Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya, Isolated from 2017–2018
Abstract
1. Introduction
2. Methods
2.1. Study Design and Patient Recruitment
2.2. Library Preparation and Whole Genome Sequencing
2.3. Read Library Quality Control, Mapping and De Novo Genome Assembly
2.4. Species Identification, Sero- and Sequence Typing, Genome Annotation and Screening for UTI Virulence/AMR Genes
2.5. Bacterial Sample Collection and Antimicrobial Susceptibility Testing
2.6. Phylogenetic Reconstruction
2.7. Pangenome Analyses
3. Results
3.1. Patient and Bacterial Strain Profiles
3.2. Genomic and Pangenomic Characterization Confirmed the Virulence Factors Present in Uropathogenic E. coli and K. pneumoniae
3.3. Prevalence of AMR Genes in E. coli and K. pneumoniae Uropathogens from KY and UG
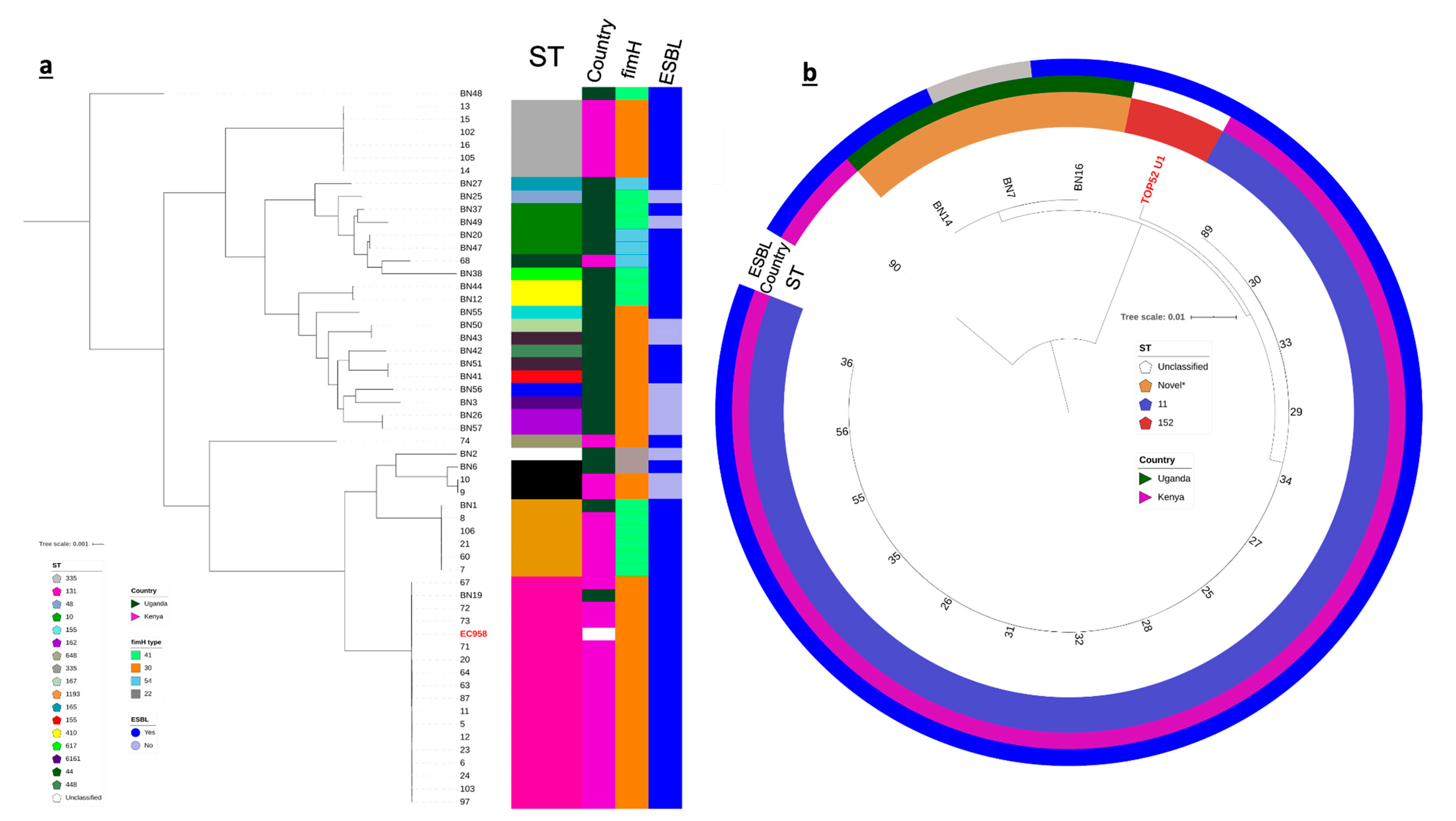
3.4. Population Structure of KY and UG Uropathogens
3.5. Plasmid Characterization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The antimicrobial resistance crisis: Causes, consequences, and management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Rather, I.A.; Kim, B.C.; Bajpai, V.K.; Park, Y.H. Self-medication and antibiotic resistance: Crisis, current challenges, and prevention. Saudi J. Biol. Sci. 2017, 24, 808–812. [Google Scholar] [CrossRef]
- Nathan, C.; Cars, O. Antibiotic resistance-problems, progress, and prospects. N. Engl. J. Med. 2014, 371, 1761–1763. [Google Scholar] [CrossRef] [PubMed]
- French, G.L. The continuing crisis in antibiotic resistance. Int. J. Antimicrob. Agents 2010, 36 (Suppl. 3), S3–S7. [Google Scholar] [CrossRef]
- Read, A.F.; Woods, R.J. Antibiotic resistance management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef] [PubMed]
- Government of Kenya. National Action Plan for the Prevention and Containment of Antimicrobial Resistance; Government of Kenya: Nairobi, Kenya, 2017. [Google Scholar]
- UNAS; CDDEP; GARP-Uganda; Mpairwe, Y.; Wamala, S. Antibiotic Resistance in Uganda: Situation Analysis and Recommendations; Uganda National Academy of Sciences: Kampala, Uganda; Center for Disease Dynamics, Economics & Policy: Washington, DC, USA, 2015; p. 107. [Google Scholar]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Kunin, C.M.; Zacha, E.; Paquin, A.J. Urinary-tract infections in schoolchildren. I. Prevalence of bacteriuria and associated urologica. N. Engl. J. Med. 1962, 266, 1287–1296. [Google Scholar] [CrossRef]
- Lindberg, U.; Hanson, L.A.; Jodal, U.; Lidin-Janson, G.; Lincoln, K.O.; lling, S. Asymptomatic bacteriuria in schoolgirls. II. Differences in Escherichia coli causing asymptomatic bacteriuria. Acta Paediatr. Scand. 1975, 64, 432–436. [Google Scholar] [CrossRef]
- Najar, M.S.; Saldanha, C.L.; Banday, K.A. Approach to urinary tract infections. Indian J. Nephrol. 2009, 19, 129–139. [Google Scholar] [CrossRef]
- Levison, M.E.; Kaye, D. Treatment of complicated urinary tract infections with an emphasis on drug-resistant gram-negative uropathogens. Curr. Infect. Dis. Rep. 2013, 15, 109–115. [Google Scholar] [CrossRef]
- Nicolle, L.E. Pivmecillinam in the treatment of urinary tract infections. J. Antimicrob. Chemother. 2000, 46 (Suppl. A), 35–39. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Didelot, X. The application of genomics to tracing bacterial pathogen transmission. Curr. Opin. Microbiol. 2015, 23, 62–67. [Google Scholar] [CrossRef]
- Goswami, C.; Fox, S.; Holden, M.; Connor, M.; Leanord, A.; Evans, T.J. Genetic analysis of invasive Escherichia coli in Scotland reveals determinants of healthcare-associated versus community-acquired infections. Microb. Genom. 2018, 4, e000190. [Google Scholar] [CrossRef]
- Argimón, S.; Masim, M.; Gayeta, J.M.; Lagrada, M.L.; Macaranas, P.; Cohen, V.; Limas, M.T.; Espiritu, H.O.; Palarca, J.C.; Chilam, J.; et al. Integrating whole-genome sequencing within the National Antimicrobial Resistance Surveillance Program in the Philippines. Nat. Commun. 2020, 11, 2719. [Google Scholar] [CrossRef]
- Halachev, M.R.; Chan, J.Z.-M.; Constantinidou, C.I.; Cumley, N.; Bradley, C.; Smith-Banks, M.; Oppenheim, B.; Pallen, M.J. Genomic epidemiology of a protracted hospital outbreak caused by multidrug-resistant Acinetobacter baumannii in Birmingham, England. Genome Med. 2014, 6, 61070. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, R.S.; Seemann, T.; Heffernan, H.; Kwong, J.C.; Gonҫalves da Silva, A.; Carter, G.P.; Woodhouse, R.; Dyet, K.H.; Bulach, D.M.; Stinear, T.P.; et al. Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J. Antimicrob. Chemother. 2018, 73, 353–364. [Google Scholar] [CrossRef]
- Ludden, C.; Decano, A.G.; Jamrozy, D.; Pickard, D.; Morris, D.; Parkhill, J.; Peacock, S.J.; Cormican, M.; Downing, T. Genomic surveillance of Escherichia coli ST131 identifies local expansion and serial replacement of subclones. Microb. Genom. 2020, 6, e000352. [Google Scholar] [CrossRef]
- Masim, M.L.; Argimón, S.; Espiritu, H.O.; Magbanua, M.A.; Lagrada, M.; Olorosa, A.M.; Cohen, V.; Gayeta, J.M.; Jeffrey, B.; Abudahab, K.; et al. Genomic surveillance of methicillin-resistant Staphylococcus aureus in the Philippines, 2013–2014. West. Pac. Surveill. Response J. 2021, 12, 6–16. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Ponstingl, H.; Ning, Z. SMALT–A New Mapper for DNA Sequencing Reads. F1000 Research 2010. Available online: https://f1000research.com/posters/327 (accessed on 18 December 2019).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Wylie, K.M.; Wylie, T.N.; Minx, P.J.; Rosen, D.A. Whole-genome sequencing of Klebsiella pneumoniae isolates to track strain progression in a single patient with recurrent urinary tract infection. Front. Cell. Infect. Microbiol. 2019, 9, 14. [Google Scholar] [CrossRef]
- Forde, B.M.; Ben Zakour, N.L.; Stanton-Cook, M.; Phan, M.-D.; Totsika, M.; Peters, K.M.; Chan, K.G.; Schembri, M.A.; Upton, M.; Beatson, S.A. The complete genome sequence of Escherichia coli EC958: A high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS ONE 2014, 9, e104400. [Google Scholar] [CrossRef]
- Centre for Genomic Pathogen Surveillance (CGPS). Pathogenwatch Private Metadata. Available online: https://cgps.gitbook.io/pathogenwatch/how-to-use-pathogenwatch/private-metadata (accessed on 31 October 2019).
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 90. [Google Scholar] [CrossRef]
- Achtman, M.; Wain, J.; Weill, F.X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Pierard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pacale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021, 49, D644–D650. [Google Scholar] [CrossRef]
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; Abudahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 30th ed.; Approved Standard; CLSI Document C24–Ed4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Didelot, X.; Wilson, D.J. ClonalFrameML: Efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 2015, 11, e1004041. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2010; Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 20 January 2020).
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Salinas, L.; Cárdenas, P.; Johnson, T.J.; Vasco, K.; Graham, J.; Trueba, G. Diverse commensal Escherichia coli clones and plasmids disseminate antimicrobial resistance genes in domestic animals and children in a semirural community in Ecuador. mSphere 2019, 4, e00316-19. [Google Scholar] [CrossRef] [PubMed]
- Kimata, K.; Shima, T.; Shimizu, M.; Tanaka, D.; Isobe, J.; Gyobu, Y.; Watahiki, M.; Nagai, Y. Rapid categorization of pathogenic Escherichia coli by multiplex PCR. Microbiol. Immunol. 2005, 49, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Montealegre, M.C.; Roy, S.; Boni, F.; Hossain, M.I.; Navab-Daneshmand, T.; Caduff, L.; Faruque, A.S.G.; Islam, M.A.; Julian, T.R. Risk factors for detection, survival, and growth of antibiotic-resistant and pathogenic Escherichia coli in household soils in rural Bangladesh. Appl. Environ. Microbiol. 2018, 84, e01978-18. [Google Scholar] [CrossRef]
- Ben Zakour, N.L.; Alsheikh-Hussain, A.S.; Ashcroft, M.M.; Khanh, N.T.; Roberts, L.W.; Stanton-Cook, M.; Schembri, M.A.; Beatson, S.A. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 2016, 7, e00347-16. [Google Scholar] [CrossRef]
- Sanjar, F.; Rusconi, B.; Hazen, T.H.; Koenig, S.S.; Mammel, M.K.; Feng, P.C.; Rasko, D.A.; Eppinger, M. Characterization of the pathogenome and phylogenomic classification of enteropathogenic Escherichia coli of the O157: Non-H7 serotypes. Pathog Dis. 2015, 73, ftv033. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Clearly, P.; Wiuff, C.; Doumith, M.; et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet. Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef]
- Valenza, G.; Werner, M.; Eisenberger, D.; Nickel, S.; Lehner-Reindl, V.; Holler, C.; Bogdan, C. First report of the new emerging global clone ST1193 among clinical isolates of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli from Germany. J. Glob. Antimicrob. Resist. 2019, 17, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Mhaya, A.; Trabelsi, R.; Begu, D.; Aillerie, S.; M’zali, F.; Tounsi, S.; Gdoura, R.; Arpin, C. Emergence of B2-ST131-C2 and A-ST617 Escherichia coli clones producing both CTX-M-15- and CTX-M-27 and ST147 NDM-1 positive Klebsiella pneumoniae in the Tunisian community. bioRxiv 2019. [Google Scholar] [CrossRef]
- Castellanos, L.R.; Donado-Godoy, P.; León, M.; Clavijo, V.; Arevalo, A.; Bernal, J.F.; Timmerman, A.J.; Mevius, D.J.; Wagenaar, J.A.; Hordijk, J. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the colombian poultry chain. PLoS ONE 2017, 12, e0170777. [Google Scholar] [CrossRef]
- Choudhury, N.A.; Paul, D.; Chakravarty, A.; Bhattacharjee, A.; Dhar Chanda, D. IncX3 plasmid mediated occurrence of blaNDM-4 within Escherichia coli ST448 from India. J. Infect. Public Health 2018, 11, 111–114. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, J.; Wang, X.; Bai, X.; Ma, J.; Dang, R.; Xiong, Y.; Fanning, S.; Bai, L.; Yang, Z. Characterization of five Escherichia coli isolates co-expressing ESBL and MCR-1 resistance mechanisms from different origins in China. Front. Microbiol. 2019, 10, 1994. [Google Scholar] [CrossRef]
- Ghenea, A.E.; Cioboată, R.; Drocaş, A.I.; Țieranu, E.N.; Vasile, C.M.; Moroşanu, A.; Țieranu, C.G.; Salan, A.I.; Popescu, M.; Turculeanu, A.; et al. Prevalence and antimicrobial resistance of Klebsiella strains isolated from a county hospital in Romania. Antibiotics 2021, 10, 868. [Google Scholar] [CrossRef]
- Yang, J.; Ye, L.; Guo, L.; Zhao, Q.; Chen, R.; Luo, Y.; Chen, Y.; Tian, S.; Zhao, J.; Shen, D.; et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: Dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin. Microbiol. Infect. 2013, 19, E509–E515. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, M.X.; Wang, H.C.; Dou, Q.Y.; Hu, Y.M.; Yan, Q.; Liu, W.E. An outbreak of infections caused by a Klebsiella pneumoniae ST11 clone coproducing Klebsiella pneumoniae carbapenemase-2 and RmtB in a Chinese Teaching Hospital. Chin. Med. J. 2016, 129, 2033–2039. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Dong, N.; Zheng, Z.; Lin, D.; Huang, M.; Wang, L.; Chan, E.W.; Shu, L.; Yu, J.; Zhang, R.; et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018, 18, 37–46. [Google Scholar] [CrossRef]
- Jayol, A.; Poirel, L.; Dortet, L.; Nordmann, P. National survey of colistin resistance among carbapenemase-producing Enterobacteriaceae and outbreak caused by colistin-resistant OXA-48-producing Klebsiella pneumoniae, France, 2014. Euro surveillance. Eur. Commun. Dis. Bull. 2016, 21, 30339. [Google Scholar] [CrossRef]
- Alqasim, A.; Abu Jaffal, A.; Alyousef, A.A. Prevalence of multidrug resistance and extended-spectrum β-lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018, 2018, 3026851. [Google Scholar] [CrossRef]
- Zhao, R.; Shi, J.; Shen, Y.; Li, Y.; Han, Q.; Zhang, X.; Gu, G.; Xu, J. Phylogenetic distribution of virulence genes among ESBL-producing uropathogenic Escherichia coli isolated from long-term hospitalized patients. J. Clin. Diagn. Res. 2015, 9, DC01–DC04. [Google Scholar] [CrossRef] [PubMed]
- Mbelle, N.M.; Osei Sekyere, J.; Amoako, D.G.; Maningi, N.E.; Modipane, L.; Essack, S.Y.; Feldman, C. Genomic analysis of a multidrug-resistant clinical Providencia rettgeri (PR002) strain with the novel integron ln1483 and an A/C plasmid replicon. Ann. N. Y. Acad. Sci. 2020, 1462, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Adefisoye, M.A.; Okoh, A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiol. Open 2016, 5, 143–151. [Google Scholar] [CrossRef] [PubMed]
| Gene | Protein Product | Strain Count and % in E. coli Collection | Strain Count and % in K. pneumoniae Collection |
|---|---|---|---|
| fimH | Type 1 fimbrin D-mannose specific adhesin | 56/56 (100) | 20/20 (100) |
| feoA/B/C | Fe(2+) transport protein A/B/C | 56/56 (100) | 20/20 (100) |
| entB | Enterobactin synthase component B | 56/56 (100) | 20/20 (100) |
| focA | Formate transporter | 56/56 (100) | 20/20 (100) |
| zapA | Cell division protein | 56/56 (100) | 20/20 (100) |
| satP | Succinate-acetate/proton symporter | 56/56 (100) | 20/20 (100) |
| chuR | Anaerobic sulfatase-maturating enzyme | 56/56 (100) | 19/20 (95) |
| mrkD | Type 3 fimbrial adhesin | 0 | 20/20 (100) |
| hlyE | Hemolysin E | 45/56 (80.4) | 2/20 (10) |
| fyuA | Pesticin receptor | 41/56 (73.2) | 3/20 (15) |
| iutA | Ferric aerobactin receptor | 25/56 (44.6) | 2/20 (10) |
| papD | Import of P pilus subunits into the periplasm | 26/56 (46.4) | 2/20 (10) |
| papA | Fimbrial major pilin protein | 23/56 (41.1) | 0 |
| kpsT | Polysialic acid transport ATP-binding protein | 15/56 (26.8) | 0 |
| pic | Serine protease pic autotransporter | 3/56 (5.4) | 0 |
| Sample Name | Serotype | Sequenc Type Pasteur | AMR Phenotype | fimH | Aminoglycosides | Macrolide | Ciplrofloxacin | β-Lactamase Inhibitors/ESBL Cephalosporins | Phenicols | Fluoroquinolones | Quinolone | Sulfonamide | Tetracycline | Folate Pathway Inhibitors | Antibiotic Effulx/Regulation | ESBL | MDR | Plasmid Replicon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | O25:H4 | 131 | AMP, CTX, CHL, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII |
| 6 | O25:H4 | 131 | AMP, CTX, CRO, CHL, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII |
| 7 | O*:H5 | 1193 | CRO, CHL, SXT, CIP, NA | H41 | aac(3)-Ib, aadA17 | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | NF | bacA, tolC, evgA | Yes | Yes | IncQ1, IncFIA, Col156, Col(BS512) |
| 8 | O75:H5 | 1193 | AMP, FOX, SXT, CIP, NA | H41 | aac(3)-Ib, aadA17 | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | NF | bacA, tolC, evgA | Yes | Yes | IncFIA, IncQ1, Col(BS512), Col156 |
| 9 | O6:H1 | 73 | AMP, FOX, SXT, CIP, NA | H30 | NF | NF | marA | NF | NF | NF | NF | NF | NF | NF | mexB | No | No | IncX1 |
| 10 | O6:H1 | 73 | AMP, CTX | H30 | NF | NF | marA | NF | NF | NF | NF | NF | NF | NF | acrB | No | No | NF |
| 11 | O25:H4 | 131 | AMP, CTX, CRO, CHL, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | NF | acrB, gadW, pmrF | Yes | Yes | IncFIA, IncFII, Col156 |
| 12 | O25:H4 | 131 | AMP, CTX, CRO, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | NF | acrB, gadW, pmrF | Yes | Yes | IncFIA, IncFII, Col156 |
| 13 | O55:H7 | 335 | AMP, SXT, NA | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | evgA, cpxA, gadW | Yes | Yes | IncQ1 |
| 14 | O55:H7 | 335 | AMP, SXT, NA | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | evgA, cpxA | Yes | Yes | IncQ1 |
| 15 | O55:H7 | 335 | AMP, CTX, SXT, NA | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | evgA, cpxA | Yes | Yes | IncQ1 |
| 16 | O55:H7 | 335 | AMP, SXT, NA | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | evgA, cpxA | Yes | Yes | IncQ1 |
| 20 | O25:H4 | 131 | AMP, CTX, FOX, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | acrB, gadW | Yes | Yes | IncFIA, IncFII, Col156 |
| 21 | O75:H5 | 1193 | AMP, FOX, SXT, CIP, NA | H41 | aac(3)-Ib | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | dfrA17 | evgA, tolC, bacA | Yes | Yes | IncQ1, IncFIA, Col156, Col(BS512) |
| 23 | O25:H4 | 131 | AMP, CTX, CRO, CAZ, FEP, CHL, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | acrB, gadW | Yes | Yes | IncFIA, IncFII, Col156 |
| 24 | O25:H4 | 131 | AMP, CTX, CRO, SXT, CIP, NA | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | acrB, gadW | Yes | Yes | IncFIA, IncFII, Col156 |
| 60 | O*:H5 | 1193 | ND | H41 | aac(3)-Ib, aadA5 | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | dfrA17 | acrS, bacA, tolC | Yes | Yes | IncQ1, IncFIA, Col156, Col(BS512) |
| 63 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFII, Col156, IncFIA |
| 64 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1, sul2 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII, Col156 |
| 67 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | NF | NF | sul1 | tet(A) | dfrA17 | acrS, gadW | Yes | Yes | IncFIA |
| 68 | O89:H4 | 44 | ND | H54 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | NF | NF | sul1 | tet(A) | dfrA17 | acrS, gadW | Yes | Yes | IncFIA, IncFII |
| 71 | O25:H4 | 131 | ND | H30 | aadA5 | NF | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | aac(3)-Ib-cr | QnrB2 | sul1 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII, IncY |
| 72 | O25:H4 | 131 | ND | H30 | aadA5 | NF | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | aac(3)-Ib-cr | NF | sul2 | NF | dfrA14 | gadW | Yes | Yes | IncFIA, IncFII |
| 73 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | NF | NF | sul1 | tet(A) | dfrA17 | acrS, gadW | Yes | Yes | IncFIA, IncFII |
| 74 | O*:H6 | 648 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | NF | NF | sul1 | tet(A) | dfrA17 | acrS, gadW | Yes | Yes | IncFIA, IncFII |
| 87 | O*:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII |
| 97 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul1 | tet(A) | dfrA17 | gadW | Yes | Yes | IncFIA, IncFII, Col156 |
| 102 | O*:H7 | 335 | ND | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | cpXxA, evgA | Yes | Yes | IncQ1 |
| 103 | O25:H4 | 131 | ND | H30 | aac(3)-Ib, aadA5 | mphA | marA | blaCTX-M-27/99 | NF | NF | NF | sul2 | tet(A) | dfrA17 | NF | Yes | Yes | IncFIA, IncFII, Col156 |
| 105 | O55:H7 | 335 | ND | H30 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | cpxA, evgA, acrB | Yes | Yes | IncQ1 |
| 106 | O*:H5 | 1193 | ND | H41 | aac(3)-Ib | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | dfrA17 | evgA, tolC, mphA | Yes | Yes | IncQ1, IncFIA, Col156 |
| BN1 | O75:H5 | 1193 | ND | H41 | aac(3)-Ib | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | dfrA17 | bacA, tolC, evgA | Yes | Yes | IncQ1, IncFIA |
| BN12 | O*:H9 | 410 | ND | H41 | aadA9, aac(3)-Ib | mphA | marA | blaTEM-220 | NF | NF | NF | NF | NF | dfrA17 | emrR | Yes | Yes | IncQ1, IncFIA |
| BN19 | O25:H4 | 131 | ND | H30 | aadA5, aac(3)-Ib, aac(3)-Iic/d/e | NF | marA | blaTEM-220, blaCTX-M-15, blaOXA-1, blaOXA-140 | catB3 | aac(6′)-Ib-cr | NF | sul1, sul2 | NF | NF | kdpE, gadW | Yes | Yes | IncFIA, IncFII |
| BN2 | O156:H7 | NF | ND | H22 | aac(3)-Ib | NF | marA | NF | NF | NF | NF | sul1 | tet(A) | dfrA17 | NF | No | Yes | Col |
| BN20 | O89:H9 | 10 | ND | H54 | aadA9, aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA1 | mdtP, msbA, acrB, baeS/R, yojI | Yes | Yes | IncQ1 |
| BN25 | O6:H11 | 48 | ND | H41 | aadA5, aac(3)-Ib | NF | marA | NF | NF | NF | NF | sul2 | NF | dfrA17 | yojI, pmrF, emrR, bacA, acrS/B/E, msbA, evgA, kdpE, mdtP, eptA | No | Yes | IncHI2A |
| BN26 | O9:H19 | 162* | ND | H30 | NF | NF | marA | NF | NF | NF | NF | NF | tet(A) | NF | emrR, mdtA | No | Yes | NF |
| BN27 | O*:H2 | 165 | ND | H54 | aac(6′)-Ib7, aac(3)-Ib, aadA9 | NF | marA | blaTEM-220 | NF | NF | qnrS1 | sul3 | tet(A) | NF | tolC, mdtO, msbA, acrB, baeS/R, acrD, gadX, evgA, pmrF | Yes | Yes | NF |
| BN3 | O18:H49 | 212 | ND | H30 | aac(6′)-Ib7, aac(3)-Ib, aadA9 | NF | marA | NF | NF | NF | NF | sul1 | tet(A) | dfrA15 | emrR | No | Yes | NF |
| BN37 | O8:H17 | 10 | ND | H41 | aac(3)-Ib, aadA5 | NF | marA | blaTEM-220 | NF | NF | NF | sul2 | tet(A) | dfrA15 | yojI, pmrF, emrR, bacA, acrS/B/E, msbA, evgA, kdpE, mdtP, eptA | Yes | Yes | IncFIA, IncFII |
| BN38 | O89:H10 | 617 | ND | H41 | NF | NF | marA | blaCTX-M-15/88, blaOXA-1/140 | catB3 | aac(6′)-Ib-cr | NF | sul1, sul2 | NF | NF | yojI, pmrF, emrR, bacA, acrS/B/E, msbA, evgA, kdpE, mdtP, eptA | Yes | Yes | IncFIA, IncFII |
| BN41 | O171:H21 | 155 | ND | H30 | aac(6′)-Ib7, aadA9 | NF | marA | blaTEM-220 | NF | NF | NF | sul2 | tet(A) | NF | emrR | Yes | Yes | IncHI1A, IncHI1B, IncFIA |
| BN42 | O29:H8 | 448 | ND | H30 | aadA13, aac(3)-Ib | NF | marA | blaCTX-M-5 | NF | NF | NF | sul2 | NF | dfrA14 | emrR, mphA | Yes | Yes | IncI, IncFII, IncFIA IncX4 |
| BN43 | O*:H4 | 6161 | ND | H30 | NF | NF | marA | NF | NF | NF | NF | sul2 | NF | dfrA14 | baeS | No | Yes | NF |
| BN44 | O8:H9 | 410 | ND | H41 | aac(6′)-Ib7, aadA9 | mphA | marA | blaCTX-M-5, blaOXA-1, blaTEM-30/220 | catB3 | aac(6′)-Ib-cr | NF | NF | NF | NF | emrR | Yes | Yes | IncFIA, IncQ1 |
| BN47 | O89:H9 | 10 | ND | H54 | acrD/E/F, aadA9, aac(3)-Ib, aac(6′)-Ib7 | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | NF | dfrA1 | gadX, tolC, mdtF/N/O, emrK | Yes | Yes | Col440II, IncQ1 |
| BN48 | O17:H11 | NF | ND | H41 | aac(3)-Ib | NF | marA | blaTEM-7/75/177 | NF | NF | NF | sul1 | NF | dfrA7 | axyY | Yes | Yes | IncQ1 |
| BN49 | O45:H11 | 10 | ND | H41 | NF | NF | marA | NF | NF | NF | qnrB19 | NF | NF | NF | yojI, pmrF, emrR, bacA, acrS/B/E, msbA, evgA, kdpE, mdtP, eptA | No | No | Col |
| BN50 | O*:H4 | 167 | ND | H30 | NF | NF | marA | NF | NF | NF | NF | sul2 | NF | dfrA14 | baeS | No | Yes | NF |
| BN51 | O171:H21 | 6161 | ND | H30 | aac(6′)-Ib7, aadA9 | NF | marA | blaTEM-220 | NF | NF | NF | sul2 | tet(A) | NF | mdtA/B, emrR | Yes | Yes | IncHI1A, IncHI1B, IncFIA |
| BN55 | O185:H8 | 155 | ND | H30 | acrB, aadA1, aac(3)-Ib, aac(6′)-Ib7 | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA1 | yojI, pmrF, emrR, bacA, acrS/B/E, msbA, evgA, kdpE, mdtP, eptA, emtK, cpxA | Yes | Yes | IncFIB, IncQ1 |
| BN56 | O*:H7 | 2163 | ND | H30 | NF | NF | marA | NF | NF | NF | NF | NF | NF | NF | emrR, baeS | No | No | NF |
| BN57 | O9:H19 | 162* | ND | H30 | NF | NF | marA | NF | NF | NF | NF | sul2 | tet(A) | NF | mdtA, emrR | No | Yes | NF |
| BN6 | O6:H1 | 73 | ND | H22 | aac(3)-Ib | NF | marA | blaTEM-220 | NF | NF | NF | sul1 | tet(A) | dfrA7 | NF | Yes | Yes | IncFIB |
| Sample Name | Sequence Type Pasteur | AMR Phenotype | Aminoglycosides | Ciplrofloxacin | Penicillins + β-lactamase Inhibitors | ESBL Cephalosporins | Phenicols | Fluoroquinolones | Quinolone | Sulfonamide | Tetracycline | Folate Pathway Inhibitors | Antibiotic Effulx/Regulation | ESBL | MDR | Plasmid Replicon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 26 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 27 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 28 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 29 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 30 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 31 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 32 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 33 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 34 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 35 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 36 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 55 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 56 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 89 | 11 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | blaLEN-4/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140, blaNDM-1 | catB3 | oqxB, aac(6′)-Ib-cr | qnrB9 | sul2 | NF | NF | NF | Yes | Yes | IncFII, IncFIB, IncR |
| 90 | NF | ND | aac(6′)-Ib7, aadA9 | NF | blaLEN-3/4/5/6 | blaSHV-28, blaCTX-M-15, blaOXA-1/140 | NF | NF | NF | sul1 | tet(A) | dfrA17 | yojI, pmrF, emrR, bacA, acrB, msbA, evgA, kdpE, mdtP, eptA, emtK, cpxA | Yes | Yes | IncN |
| BN14 | 0b8e | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | marA | blaLEN-3/4/5/6 | blaSHV-28, blaCTX-M-15, blaTEM-220 | catB3 | oqxA/B, aac(6′)-Ib-cr | qnrB6/17 | sul1 | tet(B) | dfrA27 | yojI, pmrF, emrR, bacA, acrB, msbA, evgA, kdpE, mdtP, eptA, emtK, cpxA | Yes | Yes | IncR, IncFII, IncFIA, Col, IncX4 |
| BN16 | 67b2 | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | marA | blaLEN-3/4/5/6 | blaCTX-M-15/88 | catB3 | oqxA/B, aac(6′)-Ib-cr | qnrB6/17 | sul1, sul2 | tet(B) | dfrA27 | yojI, pmrF, emrR, bacA, acrB, msbA, evgA, kdpE, mdtP, eptA, emtK, cpxA | Yes | Yes | IncR, IncFII, IncFIA, Col, IncX4 |
| BN7 | 6b6f | ND | aac(6′)-30/aac(6′)-Ib’/7/10, aadA9 | NF | NF | NF | NF | oqxA/B, aac(6′)-Ib-cr | qnrB6/17 | NF | NF | NF | NF | No | Yes | IncR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Decano, A.G.; Pettigrew, K.; Sabiiti, W.; Sloan, D.J.; Neema, S.; Bazira, J.; Kiiru, J.; Onyango, H.; Asiimwe, B.; Holden, M.T.G. Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya, Isolated from 2017–2018. Antibiotics 2021, 10, 1547. https://doi.org/10.3390/antibiotics10121547
Decano AG, Pettigrew K, Sabiiti W, Sloan DJ, Neema S, Bazira J, Kiiru J, Onyango H, Asiimwe B, Holden MTG. Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya, Isolated from 2017–2018. Antibiotics. 2021; 10(12):1547. https://doi.org/10.3390/antibiotics10121547
Chicago/Turabian StyleDecano, Arun Gonzales, Kerry Pettigrew, Wilber Sabiiti, Derek J. Sloan, Stella Neema, Joel Bazira, John Kiiru, Hellen Onyango, Benon Asiimwe, and Matthew T. G. Holden. 2021. "Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya, Isolated from 2017–2018" Antibiotics 10, no. 12: 1547. https://doi.org/10.3390/antibiotics10121547
APA StyleDecano, A. G., Pettigrew, K., Sabiiti, W., Sloan, D. J., Neema, S., Bazira, J., Kiiru, J., Onyango, H., Asiimwe, B., & Holden, M. T. G. (2021). Pan-Resistome Characterization of Uropathogenic Escherichia coli and Klebsiella pneumoniae Strains Circulating in Uganda and Kenya, Isolated from 2017–2018. Antibiotics, 10(12), 1547. https://doi.org/10.3390/antibiotics10121547





