From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use
Abstract
1. Introduction
2. Results
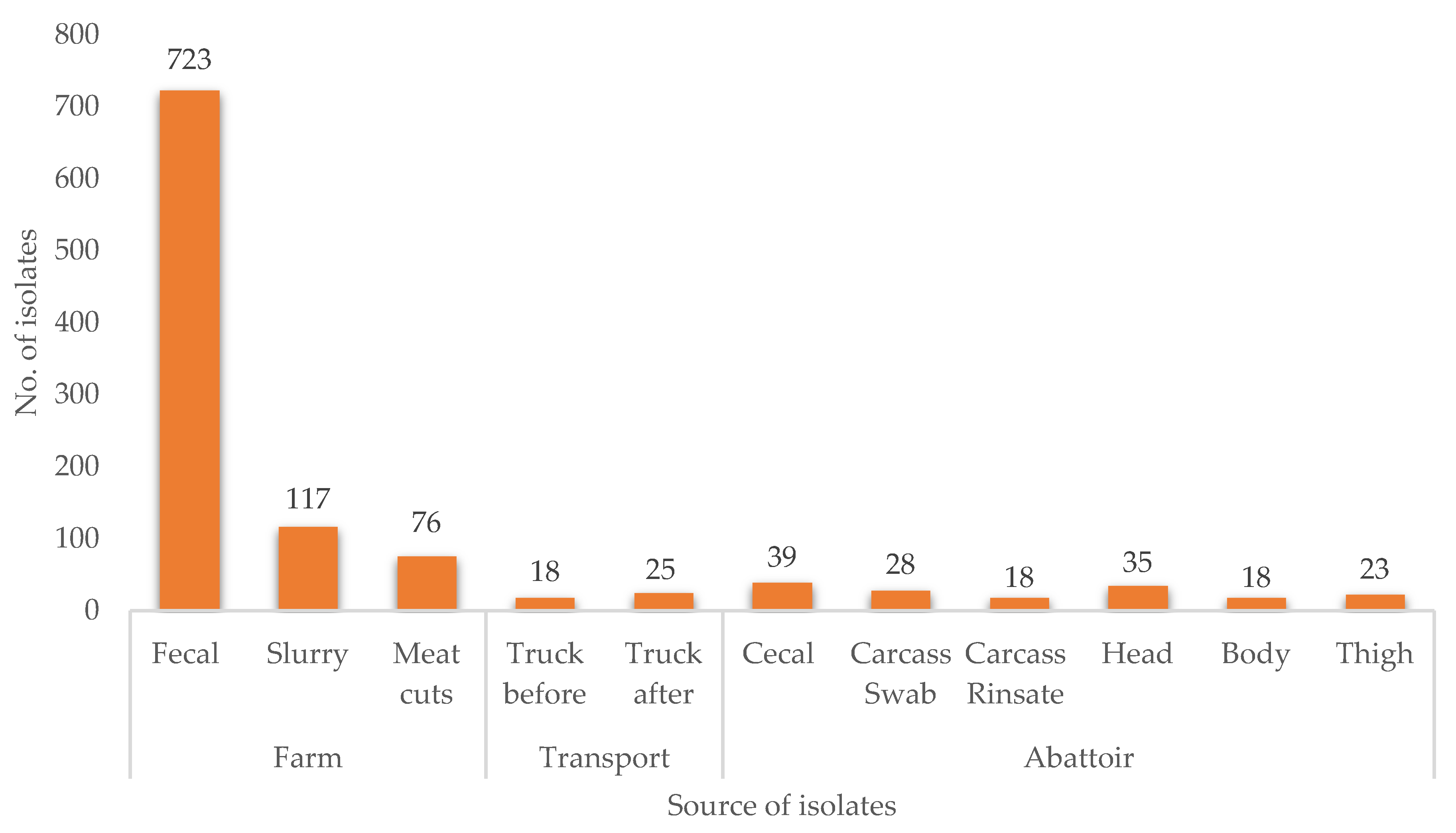
2.1. Prevalence of E. coli along the Pig Production Chain
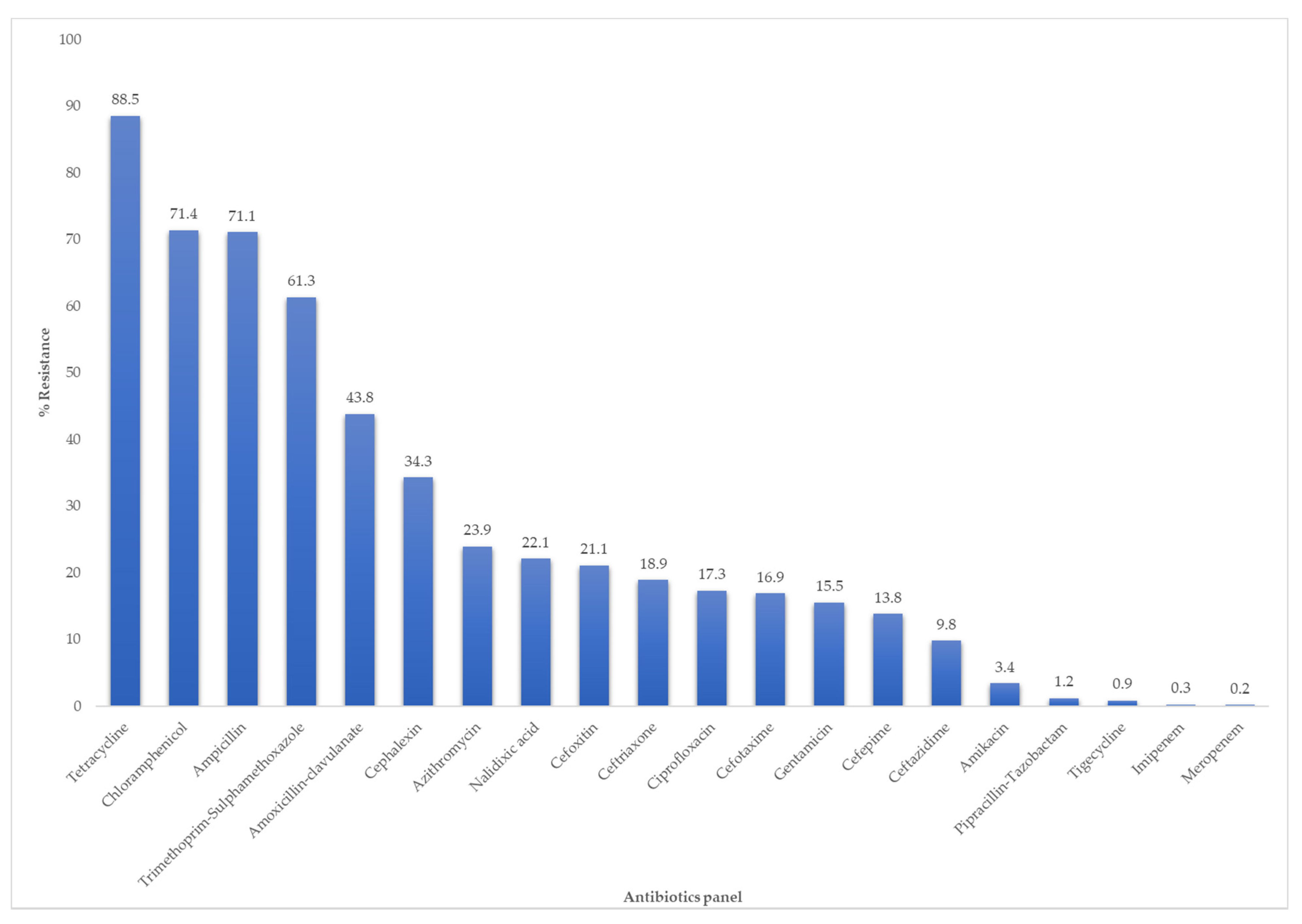
2.2. Antimicrobial Resistance Profile of E. coli Isolated from the Pig Production Chain
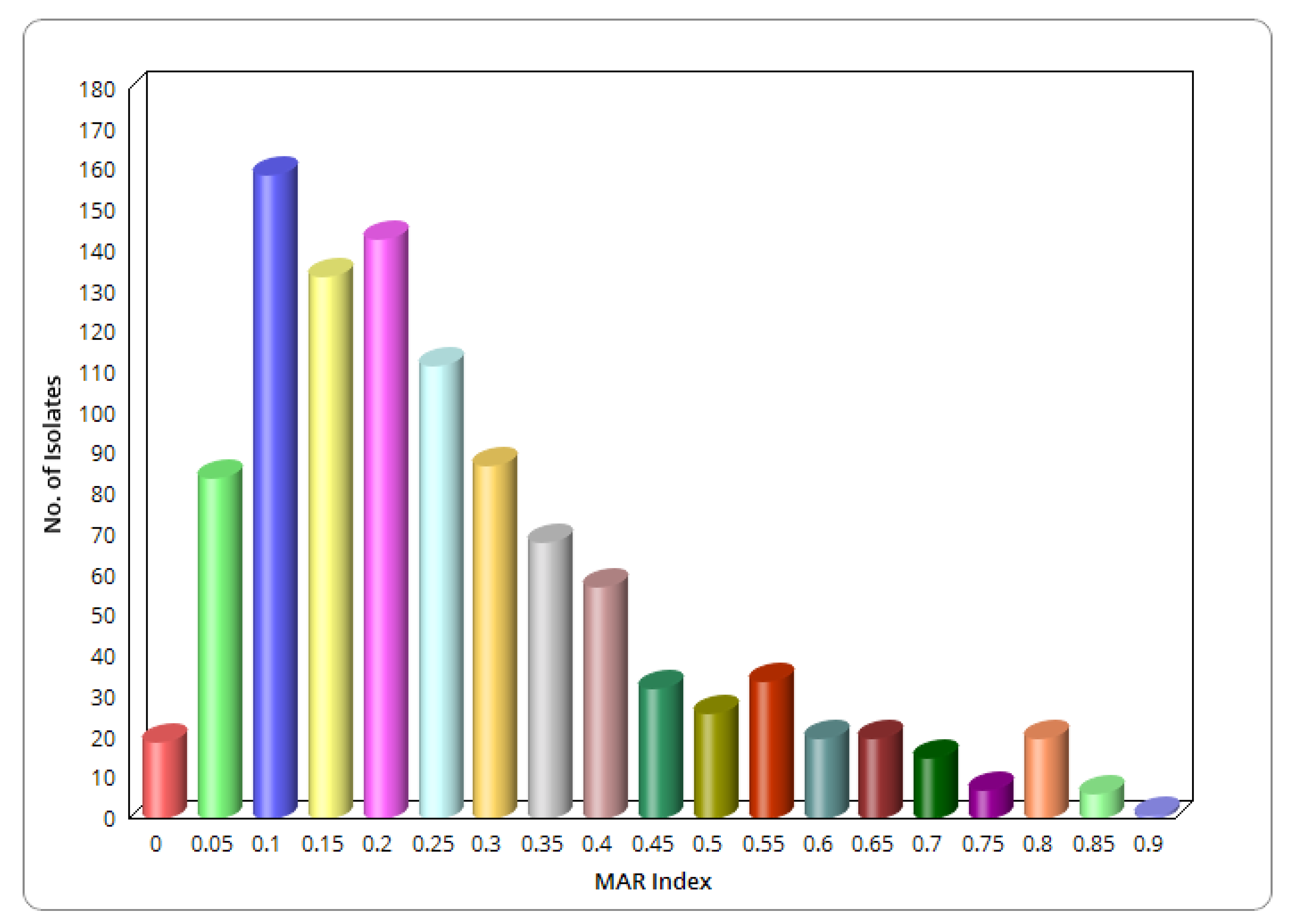
2.3. MAR Phenotypes of E. coli
3. Discussion
3.1. Prevalence of E. coli across the Pig Production System
3.2. Antimicrobial Resistance Profile of E. coli Isolated from the Pig Production Chain
3.3. Prevalence of Multidrug-Resistant and Estimation of MAR Index of E. coli Isolates in the Pig Production Chain
4. Materials and Methods
4.1. Study Clearance and Ethical Consideration
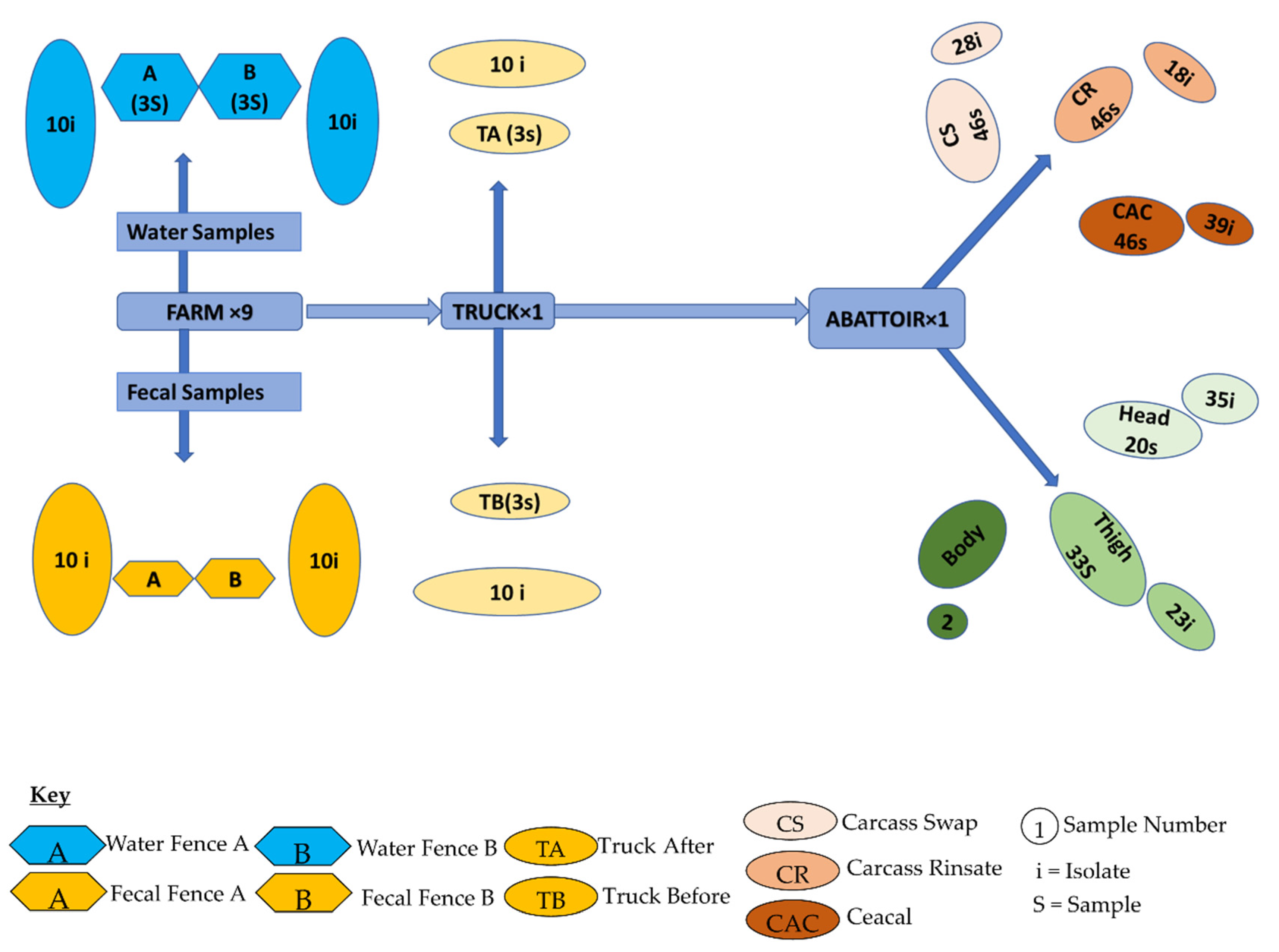
4.2. Study Site and Sample Collection
- Farm: Two groups of newborn pigs in two fences labeled A and B were selected for the study. Five fresh pig feces samples were randomly collected per fence, ensuring that each sample was from a different location. Samples were collected twice a month for 18 weeks. Additionally, slurry samples were collected in triplicate from the pipes draining the pig house at each sampling period.
- Transport: After 18 weeks, when pigs reached maturity and slaughter readiness, swab samples were taken from the transport vehicle (truck) before and after loading the pigs for transportation to the abattoir.
- Abattoir: Swabs were collected throughout the slaughter chain, viz. carcass, carcass rinsate, caeca, and pork portions before packaging (head, body, and thigh).
4.3. Sample Processing and Isolation of E. coli
4.3.1. Fecal and Slurry Samples from the Farm
4.3.2. Rinsate and Swabs (Transport and Abattoir)
4.4. Molecular Confirmation of E. coli
4.5. Antibiotic Susceptibility Testing
4.6. Determination of Multidrug-Resistant (MDR) and Multiple Antibiotic Resistance (MAR) Index
4.7. Statistical Analysis and Interpretation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Henchion, M.; McCarthy, M.; Resconi, V.C.; Troy, D. Meat consumption: Trends and quality matters. Meat Sci. 2014, 98, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Onwezen, M.C.; van der Weele, C.N. When indifference is ambivalence: Strategic ignorance about meat consumption. Food Qual. Prefer. 2016, 52, 96–105. [Google Scholar] [CrossRef]
- Alexandratos, N.; Bruinsma, J.; Alexandratos, N.; Bruinsma, J. World Agriculture towards 2030/2050: The 2012 Revision; ESA Working Paper No. 12-03; FAO: Rome, Italy, 2012. [Google Scholar]
- Happer, C.; Wellesley, L. Meat consumption, behaviour and the media environment: A focus group analysis across four countries. Food Secur. 2019, 11, 123–139. [Google Scholar] [CrossRef]
- Mathijs, E. Exploring future patterns of meat consumption. Meat Sci. 2015, 109, 112–116. [Google Scholar] [CrossRef]
- Thornton, P.K. Livestock production: Recent trends, future prospects. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 2853–2867. [Google Scholar] [CrossRef]
- OECD/FAO. OECD-FAO OECD-FAO Agricultural Outlook 2019–2028; OECD Publishing: Paris, France; Food and Agriculture Organization of the United Nations: Rome, Italy, 2019; ISBN 978-92-5-131374-9. [Google Scholar]
- Laxminarayan, R.; Heymann, D.L. Challenges of drug resistance in the developing world. BMJ 2012, 344, e1567. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Yeung, S. Patterns of antibiotic use in global pig production: A systematic review. Vet. Anim. Sci. 2019, 7, 100058. [Google Scholar] [CrossRef]
- Gonzalez Ronquillo, M.; Angeles Hernandez, J.C. Antibiotic and synthetic growth promoters in animal diets: Review of impact and analytical methods. Food Control 2017, 72, 255–267. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Shin, E.; Lee, Y. Antimicrobial resistance and integron profiles in multidrug-resistant Escherichia coli isolates from pigs. Foodborne Pathog. Dis. 2014, 11, 988–997. [Google Scholar] [CrossRef]
- Debski, B. Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials. Pol. J. Vet. Sci. 2016, 19, 917–924. [Google Scholar] [CrossRef] [PubMed]
- van der Fels-Klerx, H.J.; Puister-Jansen, L.F.; van Asselt, E.D.; Burgers, S.L. Farm factors associated with the use of antibiotics in pig production. J. Anim. Sci. 2011, 89, 1922–1929. [Google Scholar] [CrossRef]
- Lee, J.Y.; Han, G.G.; Lee, H.B.; Lee, S.M.; Kang, S.K.; Jin, G.D.; Park, J.; Chae, B.J.; Choi, Y.H.; Kim, E.B.; et al. Prohibition of antibiotic growth promoters has affected the genomic profiles of Lactobacillus salivarius inhabiting the swine intestine. PLoS ONE 2017, 12, e0186671. [Google Scholar] [CrossRef]
- Vieira, A.R.; Collignon, P.; Aarestrup, F.M.; McEwen, S.A.; Hendriksen, R.S.; Hald, T.; Wegener, H.C. Association between antimicrobial resistance in Escherichia coli isolates from food animals and blood stream isolates from humans in Europe: An ecological study. Foodborne Pathog. Dis. 2011, 8, 1295–1301. [Google Scholar] [CrossRef] [PubMed]
- Jahanbakhsh, S.; Kabore, K.P.; Fravalo, P.; Letellier, A.; Fairbrother, J.M. Impact of medicated feed along with clay mineral supplementation on Escherichia coli resistance to antimicrobial agents in pigs after weaning in field conditions. Res. Vet. Sci. 2015, 102, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Burow, E.; Simoneit, C.; Tenhagen, B.A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance in porcine E. coli—A systematic review. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef]
- OIE OIE list of antimicrobial agents of veterinary importance. OIE Int. Commitee 2015, 33, 1–9.
- World Health Organisation. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List); World Health Organization: Geneva, Swizerland, 2019. [Google Scholar]
- Mokoele, J.M.; Spencer, B.T.; van Leengoed, L.A.; Fasina, F.O. Efficiency indices and indicators of poor performance among emerging small-scale pig farmers in the Limpopo Province, South Africa. Onderstepoort J. Vet. Res. 2014, 81, 774. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muhanguzi, D.; Lutwama, V.; Mwiine, F. Factors that influence pig production in Central Uganda—Case study of Nangabo Sub-County, Wakiso district. Vet. World 2012, 5, 346–351. [Google Scholar] [CrossRef]
- McIver, K.S.; Amoako, D.G.; Abia, A.L.K.; Bester, L.A.; Chenia, H.Y.; Essack, S.Y. Molecular epidemiology of antibiotic-resistant Escherichia coli from farm-to-fork in intensive poultry production in KwaZulu-Natal, South Africa. Antibiotics 2020, 9, 850. [Google Scholar] [CrossRef]
- Birkegard, A.C.; Halasa, T.; Graesboll, K.; Clasen, J.; Folkesson, A.; Toft, N. Association between selected antimicrobial resistance genes and antimicrobial exposure in Danish pig farms. Sci. Rep. 2017, 7, 9683. [Google Scholar] [CrossRef] [PubMed]
- Frommel, U.; Lehmann, W.; Rodiger, S.; Bohm, A.; Nitschke, J.; Weinreich, J.; Gross, J.; Roggenbuck, D.; Zinke, O.; Ansorge, H.; et al. Adhesion of human and animal Escherichia coli strains in association with their virulence-associated genes and phylogenetic origins. Appl. Environ. Microbiol. 2013, 79, 5814–5829. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; Gordon, D.M.; Chin, J.; Brouwers, H.J.; Njuguna, P.; Groves, M.D.; Zhang, R.; Chapman, T.A. Molecular characterization of commensal Escherichia coli adapted to different compartments of the porcine gastrointestinal tract. Appl. Environ. Microbiol. 2012, 78, 6799–6803. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Adenipekun, E.O.; Jackson, C.R.; Oluwadun, A.; Iwalokun, B.A.; Frye, J.G.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A. Prevalence and antimicrobial resistance in Escherichia coli from food animals in Lagos, Nigeria. Microb. Drug Resist. 2015, 21, 358–365. [Google Scholar] [CrossRef]
- Alonso, C.A.; Zarazaga, M.; Ben Sallem, R.; Jouini, A.; Ben Slama, K.; Torres, C. Antibiotic resistance in Escherichia coli in husbandry animals: The African perspective. Lett. Appl. Microbiol. 2017, 64, 318–334. [Google Scholar] [CrossRef]
- Verraes, C.; Van Boxstael, S.; Van Meervenne, E.; Van Coillie, E.; Butaye, P.; Catry, B.; de Schaetzen, M.A.; Van Huffel, X.; Imberechts, H.; Dierick, K.; et al. Antimicrobial resistance in the food chain: A review. Int. J. Environ. Res. Public Health 2013, 10, 2643–2669. [Google Scholar] [CrossRef]
- Allen, H.K. Antibiotic resistance gene discovery in food-producing animals. Curr. Opin. Microbiol. 2014, 19, 25–29. [Google Scholar] [CrossRef]
- DAFF. A Profile of the South African Pork Market Value Chain 2018; South African Department of Agriculture, Forestry and Fisheries: Pretoria, South Africa, 2018. [Google Scholar]
- Moyane, J.N.; Jideani, A.I.O.; Aiyegoro, O.A. Antibiotics usage in food-producing animals in South Africa and impact on human: Antibiotic resistance. Afr. J. Microbiol. Res. 2013, 7, 2990–2997. [Google Scholar]
- Pires, D.; de Kraker, M.E.A.; Tartari, E.; Abbas, M.; Pittet, D. ‘Fight antibiotic resistance—It’s in your hands’: Call from the World Health Organization for 5th May 2017. Clin. Infect. Dis. 2017, 64, 1780–1783. [Google Scholar] [CrossRef]
- Founou, L.L.; Amoako, D.G.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in food animals in Africa: A systematic review and meta-analysis. Microb. Drug Resist. 2018, 24, 648–665. [Google Scholar] [CrossRef] [PubMed]
- Warriner, K.; Aldsworth, T.G.; Kaur, S.; Dodd, C.E.R. Cross-contamination of carcasses and equipment during pork processing. J. Appl. Microbiol. 2002, 93, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Nauta, M.; Barfod, K.; Hald, T.; Sørensen, A.H.; Emborg, H.-D.; Aabo, S. Prediction of Salmonella carcass contamination by a comparative quantitative analysis of E. coli and Salmonella during pig slaughter. Int. J. Food Microbiol. 2013, 166, 231–237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biasino, W.; De Zutter, L.; Mattheus, W.; Bertrand, S.; Uyttendaele, M.; Van Damme, I. Correlation between slaughter practices and the distribution of Salmonella and hygiene indicator bacteria on pig carcasses during slaughter. Food Microbiol. 2018, 70, 192–199. [Google Scholar] [CrossRef]
- Schwaiger, K.; Huther, S.; Holzel, C.; Kampf, P.; Bauer, J. Prevalence of antibiotic-resistant Enterobacteriaceae isolated from chicken and pork meat purchased at the slaughterhouse and at retail in Bavaria, Germany. Int. J. Food Microbiol. 2012, 154, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Shrivastava, P.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 67–76. [Google Scholar] [CrossRef]
- Zhang, P.; Shen, Z.; Zhang, C.; Song, L.; Wang, B.; Shang, J.; Yue, X.; Qu, Z.; Li, X.; Wu, L.; et al. Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Vet. Microbiol. 2017, 203, 49–55. [Google Scholar] [CrossRef]
- Góchez, D.; Raicek, M.; Ferreira, J.P.; Jeannin, M.; Moulin, G.; Erlacher-Vindel, E. OIE annual report on antimicrobial agents intended for use in animals: Methods used. Front. Vet. Sci. 2019, 6, 317. [Google Scholar] [CrossRef]
- Eagar, H.; Swan, G.; van Vuuren, M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J. S. Afr. Vet. Assoc. 2012, 83, 16. [Google Scholar] [CrossRef]
- Kidsley, A.K.; Abraham, S.; Bell, J.M.; O’Dea, M.; Laird, T.J.; Jordan, D.; Mitchell, P.; McDevitt, C.A.; Trott, D.J. Antimicrobial susceptibility of Escherichia coli and Salmonella spp. isolates from healthy pigs in Australia: Results of a pilot national survey. Front. Microbiol. 2018, 9, 1207. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobials in Agriculture and the Environment: Reducing Unnecessary Use and Waste. The Review on Antimicrobial Resist; HM Government: London, UK, 2015. [Google Scholar]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- Osterberg, J.; Wingstrand, A.; Nygaard Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic resistance in Escherichia coli from pigs in organic and conventional farming in four European countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef]
- Lay, K.K.; Koowattananukul, C.; Chansong, N.; Chuanchuen, R. Antimicrobial resistance, virulence, and phylogenetic characteristics of Escherichia coli isolates from clinically healthy swine. Foodborne Pathog. Dis. 2012, 9, 992–1001. [Google Scholar] [CrossRef]
- de Jong, A.; Thomas, V.; Simjee, S.; Godinho, K.; Schiessl, B.; Klein, U.; Butty, P.; Valle, M.; Marion, H.; Shryock, T.R. Pan-European monitoring of susceptibility to human-use antimicrobial agents in enteric bacteria isolated from healthy food-producing animals. J. Antimicrob. Chemother. 2012, 67, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, K.M.; White, D.G.; Hume, M.E.; Poole, T.L.; Nisbet, D.J. The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 2005, 243, 285–291. [Google Scholar] [CrossRef]
- Xu, X.; Cui, S.; Zhang, F.; Luo, Y.; Gu, Y.; Yang, B.; Li, F.; Chen, Q.; Zhou, G.; Wang, Y.; et al. Prevalence and characterization of cefotaxime and ciprofloxacin co-resistant Escherichia coli isolates in retail chicken carcasses and Ground Pork, China. Microb. Drug Resist. 2014, 20, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Abe, R.; Hagiya, H.; Akeda, Y.; Yamamoto, N.; Ishii, Y.; Tomono, K. Bactericidal efficacy of meropenem in combination with cefmetazole against IMP-producing carbapenem-resistant Enterobacteriaceae. BMC Res. Notes 2019, 12, 740. [Google Scholar] [CrossRef] [PubMed]
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M.A.P. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. Int. 2015, 22, 10969–10980. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Sun, Y.; Ji, X.; Du, X.; Guo, X.; Liu, J.; Zhu, L.; Zhou, B.; Zhou, W.; Liu, G.; et al. Characterization of antimicrobial resistance and extended-spectrum beta-lactamase genes in Escherichia coli isolated from chickens. Foodborne Pathog. Dis. 2015, 12, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Adenipekun, E.O.; Jackson, C.R.; Ramadan, H.; Iwalokun, B.A.; Oyedeji, K.S.; Frye, J.G.; Barrett, J.B.; Hiott, L.M.; Woodley, T.A.; Oluwadun, A. Prevalence and multidrug resistance of Escherichia coli from community-acquired infections in Lagos, Nigeria. J. Infect. Dev. Ctries. 2016, 10, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Adefisoye, M.A.; Okoh, A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiologyopen 2016, 5, 143–151. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016, 7, 1–19. [Google Scholar] [CrossRef]
- van den Honert, M.S.; Gouws, P.A.; Hoffman, L.C. Importance and implications of antibiotic resistance development in livestock and wildlife farming in South Africa: A Review. S. Afr. J. Anim. Sci. 2018, 48, 401–412. [Google Scholar] [CrossRef]
- Amoako, D.G.; Somboro, A.M.; Abia, A.L.K.; Molechan, C.; Perrett, K.; Bester, L.A.; Essack, S.Y. Antibiotic resistance in Staphylococcus aureus from poultry and poultry products in uMgungundlovu District, South Africa, using the “Farm to Fork” approach. Microb. Drug Resist. 2020, 26, 402–411. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Impact of seasonal variation on Escherichia coli concentrations in the riverbed sediments in the Apies River, South Africa. Sci. Total Environ. 2015, 537, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Molechan, C.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci. Total Environ. 2019, 692, 868–878. [Google Scholar] [CrossRef]
- Amoako, D.G.; Bester, L.A.; Somboro, A.M.; Baijnath, S.; Govind, C.N.; Essack, S.Y. Plasmid-mediated resistance and virulence mechanisms in the private health sector in KwaZulu-Natal, South Africa: An investigation of methicillin resistant Staphylococcus aureus (MRSA) clinical isolates collected during a three month period. Int. J. Infect. Dis. 2016, 46, 38–41. [Google Scholar] [CrossRef]
- Subedi, M.; Luitel, H.; Devkota, B.; Bhattarai, R.K.; Phuyal, S.; Panthi, P.; Shrestha, A.; Chaudhary, D.K. Antibiotic resistance pattern and virulence genes content in avian pathogenic Escherichia coli (APEC) from broiler chickens in Chitwan, Nepal. BMC Vet. Res. 2018, 14, 113. [Google Scholar] [CrossRef]

| Antibiograms | Frequency |
|---|---|
| AMP-SXT-TET-CHL | 49 |
| AMP-TET-CHL | 44 |
| AMP-AMC-TET-CHL | 31 |
| AMP-AMC-SXT-TET-CHL | 27 |
| AZM-SXT-TET-CHL | 27 |
| SXT-TET-CHL | 22 |
| AMP-AMC-AMK-GEN-AZM-CAZ-FOX-CTX-FEP-CRO-LEX-CIP-NAL-SXT-TET-CHL | 18 |
| AMP-AMC-GEN-CIP-NAL-SXT-TET-CHL | 16 |
| AMP-SXT-TET | 16 |
| AMP-AMC-LEX-SXT-TET-CHL | 13 |
| AMP-AMC-LEX-TET-CHL | 13 |
| AMP-GEN-CIP-NAL-SXT-TET-CHL | 11 |
| AMP-AMC-AZM-CL-SXT-TET-CHL | 10 |
| AMP-AMC-GEN-CAZ-FOX-CTX-FEP-CRO-LEX-CIP-NAL-SXT-TET-CHL | 10 |
| AMP-AZM-LEX-SXT-TET-CHL | 10 |
| AZM-TET-CHL | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use. Antibiotics 2021, 10, 178. https://doi.org/10.3390/antibiotics10020178
Abdalla SE, Abia ALK, Amoako DG, Perrett K, Bester LA, Essack SY. From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use. Antibiotics. 2021; 10(2):178. https://doi.org/10.3390/antibiotics10020178
Chicago/Turabian StyleAbdalla, Shima E., Akebe Luther King Abia, Daniel G. Amoako, Keith Perrett, Linda A. Bester, and Sabiha Y. Essack. 2021. "From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use" Antibiotics 10, no. 2: 178. https://doi.org/10.3390/antibiotics10020178
APA StyleAbdalla, S. E., Abia, A. L. K., Amoako, D. G., Perrett, K., Bester, L. A., & Essack, S. Y. (2021). From Farm-to-Fork: E. Coli from an Intensive Pig Production System in South Africa Shows High Resistance to Critically Important Antibiotics for Human and Animal Use. Antibiotics, 10(2), 178. https://doi.org/10.3390/antibiotics10020178









