Sci. 2015, 16, 29797–29814. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Rahman, A.S.; Henderson, I.; Cole, J.; Johnson, M.D.; Daudi, N.; Lund, P.; Antczak, P.; Falciani, F. A systems biology approach sheds new light on Escherichia coli acid resistance. Nucleic Acids Res. 2011, 39, 7512–7528. [Google Scholar] [CrossRef]
- De Biase, D.; Lund, P.A. The Escherichia coli Acid Stress Response and Its Significance for Pathogenesis. Adv. Appl. Microbiol. 2015, 92, 49–88. [Google Scholar] [CrossRef] [PubMed]
- De Biase, D.; Pennacchietti, E. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: Function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012, 86, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Iyer, R.; Williams, C.; Miller, C. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 2003, 185, 6556–6561. [Google Scholar] [CrossRef]
- Moreau, P.L. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J. Bacteriol. 2007, 189, 2249–2261. [Google Scholar] [CrossRef]
- Lu, P.; Ma, D.; Chen, Y.; Guo, Y.; Chen, G.Q.; Deng, H.; Shi, Y. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013, 23, 635–644. [Google Scholar] [CrossRef]
- De Biase, D.; Tramonti, A.; Bossa, F.; Visca, P. The response to stationary-phase stress conditions in Escherichia coli: Role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999, 32, 1198–1211. [Google Scholar] [CrossRef]
- Gong, S.; Richard, H.; Foster, J.W. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 2003, 185, 4402–4409. [Google Scholar] [CrossRef] [PubMed]
- Hirshfield, I.N.; Terzulli, S.; O’Byrne, C. Weak organic acids: A panoply of effects on bacteria. Sci. Prog. 2003, 86, 245–269. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Fukamachi, T.; Saito, H.; Kobayashi, H. ATP requirement for acidic resistance in Escherichia coli. J. Bacteriol. 2011, 193, 3072–3077. [Google Scholar] [CrossRef] [PubMed]
- Diez-Gonzalez, F.; Karaibrahimoglu, Y. Comparison of the glutamate-, arginine- and lysine-dependent acid resistance systems in Escherichia coli O157:H7. J. Appl. Microbiol. 2004, 96, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Richard, H.; Foster, J.W. Escherichia coli Glutamate- and Arginine-Dependent Acid Resistance Systems Increase Internal pH and Reverse Transmembrane Potential. J. Bacteriol. 2004, 186, 6032–6041. [Google Scholar] [CrossRef] [PubMed]
- Hersh, B.M.; Farooq, F.T.; Barstad, D.N.; Blankenhorn, D.L.; Slonczewski, J.L. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 1996, 178, 3978–3981. [Google Scholar] [CrossRef]
- Castanie-Cornet, M.P.; Penfound, T.A.; Smith, D.; Elliott, J.F.; Foster, J.W. Control of acid resistance in Escherichia coli. J. Bacteriol. 1999, 181, 3525–3535. [Google Scholar] [CrossRef] [PubMed]
- Bearson, B.L.; Lee, I.S.; Casey, T.A. Escherichia coli O157:H7 glutamate- and arginine-dependent acid-resistance systems protect against oxidative stress during extreme acid challenge. Microbiology 2009, 155, 805–812. [Google Scholar] [CrossRef]
- Waterman, S.R.; Small, P.L. Identification of the promoter regions and sigma(s)-dependent regulation of the gadA and gadBC genes associated with glutamate-dependent acid resistance in Shigella flexneri. FEMS Microbiol. Lett. 2003, 225, 155–160. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ryan, S.; Gahan, C.G.M.; Hill, C. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 2005, 71, 2832–2839. [Google Scholar] [CrossRef]
- Su, M.S.; Schlicht, S.; Gänzle, M.G. Contribution of glutamate decarboxylase in Lactobacillus reuteri to acid resistance and persistence in sourdough fermentation. Microb. Cell Factories 2011, 10 (Suppl. 1), S8. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, T.M.; Tarr, C.L.; Christensen, L.M.; Betting, D.J.; Whittam, T.S. Recent gene conversions between duplicated glutamate decarboxylase genes (gadA and gadB) in pathogenic Escherichia coli. Mol. Biol. Evol. 2007, 24, 2323–2333. [Google Scholar] [CrossRef] [PubMed]
- O’Byrne, C.P.; Karatzas, K.A. The role of sigma B (sigma B) in the stress adaptations of Listeria monocytogenes: Overlaps between stress adaptation and virulence. Adv. Appl. Microbiol. 2008, 65, 115–140. [Google Scholar] [CrossRef] [PubMed]
- Feehily, C.; Karatzas, K.A.G. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 2013, 114, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Castanie-Cornet, M.P.; Treffandier, H.; Francez-Charlot, A.; Gutierrez, C.; Cam, K. The glutamate-dependent acid resistance system in Escherichia coli: Essential and dual role of the His-Asp phosphorelay RcsCDB/AF. Microbiology 2007, 153, 238–246. [Google Scholar] [CrossRef]
- Pienaar, J.A.; Singh, A.; Barnard, T.G. Acid-happy: Survival and recovery of enteropathogenic Escherichia coli (EPEC) in simulated gastric fluid. Microb. Pathog. 2019, 128, 396–404. [Google Scholar] [CrossRef]
- Matter, L.B.; Ares, M.A.; Abundes-Gallegos, J.; Cedillo, M.L.; Yanez, J.A.; Martinez-Laguna, Y.; De la Cruz, M.A.; Giron, J.A. The CpxRA stress response system regulates virulence features of avian pathogenic Escherichia coli. Environ. Microbiol. 2018, 20, 3363–3377. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Yu, L.; Gao, F.; Jiang, Y.; Xu, X. Resistance of biofilm formation and formed-biofilm of Escherichia coli O157:H7 exposed to acid stress. LWT 2020, 118, 108787. [Google Scholar] [CrossRef]
- Boon, N.; Kaur, M.; Aziz, A.; Bradnick, M.; Shibayama, K.; Eguchi, Y.; Lund, P.A. The Signaling Molecule Indole Inhibits Induction of the AR2 Acid Resistance System in Escherichia coli. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Zeng, J.; Wu, L.; Liu, Z.; Lv, Y.; Feng, J.; Wang, W.; Xue, Y.; Wang, D.; Li, J.; Drlica, K.; et al. Gain-of-Function Mutations in Acid Stress Response (evgS) Protect Escherichia coli from Killing by Gallium Nitrate, an Antimicrobial Candidate. Antimicrob. Agents Chemother. 2021, 65, e01595-20. [Google Scholar] [CrossRef]
- White-Ziegler, C.A.; Um, S.; Perez, N.M.; Berns, A.L.; Malhowski, A.J.; Young, S. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiol. 2008, 154, 148–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fierke, C.A. The BaeSR regulon is involved in defense against zinc toxicity in E. coli. Metallomics 2013, 5, 372–383. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rao, S.D.; Igoshin, O.A. Overlaid positive and negative feedback loops shape dynamical properties of PhoPQ two-component system. PLoS Comput. Biol. 2021, 17, e1008130. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.M.; Kim, J.W.; Song, J.W.; Blank, L.M.; Park, J.B. Activation of the Glutamic Acid-Dependent Acid Resistance System in Escherichia coli BL21(DE3) Leads to Increase of the Fatty Acid Biotransformation Activity. PLoS ONE 2016, 11, e0163265. [Google Scholar] [CrossRef]
- Szczesny, M.; Beloin, C.; Ghigo, J.M. Increased Osmolarity in Biofilm Triggers RcsB-Dependent Lipid A Palmitoylation in Escherichia coli. mBio 2018, 9, e01415-18. [Google Scholar] [CrossRef]
- Liu, C.-J.; Lin, C.-T.; Chiang, J.-D.; Lin, C.-Y.; Tay, Y.-X.; Fan, L.-C.; Peng, K.-N.; Lin, C.-H.; Peng, H.-L. RcsB regulation of the YfdX-mediated acid stress response in Klebsiella pneumoniae CG43S3. PLoS ONE 2019, 14, e0212909. [Google Scholar] [CrossRef]
- Castanié-Cornet, M.-P.; Cam, K.; Bastiat, B.; Cros, A.; Bordes, P.; Gutierrez, C. Acid stress response in Escherichia coli: Mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 2010, 38, 3546–3554. [Google Scholar] [CrossRef]
- Burton, N.A.; Johnson, M.D.; Antczak, P.; Robinson, A.; Lund, P.A. Novel aspects of the acid response network of E. coli K-12 are revealed by a study of transcriptional dynamics. J. Mol. Biol. 2010, 401, 726–742. [Google Scholar] [CrossRef]
- Gong, S.; Ma, Z.; Foster, J.W. The Era-like GTPase TrmE conditionally activates gadE and glutamate-dependent acid resistance in Escherichia coli. Mol. Microbiol. 2004, 54, 948–961. [Google Scholar] [CrossRef]
- Eguchi, Y.; Ishii, E.; Yamane, M.; Utsumi, R. The connector SafA interacts with the multi-sensing domain of PhoQ in Escherichia coli. Mol. Microbiol. 2012, 85, 299–313. [Google Scholar] [CrossRef]
- Tu, J.; Huang, B.; Zhang, Y.; Zhang, Y.; Xue, T.; Li, S.; Qi, K. Modulation of virulence genes by the two-component system PhoP-PhoQ in avian pathogenic Escherichia coli. Pol. J. Vet. Sci. 2016, 19, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Jin, F.; Glatter, T.; Sourjik, V. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. USA 2017, 114, e10792–e10798. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.K.; Vendura, K.W.; Stevens, S.M., Jr.; Riordan, J.T. RcsB determines the locus of enterocyte effacement (LEE) expression and adherence phenotype of Escherichia coli O157:H7 spinach outbreak strain TW14359 and coordinates bicarbonate-dependent LEE activation with repression of motility. Microbiology 2013, 159, 2342–2353. [Google Scholar] [CrossRef] [PubMed]
- Giangrossi, M.; Zattoni, S.; Tramonti, A.; De Biase, D.; Falconi, M. Antagonistic role of H-NS and GadX in the regulation of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J. Biol. Chem. 2005, 280, 21498–21505. [Google Scholar] [CrossRef] [PubMed]
- Hommais, F.; Krin, E.; Coppee, J.Y.; Lacroix, C.; Yeramian, E.; Danchin, A.; Bertin, P. GadE (YhiE): A novel activator involved in the response to acid environment in Escherichia coli. Microbiology 2004, 150, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Tramonti, A.; De Canio, M.; De Biase, D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: Transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 2008, 70, 965–982. [Google Scholar] [CrossRef]
- Bordi, C.; Theraulaz, L.; Mejean, V.; Jourlin-Castelli, C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 2003, 48, 211–223. [Google Scholar] [CrossRef]
- Ma, Z.; Masuda, N.; Foster, J.W. Characterization of EvgAS-YdeO-GadE branched regulatory circuit governing glutamate-dependent acid resistance in Escherichia coli. J. Bacteriol. 2004, 186, 7378–7389. [Google Scholar] [CrossRef] [PubMed]
- Prost, L.R.; Daley, M.E.; Le Sage, V.; Bader, M.W.; Le Moual, H.; Klevit, R.E.; Miller, S.I. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 2007, 26, 165–174. [Google Scholar] [CrossRef]
- Simpson, B.W.; Trent, M.S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 2019, 17, 403–416. [Google Scholar] [CrossRef]
- Roggiani, M.; Yadavalli, S.S.; Goulian, M. Natural variation of a sensor kinase controlling a conserved stress response pathway in Escherichia coli. PLoS Genet. 2017, 13, e1007101. [Google Scholar] [CrossRef] [PubMed]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Church, G.M. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 2003, 48, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, Y.; Ishii, E.; Hata, K.; Utsumi, R. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 2011, 193, 1222–1228. [Google Scholar] [CrossRef]
- Rhouma, M.; Romero-Barrios, P.; Gaucher, M.-L.; Bhachoo, S. Antimicrobial resistance associated with the use of antimicrobial processing aids during poultry processing operations: Cause for concern? Crit. Rev. Food Sci. Nutr. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Gunn, L.; Wall, P.; Fanning, S. Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiol. 2017, 64, 23–32. [Google Scholar] [CrossRef]
- Woode, B.K.; Daliri, F.; Daliri, E.B.-M. Correlation Between Food Processing-Associated Stress Tolerance and Antimicrobial Resistance in Food Pathogens. J. Food Hyg. Saf. 2020, 35, 103–108. [Google Scholar] [CrossRef]
- Al-Nabulsi, A.A.; Osaili, T.M.; Shaker, R.R.; Olaimat, A.N.; Jaradat, Z.W.; Zain Elabedeen, N.A.; Holley, R.A. Effects of osmotic pressure, acid, or cold stresses on antibiotic susceptibility of Listeria monocytogenes. Food Microbiol. 2015, 46, 154–160. [Google Scholar] [CrossRef]
- Komora, N.; Bruschi, C.; Magalhães, R.; Ferreira, V.; Teixeira, P. Survival of Listeria monocytogenes with different antibiotic resistance patterns to food-associated stresses. Int. J. Food Microbiol. 2017, 245, 79–87. [Google Scholar] [CrossRef]
- De Sales, C.V.; De Melo, A.N.F.; Niedzwiedzka, K.M.; De Souza, E.L.; Schaffner, D.W.; Magnani, M. Changes of Antibiotic Resistance Phenotype in Outbreak-Linked Salmonella enterica Strains after Exposure to Human Simulated Gastrointestinal Conditions in Chicken Meat. J. Food Prot. 2018, 81, 1844–1850. [Google Scholar] [CrossRef]
- Karatzas, K.A.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.; Humphrey, T.J. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Ebinesh, A.; Vijaykumar, G.; Kiran, T. Exposure to stress minimizes the zone of antimicrobial action: A phenotypic demonstration with six Acinetobacter baumannii strains. MicroMedicine 2018, 6, 16–35. [Google Scholar]
- Al-Nabulsi, A.A.; Osaili, T.M.; Elabedeen, N.A.Z.; Jaradat, Z.W.; Shaker, R.R.; Kheirallah, K.A.; Tarazi, Y.H.; Holley, R.A. Impact of environmental stress desiccation, acidity, alkalinity, heat or cold on antibiotic susceptibility of Cronobacter sakazakii. Int. J. Food Microbiol. 2011, 146, 137–143. [Google Scholar] [CrossRef]
- Ma, Y.; Lan, G.; Li, C.; Cambaza, E.M.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Stress tolerance of Staphylococcus aureus with different antibiotic resistance profiles. Microb. Pathog. 2019, 133, 103549. [Google Scholar] [CrossRef] [PubMed]
- Arenas-Hernández, M.M.; Rojas-López, M.; Medrano-López, A.; Nuñez-Reza, K.J.; Puente, J.L.; Martínez-Laguna, Y.; Torres, A.G. Environmental regulation of the long polar fimbriae 2 of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol. Lett. 2014, 357, 105–114. [Google Scholar] [CrossRef] [PubMed]
- House, B.; Kus, J.V.; Prayitno, N.; Mair, R.; Que, L.; Chingcuanco, F.; Gannon, V.; Cvitkovitch, D.G.; Barnett Foster, D. Acid-stress-induced changes in enterohaemorrhagic Escherichia coli O157:H7 virulence. Microbiology (Reading, England) 2009, 155, 2907–2918. [Google Scholar] [CrossRef]
- Lajhar, S.A.; Brownlie, J.; Barlow, R. Survival capabilities of Escherichia coli O26 isolated from cattle and clinical sources in Australia to disinfectants, acids and antimicrobials. BMC Microbiol. 2017, 17, 47. [Google Scholar] [CrossRef]
- King, T.; Lucchini, S.; Hinton, J.C.; Gobius, K. Transcriptomic analysis of Escherichia coli O157:H7 and K-12 cultures exposed to inorganic and organic acids in stationary phase reveals acidulant- and strain-specific acid tolerance responses. Appl. Environ. Microbiol. 2010, 76, 6514–6528. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, M.; Horiyama, T.; Zhang, Y.; Li, X.; Nishino, K.; Yan, A. The multidrug efflux pump MdtEF protects against nitrosative damage during the anaerobic respiration in Escherichia coli. J. Biol. Chem. 2011, 286, 26576–26584. [Google Scholar] [CrossRef]
- Deng, Z.; Shan, Y.; Pan, Q.; Gao, X.; Yan, A. Anaerobic expression of the gadE-mdtEF multidrug efflux operon is primarily regulated by the two-component system ArcBA through antagonizing the H-NS mediated repression. Front. Microbiol. 2013, 4, 194. [Google Scholar] [CrossRef]
- Novoa, D.; Conroy-Ben, O. The Anaerobic Efflux Pump MdtEF-TolC Confers Resistance to Cationic Biocides. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Wierzbowski, J.; Cottarel, G.; Collins, J.J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell 2008, 135, 679–690. [Google Scholar] [CrossRef]
- Rice, A.; Wereszczynski, J. Atomistic Scale Effects of Lipopolysaccharide Modifications on Bacterial Outer Membrane Defenses. Biophys. J. 2018, 114, 1389–1399. [Google Scholar] [CrossRef]
- Nascimento, A.; Pontes, F.J.; Lins, R.D.; Soares, T.A. Hydration, ionic valence and cross-linking propensities of cations determine the stability of lipopolysaccharide (LPS) membranes. Chem. Commun. Camb. 2014, 50, 231–233. [Google Scholar] [CrossRef]
- Carpenter, T.S.; Parkin, J.; Khalid, S. The free energy of small solute permeation through the Escherichia coli outer membrane has a distinctly asymmetric profile. J. Phys. Chem. Lett. 2016, 7, 3446–3451. [Google Scholar] [CrossRef] [PubMed]
- Francis, A.-S. Bile Salts-and Ferric Iron-Induced PMRAB Dependent Resistance to CAMPs in EHEC O157:H7; Ryerson University: Toronto, ON, Canada, 2017. [Google Scholar]
- Santos, D.E.; Pol-Fachin, L.R.; Lins, R.D.; Soares, T.A. Polymyxin binding to the bacterial outer membrane reveals cation displacement and increasing membrane curvature in susceptible but not in resistant lipopolysaccharide chemotypes. J. Chem. Inf. 2017, 57, 2181–2193. [Google Scholar] [CrossRef] [PubMed]
- Laubacher, M.E.; Ades, S.E. The Rcs Phosphorelay Is a Cell Envelope Stress Response Activated by Peptidoglycan Stress and Contributes to Intrinsic Antibiotic Resistance. J. Bacteriol. 2008, 190, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.Q.; Parker, C.T.; Louie, J.W.; Huynh, S.; Fagerquist, C.K.; Mandrell, R.E. RcsB Contributes to the Distinct Stress Fitness among Escherichia coli O157:H7 Curli Variants of the 1993 Hamburger-Associated Outbreak Strains. Appl. Environ. Microbiol. 2012, 78, 7706–7719. [Google Scholar] [CrossRef]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef]
- Tierney, A.R.; Rather, P.N. Roles of two-component regulatory systems in antibiotic resistance. Future Microbiol. 2019, 14, 533–552. [Google Scholar] [CrossRef]
- Surmann, K.; Ćudić, E.; Hammer, E.; Hunke, S. Molecular and proteome analyses highlight the importance of the Cpx envelope stress system for acid stress and cell wall stability in Escherichia coli. Microbiologyopen 2016, 5, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamasaki, S.; Hayashi-Nishino, M.; Yamaguchi, A. Effect of NlpE overproduction on multidrug resistance in Escherichia coli. Antimicrob. Agents Chemother. 2010, 54, 2239–2243. [Google Scholar] [CrossRef] [PubMed]
- Weatherspoon-Griffin, N.; Zhao, G.; Kong, W.; Kong, Y.; Andrews-Polymenis, H.M.; McClelland, M.; Shi, Y. The CpxR/CpxA Two-Component System Up-Regulates Two Tat-Dependent Peptidoglycan Amidases to Confer Bacterial Resistance to Antimicrobial Peptide. J. Biol. Chem. 2011, 286, 5529–5539. [Google Scholar] [CrossRef] [PubMed]
- Rubin, E.J.; Herrera, C.M.; Crofts, A.A.; Trent, M.S. PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob. Agents Chemother. 2015, 59, 2051–2061. [Google Scholar] [CrossRef]
- Kox, L.F.; Wösten, M.M.; Groisman, E.A. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000, 19, 1861–1872. [Google Scholar] [CrossRef]
- Kato, A.; Chen, H.D.; Latifi, T.; Groisman, E.A. Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol. Cell 2012, 47, 897–908. [Google Scholar] [CrossRef]
- Herrera, C.M.; Hankins, J.V.; Trent, M.S. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010, 76, 1444–1460. [Google Scholar] [CrossRef]
- Pasqua, M.; Grossi, M.; Scinicariello, S.; Aussel, L.; Barras, F.; Colonna, B.; Prosseda, G. The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep. 2019, 9, 2906. [Google Scholar] [CrossRef]
- Wang, S.; You, C.; Memon, F.Q.; Zhang, G.; Sun, Y.; Si, H. BaeR participates in cephalosporins susceptibility by regulating the expression level of outer membrane proteins in Escherichia coli. J. Biochem. 2020, 169, 101–108. [Google Scholar] [CrossRef]
- Deininger, K.N.; Horikawa, A.; Kitko, R.D.; Tatsumi, R.; Rosner, J.L.; Wachi, M.; Slonczewski, J.L. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS ONE 2011, 6, e18960. [Google Scholar] [CrossRef]
- Baral, B.; Mozafari, M.R. Strategic Moves of “Superbugs” Against Available Chemical Scaffolds: Signaling, Regulation, and Challenges. ACS Pharmacol. Transl. Sci. 2020, 3, 373–400. [Google Scholar] [CrossRef] [PubMed]
- Greie, J.C. The KdpFABC complex from Escherichia coli: A chimeric K+ transporter merging ion pumps with ion channels. Eur. J. Cell Biol. 2011, 90, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.S.; García-Contreras, R.; Wood, T.K. YcfR (BhsA) influences Escherichia coli biofilm formation through stress response and surface hydrophobicity. J. Bacteriol. 2007, 189, 3051–3062. [Google Scholar] [CrossRef] [PubMed]
- Delhaye, A.; Collet, J.-F.; Laloux, G. Fine-Tuning of the Cpx Envelope Stress Response Is Required for Cell Wall Homeostasis in Escherichia coli. mBio 2016, 7, e00047-16. [Google Scholar] [CrossRef]
- Carone, B.R.; Xu, T.; Murphy, K.C.; Marinus, M.G. High incidence of multiple antibiotic resistant cells in cultures of in enterohemorrhagic Escherichia coli O157:H7. Mutat. Res. 2014, 759, 1–8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baranova, N.; Nikaido, H. The baeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 2002, 184, 4168–4176. [Google Scholar] [CrossRef]
- Nagakubo, S.; Nishino, K.; Hirata, T.; Yamaguchi, A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 2002, 184, 4161–4167. [Google Scholar] [CrossRef]
- Allen, K.J.; Lepp, D.; McKellar, R.C.; Griffiths, M.W. Examination of stress and virulence gene expression in Escherichia coli O157:H7 using targeted microarray analysis. Foodborne Pathog. Dis. 2008, 5, 437–447. [Google Scholar] [CrossRef]
- Pietiäinen, M.; Gardemeister, M.; Mecklin, M.; Leskelä, S.; Sarvas, M.; Kontinen, V.P. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 2005, 151, 1577–1592. [Google Scholar] [CrossRef]
- Arends, S.J.; Weiss, D.S. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 2004, 186, 880–884. [Google Scholar] [CrossRef]
- Clarke, D.J. The Rcs phosphorelay: More than just a two-component pathway. Future Microbiol. 2010, 5, 1173–1184. [Google Scholar] [CrossRef]
- Ferrières, L.; Clarke, D.J. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol. Microbiol. 2003, 50, 1665–1682. [Google Scholar] [CrossRef]
- Miller, C.; Thomsen, L.E.; Gaggero, C.; Mosseri, R.; Ingmer, H.; Cohen, S.N. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 2004, 305, 1629–1631. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Nishino, K.; Yamada, J.; Hirata, T.; Yamaguchi, A. Beta-lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Antimicrob. Chemother. 2003, 52, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Church, G.M. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 2002, 184, 6225–6234. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Inazumi, Y.; Yamaguchi, A. Global analysis of genes regulated by EvgA of the two-component regulatory system in Escherichia coli. J. Bacteriol. 2003, 185, 2667–2672. [Google Scholar] [CrossRef]
- Cole, J.A. Anaerobic Bacterial Response to Nitrosative Stress. Adv. Microb. Physiol. 2018, 72, 193–237. [Google Scholar] [CrossRef] [PubMed]
- Fernando, D.M.; Kumar, A. Resistance-Nodulation-Division Multidrug Efflux Pumps in Gram-Negative Bacteria: Role in Virulence. Antibiotics 2013, 2, 163–181. [Google Scholar] [CrossRef]
- Sharma, P.; Haycocks, J.R.J.; Middlemiss, A.D.; Kettles, R.A.; Sellars, L.E.; Ricci, V.; Piddock, L.J.V.; Grainger, D.C. The multiple antibiotic resistance operon of enteric bacteria controls DNA repair and outer membrane integrity. Nat. Commun. 2017, 8, 1444. [Google Scholar] [CrossRef] [PubMed]
- Duval, V.; Lister, I.M. MarA, SoxS and Rob of Escherichia coli—Global regulators of multidrug resistance, virulence and stress response. Int. J. Biotechnol. Wellness Ind. 2013, 2, 101–124. [Google Scholar] [CrossRef]
- White, S.; Tuttle, F.E.; Blankenhorn, D.; Dosch, D.C.; Slonczewski, J.L. pH dependence and gene structure of inaA in Escherichia coli. J. Bacteriol. 1992, 174, 1537–1543. [Google Scholar] [CrossRef]
- Barbosa, T.M.; Levy, S.B. Differential Expression of over 60 Chromosomal Genes in Escherichia coli by Constitutive Expression of MarA. J. Bacteriol. 2000, 182, 3467–3474. [Google Scholar] [CrossRef] [PubMed]
- Rossi, N.A.; Dunlop, M.J. Customized Regulation of Diverse Stress Response Genes by the Multiple Antibiotic Resistance Activator MarA. PLoS Comput. Biol. 2017, 13, e1005310. [Google Scholar] [CrossRef] [PubMed]
- Prajapat, M.K.; Jain, K.; Saini, S. Control of MarRAB Operon in Escherichia coli via Autoactivation and Autorepression. Biophys. J. 2015, 109, 1497–1508. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Krishnamoorthy, G.; Ntreh, A.; Lu, S. Mechanism and Function of the Outer Membrane Channel TolC in Multidrug Resistance and Physiology of Enterobacteria. Front. Microbiol. 2011, 2, 189. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, S.K.D.; Oates, C.W.; Raivio, T.L. Characterization of the Induction and Cellular Role of the BaeSR Two-Component Envelope Stress Response of Escherichia coli. J. Bacteriol. 2011, 193, 3367–3375. [Google Scholar] [CrossRef] [PubMed]
- Rosner, J.L.; Martin, R.G. Reduction of Cellular Stress by TolC-Dependent Efflux Pumps in Escherichia coli Indicated by BaeSR and CpxARP Activation of spy in Efflux Mutants. J. Bacteriol. 2013, 195, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Schusser, B.; Collarini, E.J.; Yi, H.; Izquierdo, S.M.; Fesler, J.; Pedersen, D.; Klasing, K.C.; Kaspers, B.; Harriman, W.D.; van de Lavoir, M.C.; et al. Immunoglobulin knockout chickens via efficient homologous recombination in primordial germ cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20170–20175. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; O’Riordan, M.X. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv. Appl. Microbiol. 2013, 85, 93–118. [Google Scholar] [CrossRef]
- Bhagwat, A.A.; Chan, L.; Han, R.; Tan, J.; Kothary, M.; Jean-Gilles, J.; Tall, B.D. Characterization of enterohemorrhagic Escherichia coli strains based on acid resistance phenotypes. Infect. Immun. 2005, 73, 4993–5003. [Google Scholar] [CrossRef]
- Dong, T.; Schellhorn, H.E. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genom. 2009, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Mand, T.D.; Döpfer, D.; Ingham, B.; Ané, C.; Kaspar, C.W. Growth and survival parameter estimates and relation to RpoS levels in serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. J. Appl. Microbiol. 2013, 114, 242–255. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, S.; Xu, Y.; Mi, X.; Xing, T.; Li, J.; Zhang, L.; Gao, F.; Jiang, Y. Acid resistance of E. coli O157:H7 and O26:H11 exposure to lactic acid revealed by transcriptomic analysis. LWT 2021, 136, 110352. [Google Scholar] [CrossRef]
- Seo, S.W.; Kim, D.; O’Brien, E.J.; Szubin, R.; Palsson, B.O. Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 2015, 6, 7970. [Google Scholar] [CrossRef]
- Kailasan Vanaja, S.; Bergholz, T.M.; Whittam, T.S. Characterization of the Escherichia coli O157:H7 Sakai GadE Regulon. J. Bacteriol. 2009, 191, 1868–1877. [Google Scholar] [CrossRef]
- Pore, D.; Hoque, K.M.; Chakrabarti, M.K. Chapter 10—Animal models in advancement of research in enteric diseases. In Animal Biotechnology, 2nd ed.; Verma, A.S., Singh, A., Eds.; Academic Press: Boston, MA, USA, 2020; pp. 199–219. [Google Scholar] [CrossRef]
- McConnell, E.L.; Basit, A.W.; Murdan, S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J. Pharm. Pharmacol. 2008, 60, 63–70. [Google Scholar] [CrossRef]
- Brenneman, K.E.; Willingham, C.; Kilbourne, J.A.; Curtiss, R., 3rd; Roland, K.L. A low gastric pH mouse model to evaluate live attenuated bacterial vaccines. PLoS ONE 2014, 9, e87411. [Google Scholar] [CrossRef]
- Price, S.B.; Wright, J.C.; DeGraves, F.J.; Castanie-Cornet, M.P.; Foster, J.W. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl. Environ. Microbiol. 2004, 70, 4792–4799. [Google Scholar] [CrossRef]
- Chingcuanco, F.; Yu, Y.; Kus, J.V.; Que, L.; Lackraj, T.; Lévesque, C.M.; Barnett Foster, D. Identification of a novel adhesin involved in acid-induced adhesion of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 2012, 158, 2399–2407. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, C.; Song, J.; Velkov, T.; Wang, L.; Zhu, Y.; Li, J. Regulating polymyxin resistance in Gram-negative bacteria: Roles of two-component systems PhoPQ and PmrAB. Future Med. 2020, 15, 445–459. [Google Scholar] [CrossRef]
- Miyawaki, S.; Uemura, Y.; Hongo, K.; Kawata, Y.; Mizobata, T. Acid-denatured small heat shock protein HdeA from Escherichia coli forms reversible fibrils with an atypical secondary structure. J. Biol. Chem. 2019, 294, 1590–1601. [Google Scholar] [CrossRef]
- Loose, M.; Naber, K.G.; Coates, A.; Wagenlehner, F.M.E.; Hu, Y. Effect of Different Media on the Bactericidal Activity of Colistin and on the Synergistic Combination With Azidothymidine Against mcr-1-Positive Colistin-Resistant Escherichia coli. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Azizoglu, R.O. Influence of Antibiotic, Acid, and Salt Stress on Resistance of Escherichia coli O157:H7; NC State University Libraries: Raleigh, NC, USA, 2006. [Google Scholar]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef]
- Schjørring, S.; Krogfelt, K.A. Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011, 2011, 312956. [Google Scholar] [CrossRef] [PubMed]
- Salyers, A.A.; Gupta, A.; Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004, 12, 412–416. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Gupta, S.S.; Nair, G.B.; Mande, S.S. In silico analysis of antibiotic resistance genes in the gut microflora of individuals from diverse geographies and age-groups. PLoS ONE 2013, 8, e83823. [Google Scholar] [CrossRef] [PubMed]
- Sparo, M.; Urbizu, L.; Solana, M.V.; Pourcel, G.; Delpech, G.; Confalonieri, A.; Ceci, M.; Sánchez Bruni, S.F. High-level resistance to gentamicin: Genetic transfer between Enterococcus faecalis isolated from food of animal origin and human microbiota. Lett. Appl. Microbiol. 2012, 54, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed]
- Trobos, M.; Lester, C.H.; Olsen, J.E.; Frimodt-Møller, N.; Hammerum, A.M. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J. Antimicrob. Chemother. 2009, 63, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Samelis, J.; Ikeda, J.S.; Sofos, J.N. Evaluation of the pH-dependent, stationary-phase acid tolerance in Listeria monocytogenes and Salmonella Typhimurium DT104 induced by culturing in media with 1% glucose: A comparative study with Escherichia coli O157:H7. J. Appl. Microbiol. 2003, 95, 563–575. [Google Scholar] [CrossRef]
- Karami, N.; Martner, A.; Enne, V.I.; Swerkersson, S.; Adlerberth, I.; Wold, A.E. Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J. Antimicrob. Chemother. 2007, 60, 1142–1145. [Google Scholar] [CrossRef]
- Fujimoto, T.; Tsuchiya, Y.; Terao, M.; Nakamura, K.; Yamamoto, M. Antibacterial effects of Chitosan solution® against Legionella pneumophila, Escherichia coli, and Staphylococcus aureus. Int. J. Food Microbiol. 2006, 112, 96–101. [Google Scholar] [CrossRef]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef]
- Capita, R.; Alvarez-Fernández, E.; Fernández-Buelta, E.; Manteca, J.; Alonso-Calleja, C. Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiol. 2013, 34, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.-B.; Seo, K.-H. Variation of antibiotic resistance in Salmonella Enteritidis, Escherichia coli O157:H7, and Listeria monocytogenes after exposure to acid, salt, and cold stress. J. Food Saf. 2020, 40, e12804. [Google Scholar] [CrossRef]
- Duffy, G.; Walsh, C.; Blair, I.S.; McDowell, D.A. Survival of antibiotic resistant and antibiotic sensitive strains of E. coli O157 and E. coli O26 in food matrices. Int. J. Food Microbiol. 2006, 109, 179–186. [Google Scholar] [CrossRef][Green Version]
- Azizoglu, R.O.; Drake, M. Impact of antibiotic stress on acid and heat tolerance and virulence factor expression of Escherichia coli O157:H7. J. Food Prot. 2007, 70, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Dai, X. High Salt Cross-Protects Escherichia coli from Antibiotic Treatment through Increasing Efflux Pump Expression. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Schembri, M.A.; Kjaergaard, K.; Klemm, P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 2003, 48, 253–267. [Google Scholar] [CrossRef]
- Beloin, C.; Valle, J.; Latour-Lambert, P.; Faure, P.; Kzreminski, M.; Balestrino, D.; Haagensen, J.A.; Molin, S.; Prensier, G.; Arbeille, B.; et al. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 2004, 51, 659–674. [Google Scholar] [CrossRef]
- Ren, D.; Bedzyk, L.A.; Thomas, S.M.; Ye, R.W.; Wood, T.K. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 2004, 64, 515–524. [Google Scholar] [CrossRef]
- DeLisa, M.P.; Valdes, J.J.; Bentley, W.E. Mapping stress-induced changes in autoinducer AI-2 production in chemostat-cultivated Escherichia coli K-12. J. Bacteriol. 2001, 183, 2918–2928. [Google Scholar] [CrossRef] [PubMed]
- Patten, C.L.; Kirchhof, M.G.; Schertzberg, M.R.; Morton, R.A.; Schellhorn, H.E. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genom. 2004, 272, 580–591. [Google Scholar] [CrossRef] [PubMed]
- Domka, J.; Lee, J.; Bansal, T.; Wood, T.K. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 2007, 9, 332–346. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Page, R.; García-Contreras, R.; Palermino, J.-M.; Zhang, X.-S.; Doshi, O.; Wood, T.K.; Peti, W. Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance. J. Mol. Biol. 2007, 373, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Mitra, A.; Fay, P.A.; Morgan, J.K.; Vendura, K.W.; Versaggi, S.L.; Riordan, J.T. Sigma factor N, liaison to an ntrC and rpoS dependent regulatory pathway controlling acid resistance and the LEE in enterohemorrhagic Escherichia coli. PLoS ONE 2012, 7, e46288. [Google Scholar] [CrossRef]
- Mathlouthi, A.; Pennacchietti, E.; Biase, D. Effect of Temperature, pH and Plasmids on In Vitro Biofilm Formation in Escherichia coli. Acta Nat. 2018, 10, 129–132. [Google Scholar] [CrossRef]
- Shayanfar, S.; Broumand, A.; Pillai, S.D. Acid stress induces differential accumulation of metabolites in Escherichia coli O26:H11. J. Appl. Microbiol. 2018, 125, 1911–1919. [Google Scholar] [CrossRef]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vázquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Siragusa, S.; De Angelis, M.; Di Cagno, R.; Rizzello, C.G.; Coda, R.; Gobbetti, M. Synthesis of γ-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007, 73, 7283–7290. [Google Scholar] [CrossRef]
- Li, H.; Cao, Y. Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 2010, 39, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.G.; Bottiglieri, T.; Snead, O.C., 3rd. GABA, gamma-hydroxybutyric acid, and neurological disease. Ann. Neurol. 2003, 54 (Suppl. 6), S3–S12. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-H.; Soh, J.; Cha, Y. Germinated brown rice extract shows a nutraceutical effect in the recovery of chronic alcohol-related symptoms. J. Med. Food 2003, 6, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Mazzacane, F.; Rizzello, C.G.; De Angelis, M.; Giuliani, G.; Meloni, M.; De Servi, B.; Gobbetti, M. Synthesis of gamma-aminobutyric acid (GABA) by Lactobacillus plantarum DSM19463: Functional grape must beverage and dermatological applications. Appl. Microbiol. Biotechnol. 2010, 86, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Charlet, R.; Bortolus, C.; Sendid, B.; Jawhara, S. Bacteroides thetaiotaomicron and Lactobacillus johnsonii modulate intestinal inflammation and eliminate fungi via enzymatic hydrolysis of the fungal cell wall. Sci. Rep. 2020, 10, 11510. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Petersen, F.C.; Shekhar, S. Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Needham, B.D.; Trent, M.S. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013, 11, 467–481. [Google Scholar] [CrossRef]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. J. mBio 2016, 7, e01250-16. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, W.; Lan, P.; Mou, X. The microbiome in inflammatory bowel diseases: From pathogenesis to therapy. Protein Cell 2020. [Google Scholar] [CrossRef]
- Chin, V.K.; Yong, V.C.; Chong, P.P.; Amin Nordin, S.; Basir, R.; Abdullah, M. Mycobiome in the Gut: A Multiperspective Review. Mediat. Inflamm. 2020, 2020, 9560684. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, F.; Massimino, L.; D’Alessio, S.; Danese, S. The gut virome in inflammatory bowel disease pathogenesis: From metagenomics to novel therapeutic approaches. United Eur. Gastroenterol. J. 2019, 7, 999–1007. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).