Burkholderia Bacteria Produce Multiple Potentially Novel Molecules that Inhibit Carbapenem-Resistant Gram-Negative Bacterial Pathogens
Abstract
1. Introduction
2. Results
2.1. Exploring the Antimicrobial Potential of Burkholderia Isolates
2.2. Determining the Spectrum of Antimicrobial Activity of Burkholderia Isolates That Inhibit Gram-Negative Pathogens
2.3. Extraction, Identification, and Dereplication of Burkholderia Specialized Metabolites
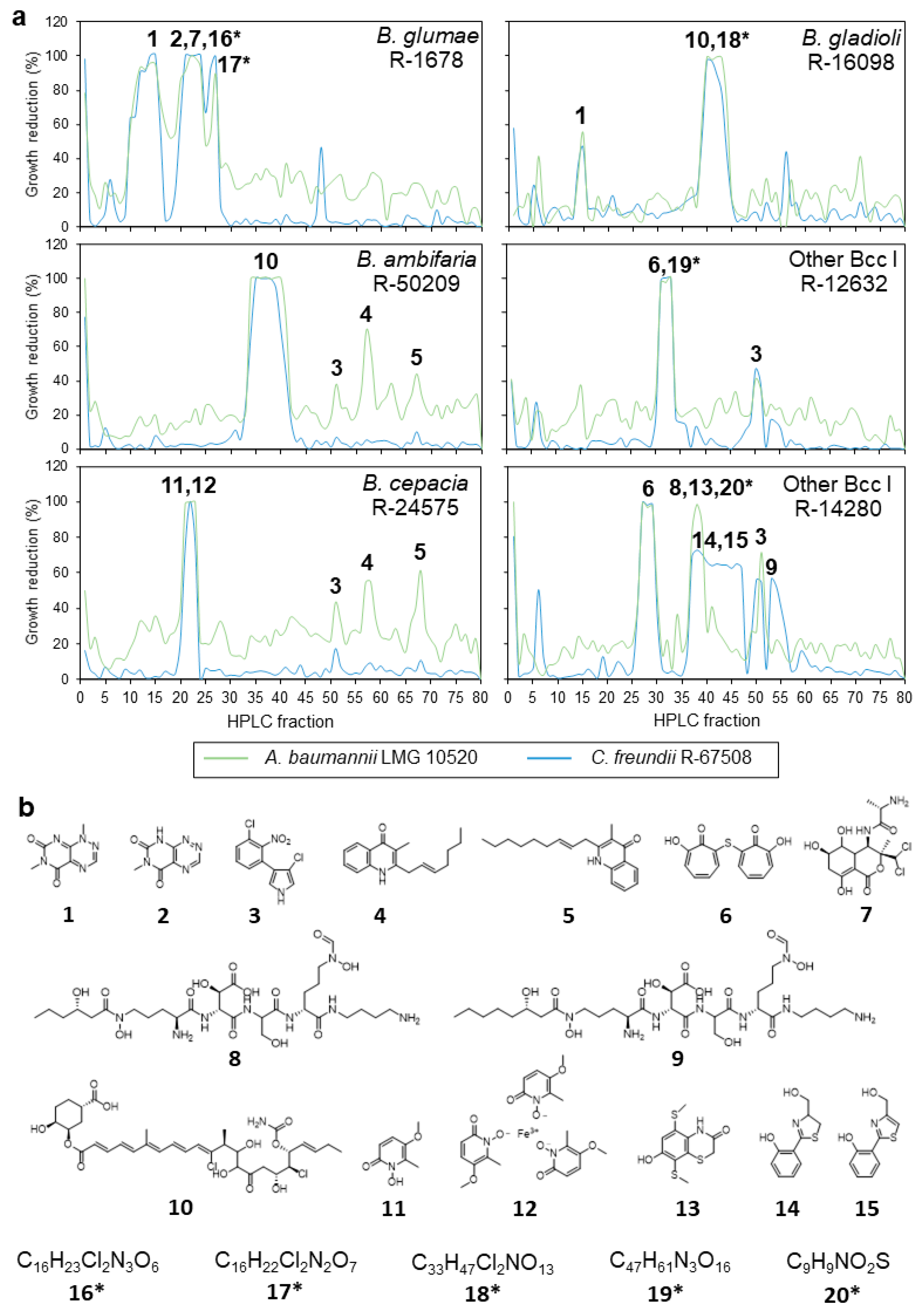
2.4. Semipreparative Fractionation of Crude Extracts and Dereplication of Active Fractions
3. Discussion
4. Materials and Methods
4.1. Strains and Growth Media
4.2. Overlay Assay to Detect Antimicrobial Activity
4.3. Extraction and Identification of Specialized Metabolites
4.4. Semipreparative Fractionation of Crude Extracts and Dereplication of Active Fractions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IACG. No Time to Wait: Securing the Future from Drug-Resistant Infections—Report to the Secretary-General of the United Nations. 2019. Available online: https://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/ (accessed on 29 January 2021).
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Spížek, J.; Novotná, J.; Rezanka, T.; Demain, A.L. Do we need new antibiotics? The search for new targets and new compounds. J. Ind. Microbiol. Biotechnol. 2010, 37, 1241–1248. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014, 41, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Depoorter, E.; Bull, M.J.; Peeters, C.; Coenye, T.; Vandamme, P.; Mahenthiralingam, E. Burkholderia: An update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 2016, 100, 5215–5229. [Google Scholar] [CrossRef]
- Kunakom, S.; Eustáquio, A.S. Burkholderia as a Source of Natural Products. J. Nat. Prod. 2019, 82, 2018–2037. [Google Scholar] [CrossRef]
- Esmaeel, Q.; Pupin, M.; Jacques, P.; Leclère, V. Nonribosomal peptides and polyketides of Burkholderia: New compounds potentially implicated in biocontrol and pharmaceuticals. Environ. Sci. Pollut. Res. 2018, 25, 29794–29807. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef]
- Eberl, L.; Vandamme, P. Members of the genus Burkholderia: Good and bad guys. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring env. Front. Genet. 2014, 5, 1–22. [Google Scholar] [CrossRef]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Santos, L.; Castro, D.B.A.; Ferreira-Tonin, M.; Corrêa, D.B.A.; Weir, B.S.; Park, D.; Ottoboni, L.M.M.; Neto, J.R.; Destéfano, S.A.L. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie Leeuwenhoek 2017, 110, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Estrada-de los Santos, P.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef] [PubMed]
- Marolda, C.L.; Hauröder, B.; John, M.A.; Michel, R.; Valvano, M.A. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 1999, 145, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Partida-Martinez, L.P.; Groth, I.; Schmitt, I.; Richter, W.; Roth, M.; Hertweck, C. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int. J. Syst. Evol. Microbiol. 2007, 57, 2583–2590. [Google Scholar] [CrossRef]
- Currie, B.J. Burkholderia pseudomallei and Burkholderia mallei. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2010; pp. 2869–2879. ISBN 9781455748013. [Google Scholar]
- Vial, L.; Chapalain, A.; Groleau, M.C.; Déziel, E. The various lifestyles of the Burkholderia cepacia complex species: A tribute to adaptation. Environ. Microbiol. 2011, 13, 1–12. [Google Scholar] [CrossRef]
- Suárez-Moreno, Z.R.; Caballero-Mellado, J.; Coutinho, B.G.; Mendonça-Previato, L.; James, E.K.; Venturi, V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef]
- LiPuma, J.J. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 2010, 23, 299–323. [Google Scholar] [CrossRef]
- Azegami, K.; Nishiyama, K.; Watanabe, Y.; Kadota, I.; Ohuchi, A.; Fukazawa, C. Pseudomonas plantarii sp. nov., the causal agent of rice seedling blight. Int. J. Syst. Bacteriol. 1987, 37, 144–152. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, J.; Kim, S.; Kang, Y.; Nagamatsu, T.; Hwang, I. Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Dis. 2003, 87, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Parke, J.L.; Gurian-Sherman, D. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 2001, 39, 225–258. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin production by Burkholderia cepacia and biocontrol of Rhizoctonia stem rot of poinsettia. Biol. Control 2002, 25, 56–63. [Google Scholar] [CrossRef]
- Wang, X.Q.; Liu, A.X.; Guerrero, A.; Liu, J.; Yu, X.Q.; Deng, P.; Ma, L.; Baird, S.M.; Smith, L.; Li, X.D.; et al. Occidiofungin is an important component responsible for the antifungal activity of Burkholderia pyrrocinia strain Lyc2. J. Appl. Microbiol. 2016, 120, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Mullins, A.J.; Murray, J.A.H.; Bull, M.J.; Jenner, M.; Jones, C.; Webster, G.; Green, A.E.; Neill, D.R.; Connor, T.R.; Parkhill, J.; et al. Genome mining identifies cepacin as a plant-protective metabolite of the biopesticidal bacterium Burkholderia ambifaria. Nat. Microbiol. 2019, 4, 996–1005. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Song, L.; Sass, A.; White, J.; Wilmot, C.; Marchbank, A.; Boaisha, O.; Paine, J.; Knight, D.; Challis, G.L. Enacyloxins are products of an unusual hybrid modular polyketide synthase encoded by a cryptic Burkholderia ambifaria genomic island. Chem. Biol. 2011, 18, 665–677. [Google Scholar] [CrossRef]
- Song, L.; Jenner, M.; Masschelein, J.; Jones, C.; Bull, M.J.; Harris, S.R.; Hartkoorn, R.C.; Vocat, A.; Romero-Canelon, I.; Coupland, P.; et al. Discovery and Biosynthesis of Gladiolin: A Burkholderia gladioli Antibiotic with Promising Activity against Mycobacterium tuberculosis. J. Am. Chem. Soc. 2017, 139, 7974–7981. [Google Scholar] [CrossRef]
- Wu, Y.; Seyedsayamdost, M.R. The Polyene Natural Product Thailandamide A Inhibits Fatty Acid Biosynthesis in Gram-Positive and Gram-Negative Bacteria. Biochemistry 2018, 57, 4247–4251. [Google Scholar] [CrossRef]
- Spilker, T.; Baldwin, A.; Bumford, A.; Dowson, C.G.; Mahenthiralingam, E.; LiPuma, J.J. Expanded Multilocus Sequence Typing for Burkholderia Species. J. Clin. Microbiol. 2009, 47, 2607–2610. [Google Scholar] [CrossRef]
- Vandamme, P.; Peeters, C. Time to revisit polyphasic taxonomy. Antonie Leeuwenhoek 2014, 106, 57–65. [Google Scholar] [CrossRef]
- Byng, G.S.; Turner, J.M. Phenazine Biosynthesis by a Pseudomonad. Biochem. Soc. Trans. 1975, 3, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, P.; Opelt, K.; Knochel, N.; Berg, C.; Schonmann, S.; De Brandt, E.; Eberl, L.; Falsen, E.; Berg, G. Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 2007, 57, 2228–2235. [Google Scholar] [CrossRef] [PubMed]
- Gasser, I.; Cardinale, M.; Müller, H.; Heller, S.; Eberl, L.; Lindenkamp, N.; Kaddor, C.; Steinbüchel, A.; Berg, G. Analysis of the endophytic lifestyle and plant growth promotion of Burkholderia terricola ZR2-12. Plant Soil 2011, 347, 125–136. [Google Scholar] [CrossRef]
- Baldwin, A.; Mahenthiralingam, E.; Kathleen, M.; Honeybourne, D.; Maiden, M.C.J.; John, R.; Speert, D.P.; Lipuma, J.J.; Vandamme, P.; Dowson, C.G.; et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 2005, 43, 4665–4673. [Google Scholar] [CrossRef] [PubMed]
- Groenhagen, U.; Baumgartner, R.; Bailly, A.; Gardiner, A.; Eberl, L.; Schulz, S.; Weisskopf, L. Production of bioactive volatiles by different Burkholderia ambifaria strains. J. Chem. Ecol. 2013, 39, 892–906. [Google Scholar] [CrossRef]
- Meyer, J.M.; Hohnadel, D.; Hallé, F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J. Gen. Microbiol. 1989, 135, 1479–1487. [Google Scholar] [CrossRef]
- Itoh, J.; Miyadoh, S.; Takahasi, S.; Amano, S.; Ezaki, N.; Yamada, Y. Studies on antibiotics BN-227 and BN-227-F., new antibiotics. I. Taxonomy, isolation and characterization. J. Antibiot. (Tokyo) 1979, 32, 1089–1095. [Google Scholar] [CrossRef]
- Deng, P.; Foxfire, A.; Xu, J.; Baird, S.M.; Jia, J.; Delgado, K.H.; Shin, R.; Smith, L.; Lu, S.-E. The Siderophore Product Ornibactin Is Required for the Bactericidal Activity of Burkholderia contaminans MS14. Appl. Environ. Microbiol. 2017, 83, e00051-17. [Google Scholar] [CrossRef]
- Adler, C.; Corbalán, N.S.; Seyedsayamdost, M.R.; Pomares, M.F.; de Cristóbal, R.E.; Clardy, J.; Kolter, R.; Vincent, P.A. Catecholate Siderophores Protect Bacteria from Pyochelin Toxicity. PLoS ONE 2012, 7, e46754. [Google Scholar] [CrossRef]
- Brüsewitz, G.; Molls, W.; Westphal, C.; Pulverer, G. Substituted Tropolones, Process for the Preparation Thereof and Pharmaceutical Compositions Containing These. DE 3149608A1, 15 December 1981. [Google Scholar]
- Lee, J.Y.; Moon, S.S.; Hwang, B.K. Isolation and Antifungal and Antioomycete Activities of Aerugine Produced by Pseudomonas fluorescens Strain MM-B16. Appl. Environ. Microbiol. 2003, 69, 2023–2031. [Google Scholar] [CrossRef]
- Carmi, R.; Carmeli, S.; Levy, E.; Gough, F.J. (+)-(S)-Dihydroaeruginoic Acid, an Inhibitor of Septoria tritici and Other Phytopathogenic Fungi and Bacteria, Produced by Pseudomonas fluorescens. J. Nat. Prod. 1994, 57, 1200–1205. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Cornelis, P.; Guillemyn, K.; Ballet, S.; Christophersen, C.; Hammerich, O. Structure revision of N-mercapto-4-formylcarbostyril produced by Pseudomonas fluorescens G308 to 2-(2-hydroxyphenyl)thiazole-4-carbaldehyde [aeruginaldehyde]. Nat. Prod. Commun. 2014, 9, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Trottmann, F.; Franke, J.; Ishida, K.; García-Altares, M.; Hertweck, C. A Pair of Bacterial Siderophores Releases and Traps an Intercellular Signal Molecule: An Unusual Case of Natural Nitrone Bioconjugation. Angew. Chem. Int. Ed. 2019, 58, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.; Opel, V.; Scherlach, K.; Hertweck, C. Biosynthesis of antifungal and antibacterial polyketides by Burkholderia gladioli in coculture with Rhizopus microsporus. Mycoses 2014, 57, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Horiuchi, Y.; Hamada, M.; Takeuchi, T.; Umezawa, H. A new antitumor antibiotic, bactobolin produced by Pseudomonas. J. Antibiot. (Tokyo) 1979, 32, 1069–1071. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Chandler, J.R.; Blodgett, J.A.V.; Lima, P.S.; Duerkop, B.A.; Oinuma, K.-I.; Greenberg, E.P.; Clardy, J. Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org. Lett. 2010, 12, 716–719. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Wang, R.; Wang, Q.; Lu, L. Toxoflavin Produced by Burkholderia gladioli from Lycoris aurea Is a New Broad-Spectrum Fungicide. Appl. Environ. Microbiol. 2019, 85. [Google Scholar] [CrossRef]
- Ma, J.; Yoneda, F.; Nagamatsu, T. Synthesis of 6-Azapurines by Transformation of Toxoflavins and Reumycins (7-Azapteridines) and their Cytotoxicities. Aust. J. Chem. 2015, 68, 203. [Google Scholar] [CrossRef]
- Butt, A.T.; Thomas, M.S. Iron Acquisition Mechanisms and Their Role in the Virulence of Burkholderia Species. Front. Cell. Infect. Microbiol. 2017, 7, 460. [Google Scholar] [CrossRef]
- Schmidt, S.; Blom, J.F.; Pernthaler, J.; Berg, G.; Baldwin, A.; Mahenthiralingam, E.; Eberl, L. Production of the antifungal compound pyrrolnitrin is quorum sensing-regulated in members of the Burkholderia cepacia complex. Environ. Microbiol. 2009, 11, 1422–1437. [Google Scholar] [CrossRef]
- Serino, L.; Reimmann, C.; Visca, P.; Beyeler, M.; Della Chiesa, V.; Haas, D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Biggins, J.B.; Liu, X.; Feng, Z.; Brady, S.F. Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J. Am. Chem. Soc. 2011, 133, 1638–1641. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, L.A.; Weightman, A.J.; Jones, T.H.; Marchbank, A.M.; Tiedje, J.M.; Mahenthiralingam, E. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ. Microbiol. 2007, 9, 1017–1034. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.D.; Vitorino, I.; De La Cruz, M.; Díaz, C.; Cautain, B.; Annang, F.; Pérez-Moreno, G.; Martinez, I.G.; Tormo, J.R.; Martín, J.M.; et al. Bioactivities and extract dereplication of actinomycetales isolated from marine sponges. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Pence, H.E.; Williams, A. Chemspider: An online chemical information resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Audoin, C.; Bonhomme, D.; Ivanisevic, J.; Cruz, M.; Cautain, B.; Monteiro, M.; Reyes, F.; Rios, L.; Perez, T.; Thomas, O. Balibalosides, an Original Family of Glucosylated Sesterterpenes Produced by the Mediterranean Sponge Oscarella balibaloi. Mar. Drugs 2013, 11, 1477–1489. [Google Scholar] [CrossRef]
| Species | Number of Isolates | Number with Antimicrobial Activity (CLIN/ENV) | ||
|---|---|---|---|---|
| (CLIN/ENV) | S. aureus | A. baumannii | C. albicans | |
| Burkholderia aenigmatica | 3 (1/2) | 0 | 0 | 0 |
| Burkholderia ambifaria | 12 (3/8) 1 | 8 (2/6) | 3 (0/3) | 8 (3/5) |
| Burkholderia anthina | 11 (2/9) | 0 | 0 | 0 |
| Burkholderia arboris | 17 (10/7) | 3 (0/3) | 0 | 10 (3/7) |
| Burkholderia catarinensis | 1 (0/1) | 0 | 0 | 1 (0/1) |
| Burkholderia cenocepacia | 12 (8/4) | 2 (2/0) | 1 (1/0) | 5 (5/0) |
| Burkholderia cepacia | 10 (5/5) | 6 (3/3) | 1 (0/1) | 3 (1/2) |
| Burkholderia contaminans | 16 (13/3) | 1 (1/0) | 0 | 9 (2/7) |
| Burkholderia diffusa | 2 (1/1) | 0 | 0 | 0 |
| Burkholderia dolosa | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia lata | 7 (2/5) | 4 (2/2) | 0 | 3 (1/2) |
| Burkholderia latens | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia metallica | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia multivorans | 2 (2/0) | 0 | 0 | 0 |
| Burkholderia pseudomultivorans | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia puraquae | 1 (0/1) | 0 | 0 | 0 |
| Burkholderia pyrrocinia | 10 (2/8) | 1 (0/1) | 0 | 4 (0/4) |
| Burkholderia seminalis | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia stabilis | 1 (1/0) | 0 | 0 | 0 |
| Burkholderia stagnalis | 1 (0/1) | 0 | 0 | 0 |
| Burkholderia territorii | 1 (0/1) | 0 | 0 | 0 |
| Burkholderia ubonensis | 1 (0/1) | 0 | 0 | 0 |
| Burkholderia vietnamiensis | 10 (2/8) | 6 (1/5) | 0 | 8 (1/7) |
| Other Bcc D | 3 (3/0) | 1 (1/0) | 0 | 1 (1/0) |
| Other Bcc E | 1 (0/1) | 0 | 0 | 1 (0/1) |
| Other Bcc I | 8 (1/7) | 7 (1/6) | 7 (1/6) | 8 (1/7) |
| Other Bcc J | 1 (1/0) | 0 | 0 | 0 |
| Other Bcc N | 1 (0/1) | 1 (0/1) | 1 (0/1) | 1 (0/1) |
| Other Bcc | 1 (1/0) | 0 | 0 | 1 (1/0) |
| Burkholderia gladioli | 11 (5/5) 1 | 5 (2/2) 1 | 3 (1/2) | 4 (2/2) |
| Burkholderia glumae | 15 (1/14) | 10 (0/10) | 2 (0/2) | 0 |
| Burkholderia plantarii | 8 (1/7) | 8 (1/7) | 0 | 2 (1/1) |
| Burkholderia oklahomensis | 1 (1/0) | 1 (1/0) | 0 | 0 |
| Burkholderia singularis | 2 (2/0) | 1 (1/0) | 1 (1/0) | 1 (1/0) |
| Burkholderia thailandensis | 1 (0/1) | 1 (0/1) | 0 | 0 |
| Paraburkholderia bryophila | 6 (0/6) | 1 (0/1) | 0 | 1 (0/1) |
| Paraburkholderia phenazinium | 1 (0/1) | 1 (0/1) | 0 | 1 (0/1) |
| Paraburkholderia terricola | 6 (0/6) | 1 (0/1) | 0 | 0 |
| Paraburkholderia spp. | 39 (0/39) | 0 | 0 | 0 |
| Caballeronia spp. | 25 (2/23) | 0 | 0 | 0 |
| Robbsia andropogonis | 1 (0/1) | 0 | 0 | 0 |
| Trinickia caryophyllii | 5 (0/5) | 3 (0/3) | 0 | 3 (0/3) |
| Trinickia spp. | 2 (0/2) | 0 | 0 | 0 |
| Mycetohabitans spp. | 2 (0/2) | 0 | 0 | 0 |
| Total isolates | 263 (75/186) 1 | 72 (18/53) | 19(4/15) | 75 (23/52) |
| Results of First Screen | Number of Strains with Antimicrobial Activity | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sa | Ab | Ca | ||||||||||||||
| Strain | ATCC 29213 | LMG 10520 | SC 50314 | Sa | Ef | Cd | Ab | Pa | Eb | Kp | Cf | Ec | Mm | Cg | Ck | Af |
| Burkholderia ambifaria LMG 19182T t1 1 | + | + | + | 1 | 3 | 3 | 3 | 0 | 2 | 0 | 1 | 2 | 1 | 3 | 3 | 3 |
| Burkholderia ambifaria LMG 19182T t2 1 | + | + | − | 3 | 2 | 3 | 2 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 |
| Burkholderia ambifaria R-50209 | + | + | + | 2 | 3 | 3 | 2 | 0 | 2 | 0 | 1 | 2 | 1 | 3 | 3 | 3 |
| Burkholderia cenocepacia R-1474 | + | + | + | 3 | 3 | 3 | 3 | 0 | 3 | 3 | 1 | 2 | 1 | 2 | 1 | 2 |
| Burkholderia cenocepacia R-49069 | − | − | + | 2 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 |
| Other Bcc I R-12632 | + | + | + | 3 | 3 | 3 | 2 | 0 | 3 | 3 | 1 | 2 | 1 | 1 | 0 | 3 |
| Other Bcc I R-14268 | + | + | + | 3 | 3 | 3 | 2 | 0 | 3 | 3 | 1 | 2 | 1 | 1 | 0 | 3 |
| Other Bcc I R-14280 | + | + | + | 3 | 3 | 3 | 2 | 0 | 3 | 3 | 1 | 2 | 1 | 2 | 0 | 3 |
| Other Bcc I R-14352 | + | + | + | 3 | 3 | 3 | 1 | 0 | 3 | 3 | 1 | 2 | 1 | 1 | 0 | 3 |
| Other Bcc I R-14356 | + | + | + | 3 | 3 | 3 | 1 | 0 | 3 | 3 | 1 | 2 | 1 | 1 | 0 | 3 |
| Other Bcc I R-10741 | + | + | + | 3 | 3 | 2 | 0 | 0 | 3 | 3 | 1 | 2 | 1 | 1 | 1 | 3 |
| Other Bcc I R-52250 | + | + | + | 3 | 3 | 3 | 2 | 0 | 3 | 3 | 1 | 2 | 1 | 0 | 0 | 0 |
| Other Bcc N R-52245 | + | + | + | 3 | 3 | 3 | 1 | 0 | 2 | 2 | 1 | 2 | 1 | 0 | 0 | 0 |
| Burkholderia cepacia R-49076 | + | − | + | 3 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 3 |
| Burkholderia cepacia R-24575 | + | − | − | 1 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | 2 |
| Burkholderia cepacia LMG 1222T | + | + | + | 3 | 3 | 3 | 2 | 0 | 3 | 3 | 1 | 2 | 1 | 2 | 0 | 0 |
| Burkholderia arboris R-8833 | − | − | + | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 3 |
| Burkholderia vietnamiensis R-24454 | + | − | + | 2 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 3 | 3 |
| Burkholderia gladioli R-11809 | + | − | + | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 | 3 | 3 |
| Burkholderia gladioli LMG 2216T | + | + | + | 3 | 3 | 3 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 |
| Burkholderia gladioli R-16098 | + | + | + | 3 | 3 | 3 | 3 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Burkholderia gladioli R-20794 | − | + | − | 2 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| Burkholderia glumae R-1678 | + | + | − | 3 | 3 | 3 | 1 | 0 | 2 | 0 | 1 | 2 | 1 | 0 | 0 | 0 |
| Burkholderia glumae R-8618 | + | − | − | 2 | 1 | 2 | 0 | 0 | 2 | 0 | 1 | 2 | 1 | 0 | 0 | 0 |
| Burkholderia glumae LMG 2196T | + | + | − | 3 | 3 | 2 | 2 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Burkholderia plantarii LMG 10908 | + | − | − | 3 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Burkholderia singularis LMG 28155 | + | + | + | 3 | 3 | 3 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | 2 |
| Molecular Formula | Common Name | R-50209 | R-16098 | R-20794 | R-1678 | R-8618 | R-24575 | LMG 1222T | R-12632 | R-10741 | R-14280 | R-52250 | R-52245 | R-1474 | LMG 28155 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C33H45Cl2NO11 | Enacyloxin | + | + | + | − | − | − | − | − | − | − | − | − | − | − |
| C7H7N5O2 | Toxoflavin | − | + | − | + | + | − | − | − | − | − | − | − | − | − |
| C6H5N5O2 | Reumycin | − | − | − | + | + | − | − | − | − | − | − | − | − | − |
| C14H20Cl2N2O6 | Bactobolin A | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| C52H85N11O22 | Cepacidin A1 | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| C19H25NO | Antibiotic SF 2420B | + | − | − | − | − | + | − | − | − | − | − | − | − | − |
| C17H21NO | + | − | − | − | − | + | − | − | − | − | − | − | − | − | |
| C10H12N5O7P | cGMP | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| C8H11NO3 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| C10H6Cl2N2O2 | Pyrrolnitrin | − | − | − | − | − | + | − | + | + | + | + | − | − | − |
| C10H8Cl2N2 | Aminopyrrolnitrin | − | − | − | − | − | + | − | − | + | − | + | − | − | − |
| C21H24FeN3O9 | Antibiotic BN-227-F | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| C7H9NO3 | Cepabactin | − | − | − | − | − | + | + | − | − | − | − | − | − | − |
| C10H7NO3S | Aeruginoic acid | − | − | − | − | − | + | + | + | − | + | + | − | − | − |
| C10H9NO3S | Dihydroaeruginoic acid | − | − | − | − | − | − | + | + | − | + | + | − | − | − |
| C10H11NO2S | Aerugine | − | − | − | − | − | − | + | + | − | + | + | − | − | − |
| C14H16N2O3S2 | Pyochelin | − | − | − | − | − | − | + | + | − | + | + | − | − | − |
| C14H10O4S | Ditropolonyl sulfide | − | − | − | − | − | − | + | − | + | + | + | + | + | − |
| C30H56N8O13 | Ornibactin C8 | − | − | − | − | − | − | + | + | + | + | + | + | + | − |
| C28H52N8O13 | Ornibactin C6 | − | − | − | − | − | − | − | + | + | + | + | + | + | − |
| C26H48N8O13 | Ornibactin C4 | − | − | − | − | − | − | − | + | − | − | − | + | + | − |
| C11H8Cl2N2O4 | − | − | − | − | − | − | − | + | + | − | − | − | − | − | |
| C12H12O4 | Differolide | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| C33H47Cl2NO13 1 | + | − | + | − | − | − | − | − | − | − | − | − | − | − | |
| C36H64N4O10 1 | − | + | − | − | − | − | − | − | − | − | − | − | − | − | |
| C17H27Cl2NO7 1 | − | − | + | − | − | − | − | − | − | − | − | − | − | − | |
| C7H9N5O3 1 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| C9H21NO3 1 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| C17H25N3O13 1 | − | − | − | + | − | − | − | − | − | − | − | − | − | − | |
| C10H9NO2 1 | − | − | − | − | − | + | − | − | − | − | − | − | − | − | |
| C19H13N5O12S 1 | − | − | − | − | − | + | − | − | − | − | − | − | − | − | |
| C9H9NO3 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |
| C15H12O6S 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |
| C18H33N3O12 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |
| C26H42O7 1 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | |
| C35H39N9O11 1 | − | − | − | − | − | − | − | + | − | − | − | + | + | − | |
| C37H43N9O11 1 | − | − | − | − | − | − | − | + | − | − | − | + | + | − | |
| C26H48N8O14 1 | − | − | − | − | − | − | − | − | − | − | − | + | + | − | |
| C33H35N9O11 1 | − | − | − | − | − | − | − | − | − | − | − | + | + | − | |
| C21H25FeNO15S2 1 | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| C14H14O3 1 | − | − | − | − | − | − | − | − | − | − | − | − | − | + | |
| C16H22O3 1 | − | − | − | − | − | − | − | − | − | − | − | − | − | + |
| Strain | Fraction | Metabolite | Growth Reduction (%) | |
|---|---|---|---|---|
| C. freundii | A. baumannii | |||
| Burkholderia ambifaria R-50209 | ||||
| 36 | Enacyloxin IIa or IIb | 100 | 100 | |
| 38 | Enacyloxin IIa or IIb | 100 | 99 | |
| 51 | Pyrrolnitrin | 7 | 38 | |
| 57 | C17H21NO | 5 | 70 | |
| 67 | Antibiotic SF 2420B | 10 | 44 | |
| Burkholderia gladioli R-16098 | ||||
| 15 | Reumycin | 47 | 55 | |
| 40 | C33H47Cl2NO13 1 | 98 | 100 | |
| 42 | Enacyloxin IIa or IIb | 89 | 100 | |
| 43 | Enacyloxin IIa or IIb | 78 | 99 | |
| Burkholderia glumae R-1678 | ||||
| 14 | Reumycin | 100 | 96 | |
| 15 | Reumycin | 100 | 95 | |
| 20 | Toxoflavin | 64 | 86 | |
| 22 | Bactobolin A | 100 | 100 | |
| 23 | C16H23Cl2N3O6 1 | 100 | 99 | |
| 26 | C16H22Cl2N2O7 1 | 93 | 55 | |
| 27 | C16H22Cl2N2O7 1 | 100 | 90 | |
| Burkholderia cepacia R-24575 | ||||
| 22 | Cepabactin | 100 | 99 | |
| Antibiotic BN-227-F | ||||
| 51 | Pyrrolnitrin | 18 | 43 | |
| 58 | C17H21NO | 10 | 55 | |
| 68 | Antibiotic SF 2420B | 11 | 61 | |
| Other Bcc I R-12632 | ||||
| 31 | Ditropolonyl sulfide | 99 | 97 | |
| C47H61N3O16 1 | ||||
| 51 | Pyrrolnitrin | 39 | 36 | |
| Other Bcc I R-14280 | ||||
| 28 | Ditropolonyl sulfide | 98 | 97 | |
| 38 | Ornibactin C6 | 73 | 99 | |
| C10H11NO2S3 | ||||
| C9H9NO2S 1 | ||||
| 39 | Ornibactin C6 | 70 | 88 | |
| Aerugine | ||||
| Aeruginol | ||||
| 51 | Ornibactin C8 | 54 | 72 | |
| Pyrrolnitrin | ||||
| 54 | Ornibactin C8 | 52 | 22 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Depoorter, E.; De Canck, E.; Coenye, T.; Vandamme, P. Burkholderia Bacteria Produce Multiple Potentially Novel Molecules that Inhibit Carbapenem-Resistant Gram-Negative Bacterial Pathogens. Antibiotics 2021, 10, 147. https://doi.org/10.3390/antibiotics10020147
Depoorter E, De Canck E, Coenye T, Vandamme P. Burkholderia Bacteria Produce Multiple Potentially Novel Molecules that Inhibit Carbapenem-Resistant Gram-Negative Bacterial Pathogens. Antibiotics. 2021; 10(2):147. https://doi.org/10.3390/antibiotics10020147
Chicago/Turabian StyleDepoorter, Eliza, Evelien De Canck, Tom Coenye, and Peter Vandamme. 2021. "Burkholderia Bacteria Produce Multiple Potentially Novel Molecules that Inhibit Carbapenem-Resistant Gram-Negative Bacterial Pathogens" Antibiotics 10, no. 2: 147. https://doi.org/10.3390/antibiotics10020147
APA StyleDepoorter, E., De Canck, E., Coenye, T., & Vandamme, P. (2021). Burkholderia Bacteria Produce Multiple Potentially Novel Molecules that Inhibit Carbapenem-Resistant Gram-Negative Bacterial Pathogens. Antibiotics, 10(2), 147. https://doi.org/10.3390/antibiotics10020147






