Rapid and High-Throughput Evaluation of Diverse Configurations of Engineered Lysins Using the VersaTile Technique
Abstract
1. Introduction
2. Results
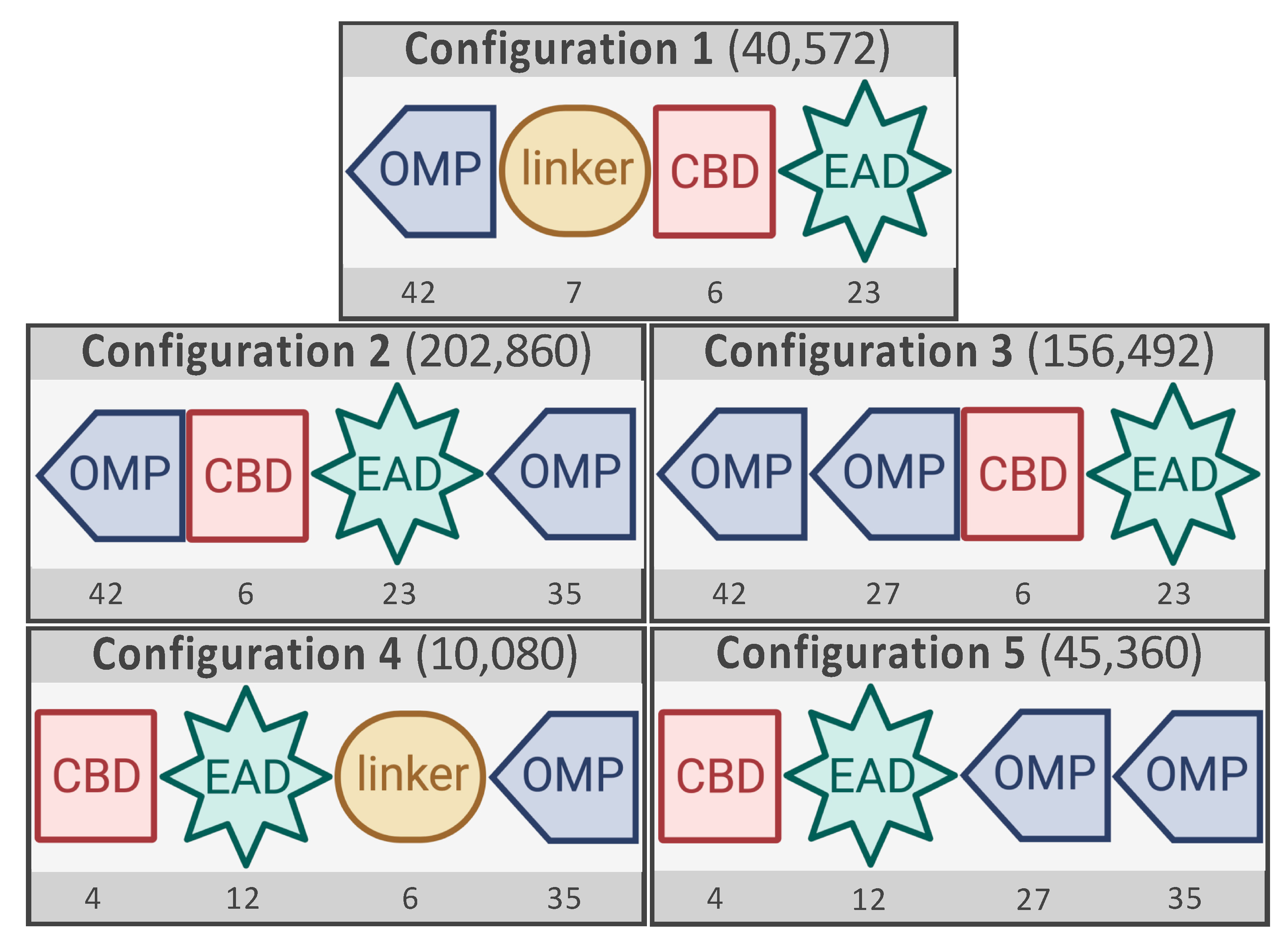
2.1. The Construction of Five Libraries with Different Configurations Using VersaTile
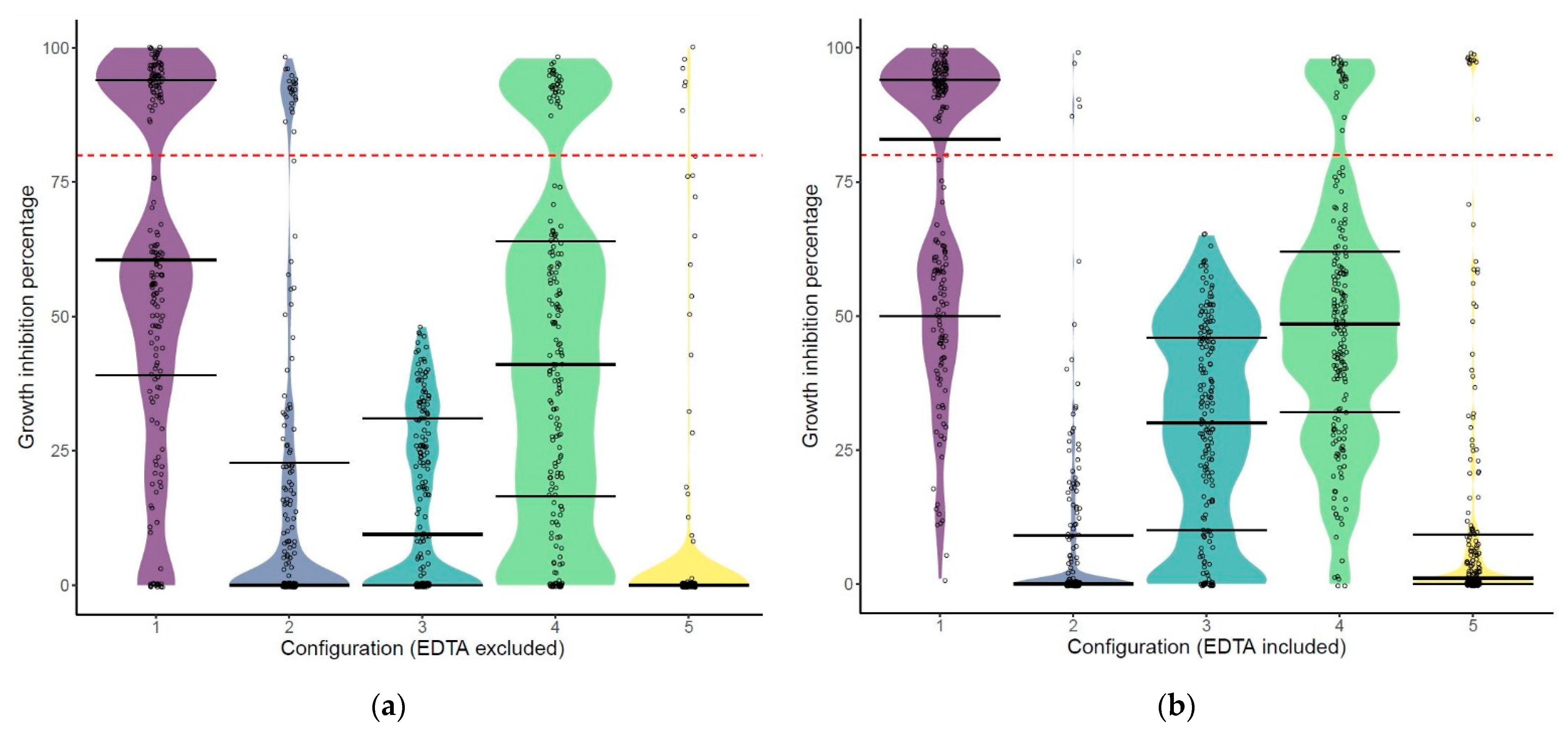
2.2. The Hit Rate of the Libraries Depends on the Configuration of the Engineered Lysins
2.3. Sequencing of the Most Active Hits Gives Design Rules
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Media
4.2. Expanding of Tile Repository: VersaTile Cloning
4.3. Construction of Libraries: VersaTile Shuffling
4.4. High-Throughput Growth Inhibitory Assay
4.5. Statistics
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gerstmans, H.; Criel, B.; Briers, Y. Synthetic biology of modular endolysins. Biotechnol. Adv. 2018, 36, 624–640. [Google Scholar] [CrossRef] [PubMed]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Defraine, V.; Schuermans, J.; Grymonprez, B.; Govers, S.K.; Aertsen, A.; Fauvart, M.; Michiels, J.; Lavigne, R.; Briers, Y. Efficacy of Artilysin Art-175 against resistant and persistent Acinetobacter baumannii. Antimicrob. Agents Chemother. 2016, 60, 3480–3488. [Google Scholar] [CrossRef] [PubMed]
- Fenton, M.; Ross, P.; McAuliffe, O.; O’Mahony, J.; Coffey, A. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs. 2010, 1, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Gerstmans, H.; Grimon, D.; Gutiérrez, D.; Lood, C.; Rodríguez, A.; van Noort, V.; Lammertyn, J.; Lavigne, R.; Briers, Y. A VersaTile driven platform for rapid hit-to-lead development of engineered lysins. Sci. Adv. 2020, 6, eaaz1136. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Walmagh, M.; Van Puyenbroeck, V.; Cornelissen, A.; Cenens, W.; Aertsen, A.; Oliveira, H.; Azeredo, J.; Verween, G.; Pimay, J.P.; et al. Engineered endolysin-based “Artilysins” to combat multidrug-resistant Gram-negative pathogens. Mbio 2014, 5, e01379-14. [Google Scholar] [CrossRef] [PubMed]
- Röhrig, C.; Huemer, M.; Lorgé, D.; Luterbacher, S.; Phothaworn, P.; Schefer, C.; Sobieraj, A.M.; Zinsli, L.V.; Shambat, S.M.; Leimer, N.; et al. Targeting hidden pathogens: Cell-penetrating enzybiotics eradicate intracellular drug-resistant Staphylococcus aureus. Mbio 2020, 11, e00209-20. [Google Scholar] [CrossRef] [PubMed]
- Seijsing, J.; Sobieraj, A.M.; Keller, N.; Shen, Y.; Zinkernagel, A.S.; Loessner, M.J.; Schmelcher, M. Improved biodistribution and extended serum half-life of a bacteriophage endolysin by albumin binding domain fusion. Front. Microbiol. 2018, 9, 2927. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, H.; Wang, J.; Yu, J.; Wei, H. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Sci. Rep. 2017, 7, 40182. [Google Scholar] [CrossRef] [PubMed]
- Verbree, C.T.; Dätwyler, S.M.; Meile, S.; Eichenseher, F.; Donovan, D.M.; Loessner, M.J.; Schmelcher, M. Identification of peptidoglycan hydrolase constructs with synergistic staphylolytic activity in cow milk. Appl. Environ. Microbiol. 2017, 83, e03445-16. [Google Scholar] [CrossRef] [PubMed]
- Antibiotic Resistance Threats in the United States. 2019. Available online: www.cdc.gov/DrugResistance/Biggest-Threats.html (accessed on 23 April 2020).
- Healthcare-associated Infections (HAI)—Diseases and Organisms: Klebsiella. Available online: https://www.cdc.gov/hai/organisms/klebsiella/klebsiella.html (accessed on 23 April 2020).
- Lam, N.H.; Ma, Z.; Ha, B. Electrostatic modification of the lipopolysaccharide layer: Competing effects of divalent cations and polycationic or polyanionic molecules. Soft Matter. 2014, 10, 7528–7544. [Google Scholar] [CrossRef] [PubMed]
- Lood, C.; Gerstmans, H.; Briers, Y.; van Noort, V.; Lavigne, R. Quality control and statistical evaluation of combinatorial DNA libraries using nanopore sequencing. BioTechniques 2020, 69, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Walmagh, M.; Briers, Y.; dos Santos, S.B.; Azeredo, J.; Lavigne, R. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201φ2-1 and PVP-SE1. PLoS ONE 2012, 7, e36991. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zaro, J.L.; Shen, W. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Briers, Y.; Miroshnikov, K.; Chertkov, O.; Nekrasov, A.; Mesyanzhinov, V.; Volckaert, G.; Lavigne, R. The structural peptidoglycan hydrolase gp181 of bacteriophage phiKZ. Biochem. Biophys. Res. Commun. 2008, 374, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Gerhardsson, L.; Kazantzis, G. Diagnosis and Treatment of Metal Poisoning: General Aspects. In Handbook on the Toxicology of Metals, 4th ed.; Nordberg, G.F., Fowler, B.A., Nordberg, M., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 1, pp. 487–505. [Google Scholar]
- Schmidt-Dannert, C.; Arnold, F.H. Directed evolution of industrial enzymes. Trends Biotechnol. 1999, 17, 135–136. [Google Scholar] [CrossRef]
| Configuration | Position | Type Building Block | p-Value (no EDTA) | p-Value (EDTA) |
|---|---|---|---|---|
| 1 | 1 | OMP | 0.00035 * | 0.0074 * |
| 1 | 2 | Linker | 1 | 1 |
| 1 | 3 | CBD | 1 | 1 |
| 1 | 4 | EAD | 0.021 * | 0.047 * |
| 4 | 1 | CBD | 1 | 1 |
| 4 | 2 | EAD | 0.19677 | 0.39673 |
| 4 | 3 | Linker | 0.68365 | 0.04107 * |
| 4 | 4 | OMP | 0.01894 * | 0.00001 * |
| Configuration | Type Building Block | Tile | Without EDTA | With EDTA | ||
|---|---|---|---|---|---|---|
| N° | p-Value | N° | p-Value | |||
| 1 | OMP | Protegrin (OMP3) | 7 | 8.4 × 10−7 | 10 | 4.8 × 10−10 |
| 1 | OMP | Chrysophsin-1 (OMP20) | 4 | 0.0010 | 3 | NS (>0.0024) |
| 1 | EAD | 201ϕ2-1gp229-EAD (EAD12) | 5 | 0.0027 | 4 | NS (>0.0043) |
| 1 | EAD | KZgp181 (EAD19) | 3 | NS (>0.0043) | 7 | 0.00013 |
| 4 | OMP | Cathelicidin-BF antimicrobial peptide (OMP35) | 5 | 2.2 × 10−7 | 5 | 4.3 × 10−6 |
| 4 | Linker | Rigid long helix (Linker 4) | 4 | NS (>0.017) | 5 | 0.0080 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duyvejonck, L.; Gerstmans, H.; Stock, M.; Grimon, D.; Lavigne, R.; Briers, Y. Rapid and High-Throughput Evaluation of Diverse Configurations of Engineered Lysins Using the VersaTile Technique. Antibiotics 2021, 10, 293. https://doi.org/10.3390/antibiotics10030293
Duyvejonck L, Gerstmans H, Stock M, Grimon D, Lavigne R, Briers Y. Rapid and High-Throughput Evaluation of Diverse Configurations of Engineered Lysins Using the VersaTile Technique. Antibiotics. 2021; 10(3):293. https://doi.org/10.3390/antibiotics10030293
Chicago/Turabian StyleDuyvejonck, Lisa, Hans Gerstmans, Michiel Stock, Dennis Grimon, Rob Lavigne, and Yves Briers. 2021. "Rapid and High-Throughput Evaluation of Diverse Configurations of Engineered Lysins Using the VersaTile Technique" Antibiotics 10, no. 3: 293. https://doi.org/10.3390/antibiotics10030293
APA StyleDuyvejonck, L., Gerstmans, H., Stock, M., Grimon, D., Lavigne, R., & Briers, Y. (2021). Rapid and High-Throughput Evaluation of Diverse Configurations of Engineered Lysins Using the VersaTile Technique. Antibiotics, 10(3), 293. https://doi.org/10.3390/antibiotics10030293







