Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians
Abstract
1. Introduction
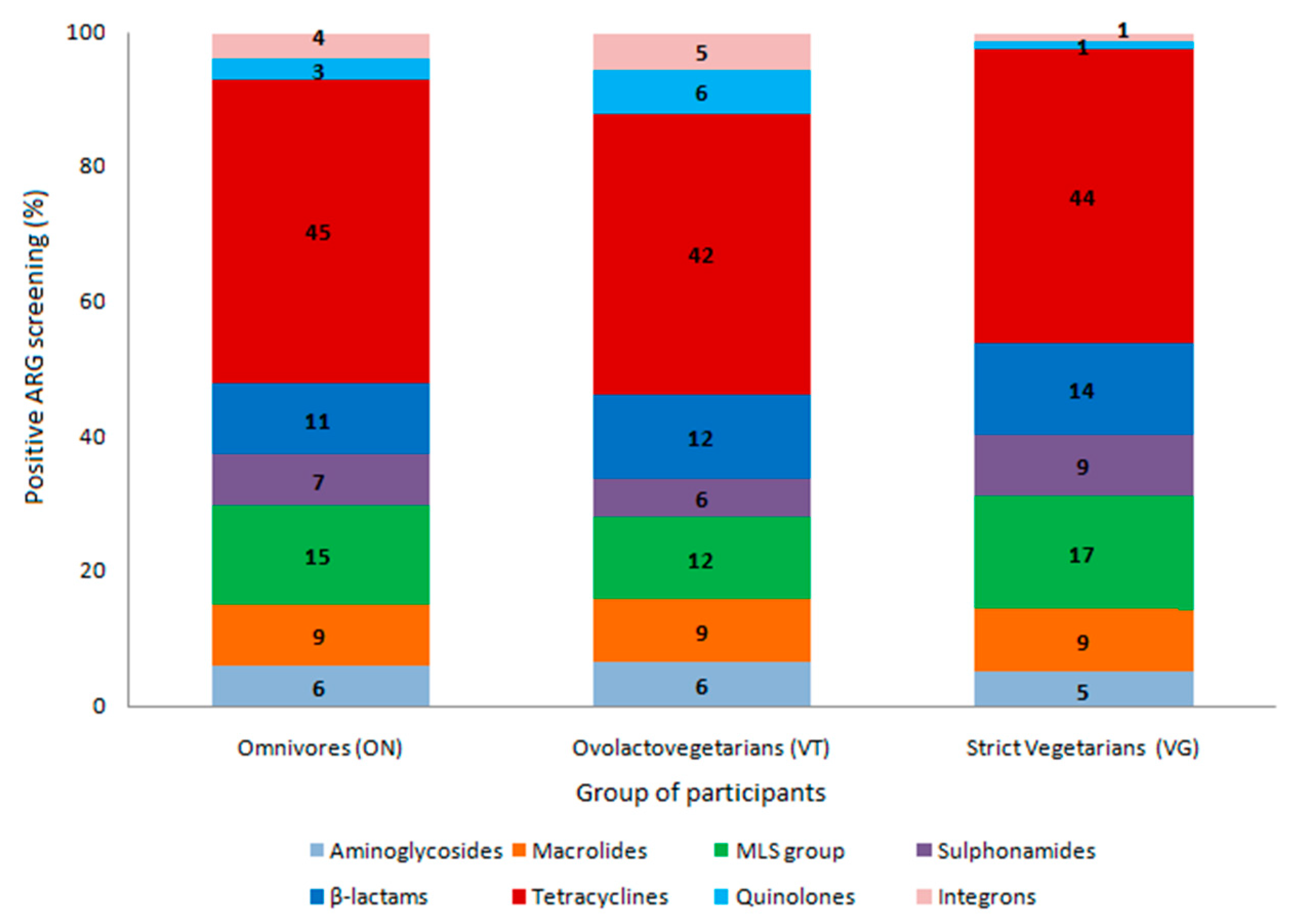
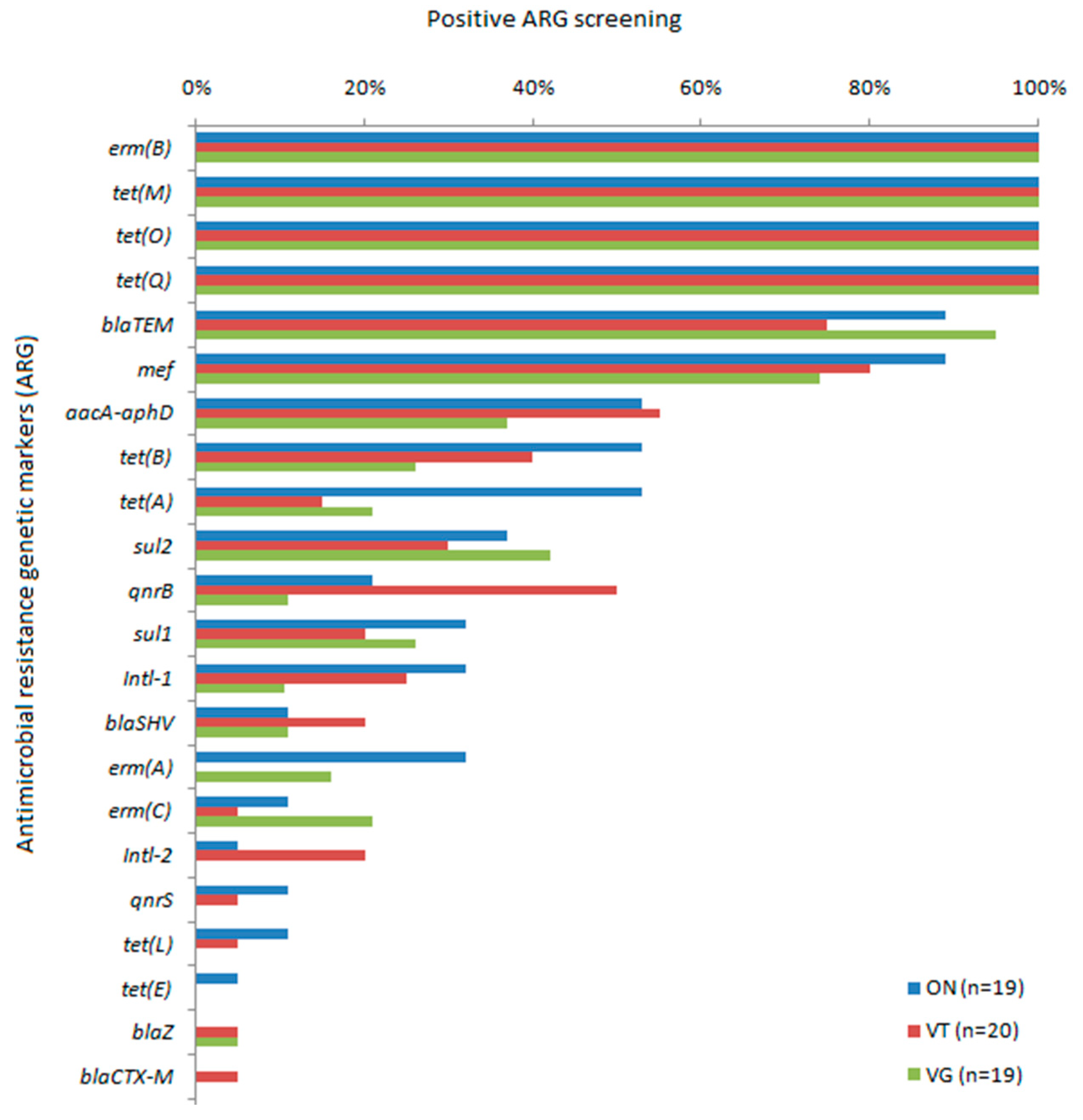
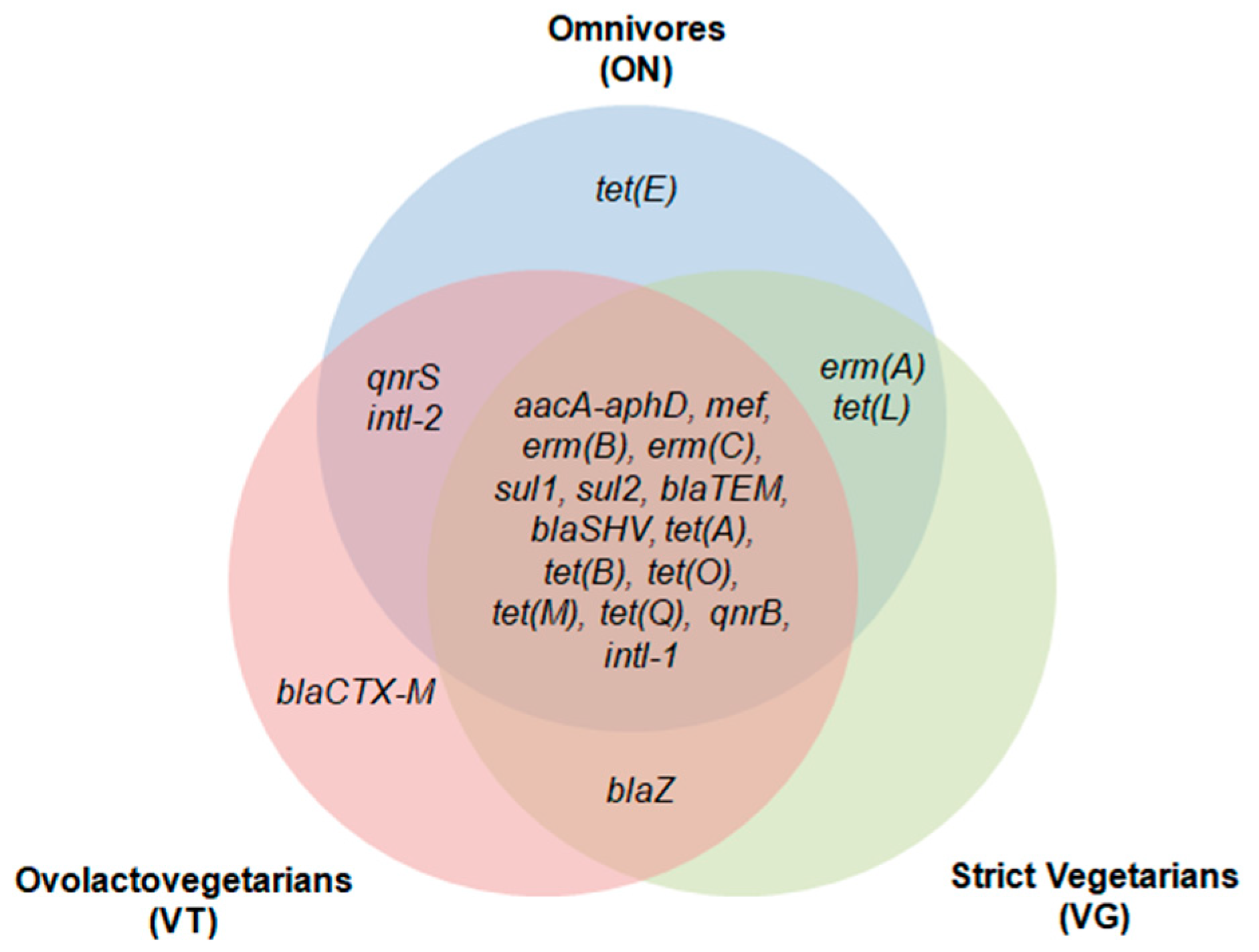
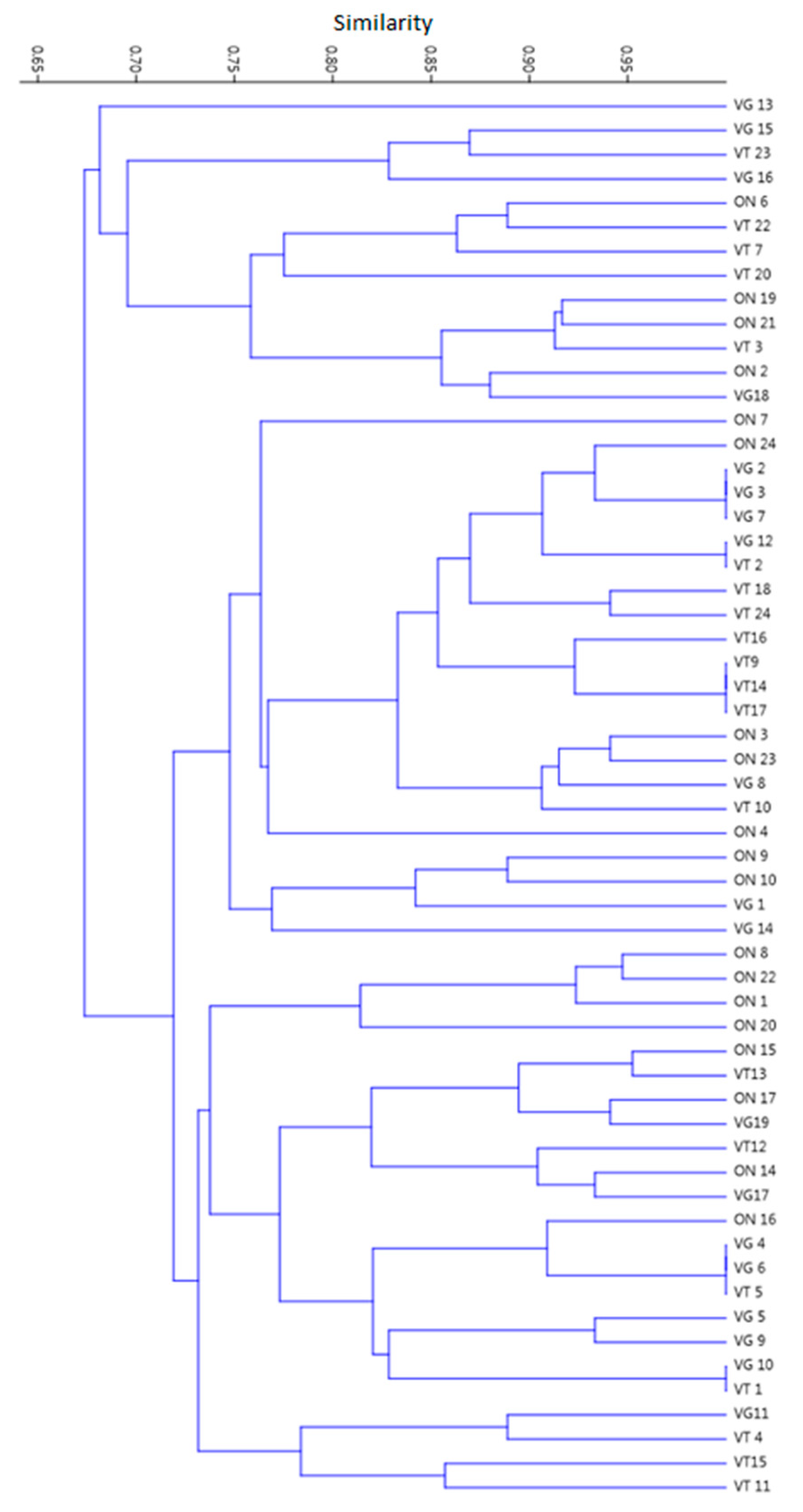
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Participants’ Anthropometric Data and Dietary Assessment
4.3. Fecal Specimen Collection and Storage
4.4. Extraction, Quantitation, and Integrity of Metagenomic DNA
4.5. Screening of Antimicrobial Resistance Genes (ARG)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Gillings, M.R. Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Front. Microbiol. 2013, 4, 4. [Google Scholar] [CrossRef]
- Ho, J.; Yeoh, Y.K.; Barua, N.; Chen, Z.; Lui, G.; Wong, S.H.; Yang, X.; Chan, M.C.; Chan, P.K.; Hawkey, P.M.; et al. Systematic review of human gut resistome studies revealed variable definitions and approaches. Gut Microbes 2020, 12, 1–12. [Google Scholar] [CrossRef]
- Baquero, F. Metagenomic epidemiology: A public health need for the control of antimicrobial resistance. Clin. Microbiol. Infect. 2012, 18, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- Destoumieux-Garzón, D.; Mavingui, P.; Boetsch, G.; Boissier, J.; Darriet, F.; Duboz, P.; Fritsch, C.; Giraudoux, P.; Le Roux, F.; Morand, S.; et al. The One Health Concept: 10 Years Old and a Long Road Ahead. Front. Vet. Sci. 2018, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Cha, C.J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 1–9. [Google Scholar] [CrossRef]
- Milanović, V.; Osimani, A.; Aquilanti, L.; Tavoletti, S.; Garofalo, C.; Polverigiani, S.; Litta-Mulondo, A.; Cocolin, L.; Ferrocino, I.; Di Cagno, R.; et al. Occurrence of antibiotic re-sistance genes in the fecal DNA of healthy omnivores, ovo-lacto vegetarians and vegans. Mol. Nutr. Food Res. 2017, 61, 1601098. [Google Scholar] [CrossRef]
- Gillings, M.R.; Paulsen, I.T.; Tetu, S.G. Ecology and Evolution of the Human Microbiota: Fire, Farming and Antibiotics. Genes 2015, 6, 841–857. [Google Scholar] [CrossRef]
- Swift, B.M.C.; Bennett, M.; Waller, K.; Dodd, C.; Murray, A.; Gomes, R.L.; Humphreys, B.; Hobman, J.L.; Jones, M.A.; Whitlock, S.E.; et al. Anthropogenic environmental drivers of anti-microbial resistance in wildlife. Sci. Total Environ. 2019, 649, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.J.; Hungate, B.A.; Price, L.B. Food-animal production and the spread of antibiotic resistance: The role of ecology. Front. Ecol. Environ. 2017, 15, 309–318. [Google Scholar] [CrossRef]
- Levy, S.B.; Fitzgerald, G.B.; Macone, A.B. Changes in Intestinal Flora of Farm Personnel after Introduction of a Tetracycline-Supplemented Feed on a Farm. N. Engl. J. Med. 1976, 295, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Fitzgerald, G.B.; Macone, A.B. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nat. Cell Biol. 1976, 260, 40–42. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal micro-biome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Knapp, C.W.; Gálvez, A.; Benomar, N. Chapter 29—Antibiotic Resistance Profile of Microbes from Traditional Fermented Foods. In Fermented Foods in Health and Disease Prevention; Frias, J., Martinez-Villaluenga, C., Peñas, E., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 675–704. [Google Scholar]
- Forslund, K.; Sunagawa, S.; Kultima, J.R.; Mende, D.R.; Arumugam, M.; Typas, A.; Bork, P. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 2013, 23, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, D.-W.; Lee, D.-H.; Kim, Y.-S.; Bu, J.-H.; Cha, J.-H.; Thawng, C.N.; Hwang, E.-M.; Seong, H.J.; Sul, W.J.; et al. Mobile resistome of human gut and pathogen drives anthro-pogenic bloom of antibiotic resistance. Microbiome 2020, 8, 2. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. The structure and diversity of human, animal and environmental resistomes. Microbiome 2016, 4, 1–15. [Google Scholar] [CrossRef]
- Nobles, M. History counts: A comparative analysis of racial/color categorization in US and Brazilian censuses. Am. J. Public Health 2000, 90, 1738–1745. [Google Scholar] [CrossRef]
- LoSasso, C.; Di Cesare, A.; Mastrorilli, E.; Patuzzi, I.; Cibin, V.; Eckert, E.M.; Fontaneto, D.; Vanzo, A.; Ricci, A.; Corno, G.; et al. Assessing antimicrobial resistance gene load in vegan, vegetarian and omnivore human gut microbiota. Int. J. Antimicrob. Agents 2018, 52, 702–705. [Google Scholar] [CrossRef]
- Versluis, D.; D’Andrea, M.M.; Garcia, J.R.; Leimena, M.M.; Hugenholtz, F.; Zhang, J.; Ozturk, B.; Nylund, L.; Sipkema, D.; Van Schaik, W.; et al. Mining microbial metatranscriptomes for expression of antibiotic resistance genes under natural conditions. Sci. Rep. 2015, 5, 11981. [Google Scholar] [CrossRef]
- Singh, S.; Verma, N.; Taneja, N. The human gut resistome: Current concepts & future prospects. Indian J. Med Res. 2019, 150, 345–358. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Liu, Y.; Liu, K.; Zhang, L.; Huang, F.; Wang, L.; Rashid, A.; Hu, A.; Yu, C. Horizontal and ver-tical gene transfer drive sediment antibiotic resistome in an urban lagoon system. J. Environ. Sci. 2021, 102, 11–23. [Google Scholar] [CrossRef]
- Chen, Q.-L.; Cui, H.-L.; Su, J.-Q.; Penuelas, J.; Zhu, Y.-G. Antibiotic Resistomes in Plant Microbiomes. Trends Plant Sci. 2019, 24, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kinkelaar, D.; Huang, Y.; Li, Y.; Li, X.; Wang, H.H. Acquired Antibiotic Resistance: Are We Born with It? Appl. Environ. Microbiol. 2011, 77, 7134–7141. [Google Scholar] [CrossRef] [PubMed]
- Verraes, C.; VanBoxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; De Schaetzen, M.-A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicro-bial Resistance in the Food Chain: A Review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Su, J.-Q.; Wei, B.; Ou-Yang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Xu, H.-J.; Zhu, Y.-G. Antibiotic Resistome and Its As-sociation with Bacterial Communities during Sewage Sludge Composting. Environ. Sci. Technol. 2015, 49, 7356–7363. [Google Scholar] [CrossRef]
- Zhou, X.; Qiao, M.; Wang, F.H.; Zhu, Y.G. Use of commercial organic fertilizer increases the abundance of an-tibiotic resistance genes and antibiotics in soil. Environ. Sci. Pollut. Res. Int. 2017, 24, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Chapman, S.J.; Freitag, T.E.; Kyle, C.; Ma, J.; Yang, Y.; Zhang, Z. Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers. Ecotoxicol. Environ. Saf. 2019, 170, 39–46. [Google Scholar] [CrossRef]
- Kumar, K.; Gupta, S.C.; Baidoo, S.K.; Chander, Y.; Rosen, C.J. Antibiotic Uptake by Plants from Soil Fertilized with Animal Manure. J. Environ. Qual. 2005, 34, 2082–2085. [Google Scholar] [CrossRef]
- Sarmiento, M.R.A.; de Paula, T.O.; Borges, F.M.; Ferreira-Machado, A.B.; Resende, J.A.; Moreira, A.P.B.; Du-tra Luquetti, S.C.P.; Cesar, D.E.; da Silva, V.L.; Diniz, C.G. Obesity, Xenobiotic Intake and Antimicrobi-al-Resistance Genes in the Human Gastrointestinal Tract: A Comparative Study of Eutrophic, Overweight and Obese Individuals. Genes 2019, 10, 349. [Google Scholar] [CrossRef]
- Sánchez-Osuna, M.; Cortés, P.; Barbé, J.; Erill, I. Origin of the Mobile Di-Hydro-Pteroate Synthase Gene Deter-mining Sulfonamide Resistance in Clinical Isolates. Front. Microbiol. 2019, 9, 3332. [Google Scholar] [CrossRef] [PubMed]
- de Souza, M.M.S.; Coelho, S.D.M.; Coelho, I.D.S.; Soares, B.D.S.; da Motta, C.C.; de Melo, D.A.; Dubenczuk, F.C.; Santiago, G.S.; Pimenta, R.L.; Marques, V.F.; et al. Antimicrobial Resistance in Animal Production: An Overview. Braz. J. Vet. Med. 2016, 38, 68–74. [Google Scholar]
- Koike, S.; Mackie, R.; Aminov, R.; Mirete, S.; Pérez, M.L. Agricultural use of antibiotics and antibiotic re-sistance. Antibiot. Resist. Genes Nat. Environ. Long Term Eff. 2017, 217–250. [Google Scholar]
- Marosevic, D.; Kaevska, M.; Jaglic, Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage—A review. Ann. Agric. Environ. Med. 2017, 24, 338–344. [Google Scholar] [CrossRef]
- Niestępski, S.; Harnisz, M.; Korzeniewska, E.; Aguilera-Arreola, M.G.; Contreras-Rodríguez, A.; Filipkowska, Z.; Osińska, A. The emergence of antimicrobial resistance in environmental strains of the Bacteroides fragilis group. Environ. Int. 2019, 124, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Pazhani, G.P.; Dharanidharan, R.; Ghosh, A.; Ramamurthy, T. Detection of integron-associated gene cassettes and other antimicrobial resistance genes in enterotoxigenic Bacteroides fragilis. Anaerobe 2015, 33, 18–24. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Fornaciari, G.; Luciani, S.; Toranzos, G.A.; Marota, I.; Giuffra, V.; Sangwan, N.; Cano, R.J. Tetracycline-like resistome of ancient human guts. Hum. Microbiome J. 2018, 10, 21–26. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Fornaciari, G.; Luciani, S.; Dowd, S.E.; Toranzos, G.A.; Marota, I.; Cano, R.J. Gut Microbiome of an 11th Century A.D. Pre-Columbian Andean Mummy. PLoS ONE 2015, 10, e0138135. [Google Scholar] [CrossRef]
- Feng, J.; Li, B.; Jiang, X.; Yang, Y.; Wells, G.F.; Zhang, T.; Li, X. Antibiotic resistome in a large-scale healthy hu-man gut microbiota deciphered by metagenomic and network analyses. Environ. Microbiol. 2018, 20, 355–368. [Google Scholar] [CrossRef]
- Hu, Y.; Yang, X.; Qin, J.; Lu, N.; Cheng, G.; Wu, N.; Pan, Y.; Li, J.; Zhu, L.; Wang, X.; et al. Meta-genome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat. Commun. 2013, 4, 2151. [Google Scholar] [CrossRef]
- Roberts, M. Environmental Macrolide–Lincosamide–Streptogramin and Tetracycline Resistant Bacteria. Front. Microbiol. 2011, 2, 40. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile macrolide resistance genes in staphylococci. Plasmid 2018, 99, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Tekiner, S.H.; Özpinar, H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. Braz. J. Microbiol. 2016, 47, 444–451. [Google Scholar] [CrossRef] [PubMed]
- De Paula, A.C.L.; Medeiros, J.D.; De Azevedo, A.C.; De Assis Chagas, J.M.; Da Silva, V.L.; Diniz, C.G. Antibiotic Resistance Genetic Markers and Integrons in White Soft Cheese: Aspects of Clinical Resistome and Potentiality of Horizontal Gene Transfer. Genes 2018, 9, 106. [Google Scholar] [CrossRef]
- Zieliński, W.; Buta, M.; Hubeny, J.; Korzeniewska, E.; Harnisz, M.; Nowrotek, M.; Płaza, G. Prevalence of Beta Lactamases Genes in Sewage and Sludge Treated in Mechanical-Biological Wastewater Treatment Plants. J. Ecol. Eng. 2019, 20, 80–86. [Google Scholar] [CrossRef]
- van den Bunt, G.; van Pelt, W.; Hidalgo, L.; Scharringa, J.; de Greeff, S.C.; Schürch, A.C.; Mughini-Gras, L.; Bonten, M.J.M.; Fluit, A.C. Prevalence, Risk Factors and Genetic Characterisation of Extended-Spectrum Beta-Lactamase and Carbapenemase-Producing Enterobacteriaceae (ESBL-E and CPE): A community-Based Cross-Sectional Study, The Netherlands. Eurosurveillance 2019, 24, 1800594. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic Resistance and Virulence Traits in Vancomy-cin-Resistant Enterococci (VRE) and Extended-Spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) Enter-obacteriaceae from Humans and Pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Schill, F.; Abdulmawjood, A.; Klein, G.; Reich, F. Prevalence and characterization of extended-spectrum β-lactamase (ESBL) and AmpC β-lactamase producing Enterobacteriaceae in fresh pork meat at processing level in Germany. Int. J. Food Microbiol. 2017, 257, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Angelin, M.; Huss, M.; Kjellqvist, S.; Kristiansson, E.; Palmgren, H.; Larsson, D.G.J.; Johansson, A. The Human Gut Microbiome as a Transporter of Antibiotic Resistance Genes between Con-tinents. Antimicrob. Agents Chemother. 2015, 59, 6551–6560. [Google Scholar] [CrossRef] [PubMed]
- Adesoji, A.T.; Ogunjobi, A.A.; Olatoye, I.O. Characterization of Integrons and Sulfonamide Resistance Genes among Bacteria from Drinking Water Distribution Systems in Southwestern Nigeria. Chemotherapy 2016, 62, 34–42. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Zhenling, Z.; Wang, M.; Zeng, Z. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Mu, Q.; Li, J.; Sun, Y.; Mao, D.; Wang, Q.; Luo, Y. Occurrence of sulfonamide-, tetracycline-, plasmid-mediated quinolone- and macrolide-resistance genes in livestock feedlots in Northern China. Environ. Sci. Pollut. Res. 2015, 22, 6932–6940. [Google Scholar] [CrossRef]
- Emachado, E.; Coque, T.M.; Cantón, R.; Sousa, J.C.; Peixe, L. Commensal Enterobacteriaceae as reservoirs of extended-spectrum beta-lactamases, integrons, and sul genes in Portugal. Front. Microbiol. 2013, 4, 80. [Google Scholar] [CrossRef]
- Caruso, G.; Giammanco, A.; Cardamone, C.; Oliveri, G.; Mascarella, C.; Capra, G.; Fasciana, T. Extra-Intestinal Fluoroquinolone-Resistant Escherichia coli Strains Isolated from Meat. BioMed Res. Int. 2018, 2018, 8714975. [Google Scholar] [CrossRef]
- Chenia, H.Y. Prevalence and characterization of plasmid-mediated quinolone resistance genes in Aeromonas spp. isolated from South African freshwater fish. Int. J. Food Microbiol. 2016, 231, 26–32. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.; Walsh, F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol. Ecol. 2016, 92, fiv168. [Google Scholar] [CrossRef]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef]
- WHO. Obesity: Preventing and Managing the Global Epidemic; Report of a World Health Organization Consultation; World Health Organization: Geneva, Switzerland, 2000. [Google Scholar]
- Cardoso, M.A.; Stocco, P.R. Development of a dietary assessment method for people of Japanese descent living in São Paulo. Brazil. Cad. Saude Publica 2000, 16, 107–114. [Google Scholar] [CrossRef][Green Version]
- Sales, R.L.; Silva, M.M.S.; Costa, N.M.B.; Euclydes, M.P.; Eckhardt, V.F.; Rodrigues, C.M.A.; Tinoco, A.L.A. Development of a questionnaire to assess food intake of population groups. Rev. Nutr. 2006, 19, 539–552. [Google Scholar] [CrossRef][Green Version]
| Characteristics | Group of Participants According to Their Eating Habits | p < 0.05 * | |||
|---|---|---|---|---|---|
| ON (n = 19) | VT (n = 20) | VG (n = 19) | |||
| Gender (%; male/female) | 15.8/84.2 | 20.0/80.0 | 42.1/57.9 | na | |
| Average age (years ± SD) | 28.47 ± 6.02 | 31.63 ± 9.72 | 25.89 ± 6.24 | na | |
| Ethnic Group (%) | White | 52.6 | 78.9 | 89.5 | na |
| Pardo ** | 31.6 | 10.6 | 10.5 | na | |
| Black | 10.5 | 10.5 | na | ||
| East Asian | 5.3 | na | |||
| Average BMI (mean ± SD) | 21.46 ± 1.96 | 22.13 ± 1.95 | 22.12 ± 1.72 | ||
| Mean daily calorie intake (% ± SD) | 2049.3 ± 836.1 | 2092.4 ± 738.9 | 2522.7 ± 955.2 | ||
| Mean daily lipid intake (% ± SD) | 33.39 ± 6.07 | 33.64 ± 14.41 | 25.10 ± 12.11 | c | |
| Mean daily protein intake (% ± SD) | 19.22 ± 4.80 | 12.10 ± 3.16 | 10.17 ± 1.91 | a, b, c | |
| Mean daily CARB intake (% ± SD) | 47.39 ± 7.35 | 54.26 ± 13.95 | 64.73 ± 11.34 | b, c | |
| Mean daily TF intake (g ± SD) | 23.01 ± 9.04 | 41.98 ± 25.12 | 63.78 ± 35.79 | a, b, c | |
| Mean daily SF intake (g ± SD) | 2.53 ± 1.56 | 4.98 ± 2.53 | 13.58 ± 14.65 | a, b, c | |
| Mean daily IF intake (g ± SD) | 5.14 ± 3.27 | 8.88 ± 5.41 | 21.42 ± 19.36 | a, b, c | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Silva, S.F.; Reis, I.B.; Monteiro, M.G.; Dias, V.C.; Machado, A.B.F.; da Silva, V.L.; Diniz, C.G. Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians. Antibiotics 2021, 10, 276. https://doi.org/10.3390/antibiotics10030276
da Silva SF, Reis IB, Monteiro MG, Dias VC, Machado ABF, da Silva VL, Diniz CG. Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians. Antibiotics. 2021; 10(3):276. https://doi.org/10.3390/antibiotics10030276
Chicago/Turabian Styleda Silva, Suzane Fernandes, Isabela Brito Reis, Melina Gabriela Monteiro, Vanessa Cordeiro Dias, Alessandra Barbosa Ferreira Machado, Vânia Lúcia da Silva, and Cláudio Galuppo Diniz. 2021. "Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians" Antibiotics 10, no. 3: 276. https://doi.org/10.3390/antibiotics10030276
APA Styleda Silva, S. F., Reis, I. B., Monteiro, M. G., Dias, V. C., Machado, A. B. F., da Silva, V. L., & Diniz, C. G. (2021). Influence of Human Eating Habits on Antimicrobial Resistance Phenomenon: Aspects of Clinical Resistome of Gut Microbiota in Omnivores, Ovolactovegetarians, and Strict Vegetarians. Antibiotics, 10(3), 276. https://doi.org/10.3390/antibiotics10030276







