Characterization of 475 Novel, Putative Small RNAs (sRNAs) in Carbon-Starved Salmonella enterica Serovar Typhimurium
Abstract
1. Introduction
2. Results
2.1. Identification of 475 Novel Salmonella sRNAs Dynamically Expressed during C-Starvation
2.2. Characterization of sRNAs as Inter- or Intragenic
2.3. sRNAs and Related Loci Are Highly Conserved
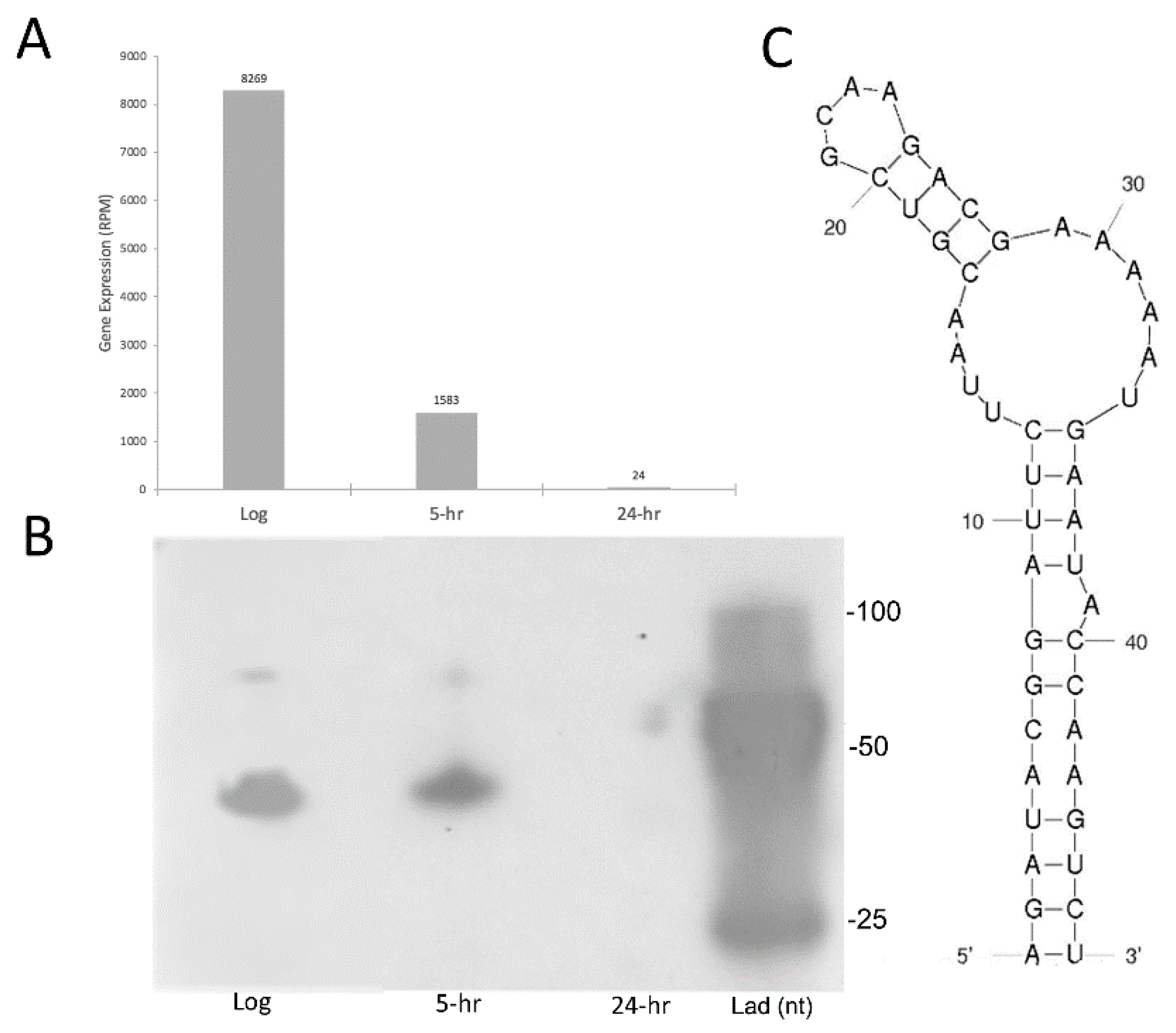
2.4. sRNA Profiles Associated with Distinct States of C-Starvation Are Highly Distinct
2.5. Identification of Putative sRNA Targets
2.6. Deletion of Novel sRNAs Can Significantly Impair the Salmonella Starvation Response
3. Discussion
4. Materials and Methods
4.1. Bacteria Strains and Media
4.2. Cultivation of Non-Starved Log-Phase and C-Starved Cell Cultures
4.3. RNA Isolation and Small RNA Sequencing
4.4. Identification of sRNA Consensus Sequences
4.5. sRNA Computational Analyses and Target Gene Prediction
4.6. Small Transcript Northern Blotting
- Probe4372453 5pBio-TGCGACGTTAAGAATCCGTATCT
- Probe4372453RevComp 5pBio-AGATACGGATTCTTAACGTCGCA
- Probe4372453SCRAMBLE 5pBio-CGTAGTGAATACCCGTTTATACG
4.7. Deletions via Lambda-Red Protocol
4.8. Quantitative RT-PCR
4.9. Phenotypic Stress Challenges
4.9.1. Biofilm Formation and Curli Fimbriae Production
4.9.2. Growth Curve Assay
4.9.3. Peroxide/Poly B Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Fischer, J.W.; Leung, A.K.L. CircRNAs: A regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Hollins, S.L.; Cairns, M.J. MicroRNA: Small RNA mediators of the brains genomic response to environmental stress. Prog. Neurobiol. 2016, 143, 61–81. [Google Scholar] [CrossRef]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell. Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef]
- Sun, X.; Zheng, H.; Sui, N. Regulation mechanism of long non-coding RNA in plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, S.S.; Karimian, A.; Parsian, H.; Majidinia, M.; Yousefi, B. Multiple Functions of Long Non-Coding RNAs in Oxidative Stress, DNA Damage Response and Cancer Progression. J. Cell. Biochem. 2018, 119, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Cavanagh, A.T.; Wassarman, K.M. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu. Rev. Microbiol. 2014, 68, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Hoe, C.H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Holmqvist, E.; Wagner, E.G.H. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef]
- Amin, S.V.; Roberts, J.T.; Patterson, D.G.; Coley, A.B.; Allred, J.A.; Denner, J.M.; Johnson, J.P.; Mullen, G.E.; O’Neal, T.K.; Smith, J.T.; et al. Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium. RNA Biol. 2016, 13, 331–342. [Google Scholar] [CrossRef]
- Deng, X.; Li, Z.; Zhang, W. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 2012, 30, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Cardin, S.E.; Borchert, G.M. Viral MicroRNAs, Host MicroRNAs Regulating Viruses, and Bacterial MicroRNA-Like RNAs. Methods Mol. Biol. 2017, 1617, 39–56. [Google Scholar] [PubMed]
- Vogel, J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009, 71, 1–11. [Google Scholar] [CrossRef]
- Barnhill, E.C.; Crucello, A.; Houserova, D.; King, V.M.; Amin, S.V.; Roberts, J.T.; Zambrano, M.E.; DeMeis, J.D.; Dahmer, D.J.; Ijaz, Z.; et al. Characterization of novel small RNAs (sRNAs) contributing to the desiccation response of Salmonella enterica serovar Typhimurium. RNA Biol. 2019, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hébrard, M.; Kröger, C.; Srikumar, S.; Colgan, A.; Händler, K.; Hinton, J.C. sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol. 2012, 9, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Kröger, C.; Dillon, S.C.; Cameron, A.D.S.; Papenfort, K.; Sivasankaran, S.K.; Hokamp, K.; Chao, Y.; Sittka, A.; Hébrard, M.; Händler, K.; et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. USA 2012, 109, E1277–E1286. [Google Scholar] [CrossRef]
- Kröger, C.; Colgan, A.; Srikumar, S.; Händler, K.; Sivasankaran, S.K.; Hammarlöf, D.L.; Canals, R.; Grissom, J.E.; Conway, T.; Hokamp, K.; et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 2013, 14, 683–695. [Google Scholar] [CrossRef]
- Ryan, D.; Mukherjee, M.; Nayak, R.; Dutta, R.; Suar, M. Biological and regulatory roles of acid-induced small RNA RyeC in Salmonella Typhimurium. Biochimie 2018, 150, 48–56. [Google Scholar] [CrossRef]
- Dar, D.; Sorek, R. Bacterial Noncoding RNAs Excised from within Protein-Coding Transcripts. MBio 2018, 9, e01730-18. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Melamed, S.; Peer, A.; Faigenbaum-Romm, R.; Gatt, Y.E.; Reiss, N.; Bar, A.; Altuvia, Y.; Argaman, L.; Margalit, H. Global Mapping of Small RNA-Target Interactions in Bacteria. Mol. Cell 2016, 63, 884–897. [Google Scholar] [CrossRef]
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L.; et al. Ensembl 2016. Nucleic Acids Res. 2016, 44, D710–D716. [Google Scholar] [CrossRef]
- Murphy, K.C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 1998, 180, 2063–2071. [Google Scholar] [CrossRef]
- Evans, D.J.; Brown, M.R.; Allison, D.G.; Gilbert, P. Susceptibility of bacterial biofilms to tobramycin: Role of specific growth rate and phase in the division cycle. J. Antimicrob. Chemother. 1990, 25, 585–591. [Google Scholar] [CrossRef]
- Raghavan, R.; Groisman, E.A.; Ochman, H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011, 21, 1487–1497. [Google Scholar] [CrossRef]
- Vulin, C.; Leimer, N.; Huemer, M.; Ackermann, M.; Zinkernagel, A.S. Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat. Commun. 2018, 9, 4074. [Google Scholar] [CrossRef]
- Chen, C.; Khaleel, S.S.; Huang, H.; Wu, C.H. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol. Med. 2014, 9, 8. [Google Scholar] [CrossRef]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2015, 43, D662–D669. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.S.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Milne, I.; Bayer, M.; Cardle, L.; Shaw, P.; Stephen, G.; Wright, F.; Marshall, D. Tablet--next generation sequence assembly visualization. Bioinformatics 2010, 26, 401–402. [Google Scholar] [CrossRef]
- Flicek, P.; Ahmed, I.; Amode, M.R.; Barrell, D.; Beal, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fairley, S.; et al. Ensembl 2013. Nucleic Acids Res. 2013, 41, D48–D55. [Google Scholar] [CrossRef]
- Cherepanov, P.P.; Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Ćirković, I.; Ranin, L.; Švabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houserova, D.; Dahmer, D.J.; Amin, S.V.; King, V.M.; Barnhill, E.C.; Zambrano, M.E.; Dean, M.A.; Crucello, A.; Aria, K.M.; Spector, M.P.; et al. Characterization of 475 Novel, Putative Small RNAs (sRNAs) in Carbon-Starved Salmonella enterica Serovar Typhimurium. Antibiotics 2021, 10, 305. https://doi.org/10.3390/antibiotics10030305
Houserova D, Dahmer DJ, Amin SV, King VM, Barnhill EC, Zambrano ME, Dean MA, Crucello A, Aria KM, Spector MP, et al. Characterization of 475 Novel, Putative Small RNAs (sRNAs) in Carbon-Starved Salmonella enterica Serovar Typhimurium. Antibiotics. 2021; 10(3):305. https://doi.org/10.3390/antibiotics10030305
Chicago/Turabian StyleHouserova, Dominika, Donovan J. Dahmer, Shivam V. Amin, Valeria M. King, Emmaline C. Barnhill, Mike E. Zambrano, Meghan A. Dean, Aline Crucello, Kevin M. Aria, Michael P. Spector, and et al. 2021. "Characterization of 475 Novel, Putative Small RNAs (sRNAs) in Carbon-Starved Salmonella enterica Serovar Typhimurium" Antibiotics 10, no. 3: 305. https://doi.org/10.3390/antibiotics10030305
APA StyleHouserova, D., Dahmer, D. J., Amin, S. V., King, V. M., Barnhill, E. C., Zambrano, M. E., Dean, M. A., Crucello, A., Aria, K. M., Spector, M. P., & Borchert, G. M. (2021). Characterization of 475 Novel, Putative Small RNAs (sRNAs) in Carbon-Starved Salmonella enterica Serovar Typhimurium. Antibiotics, 10(3), 305. https://doi.org/10.3390/antibiotics10030305






