Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance
Abstract
1. Introduction
2. Results
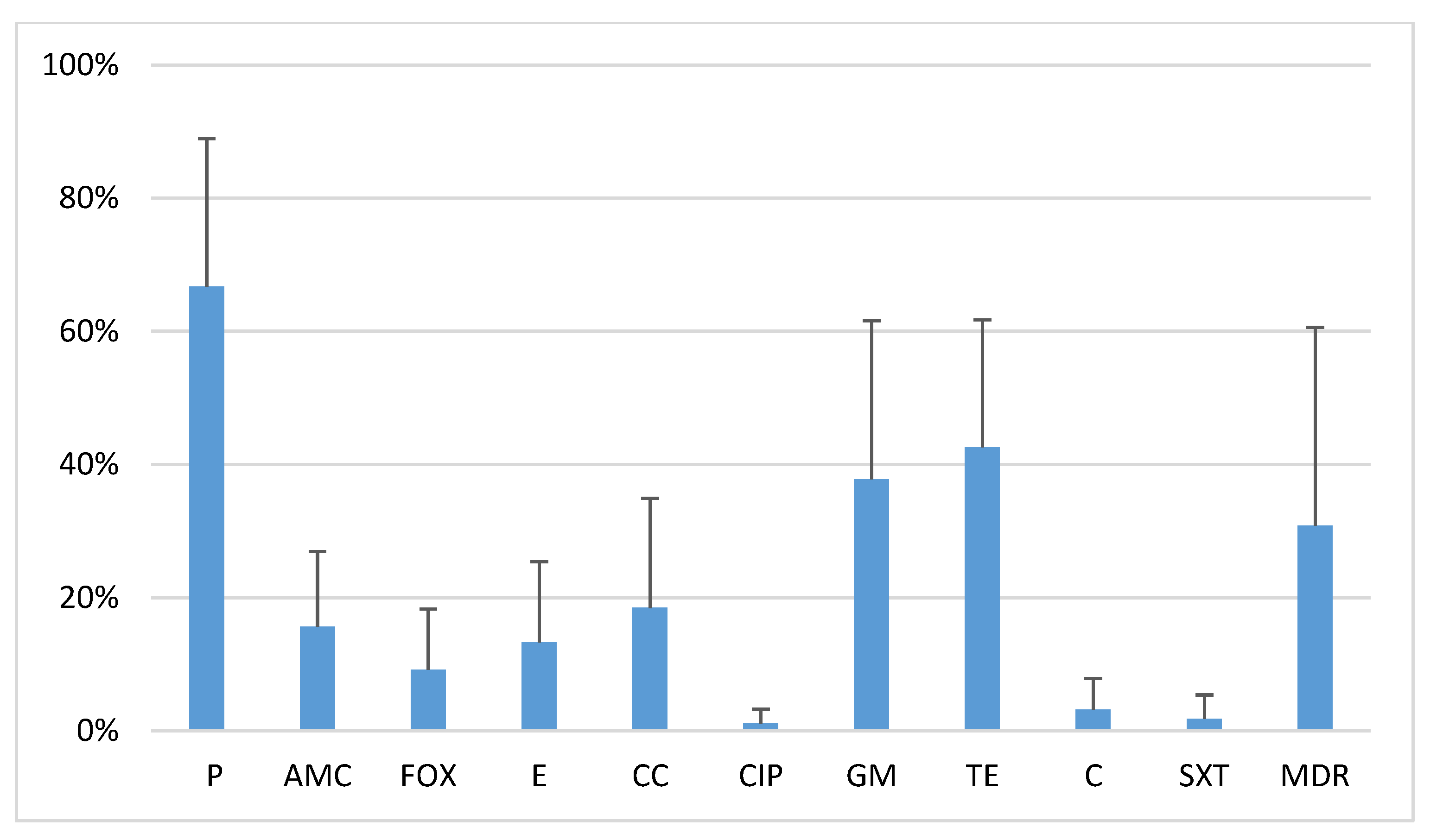
2.1. Antibiotic Resistance of Oral S. Aureus Strains
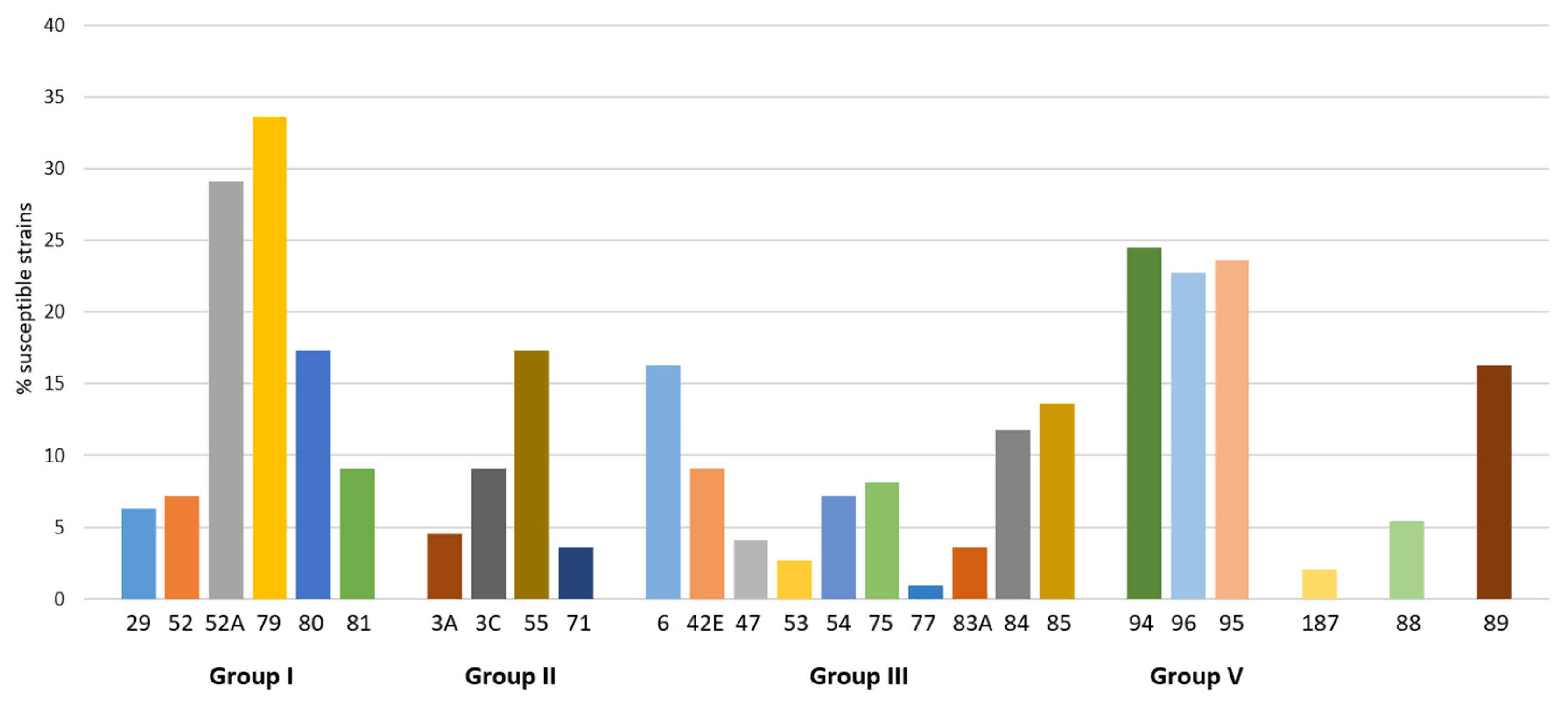
2.2. Activity of Phages from Lytic Groups
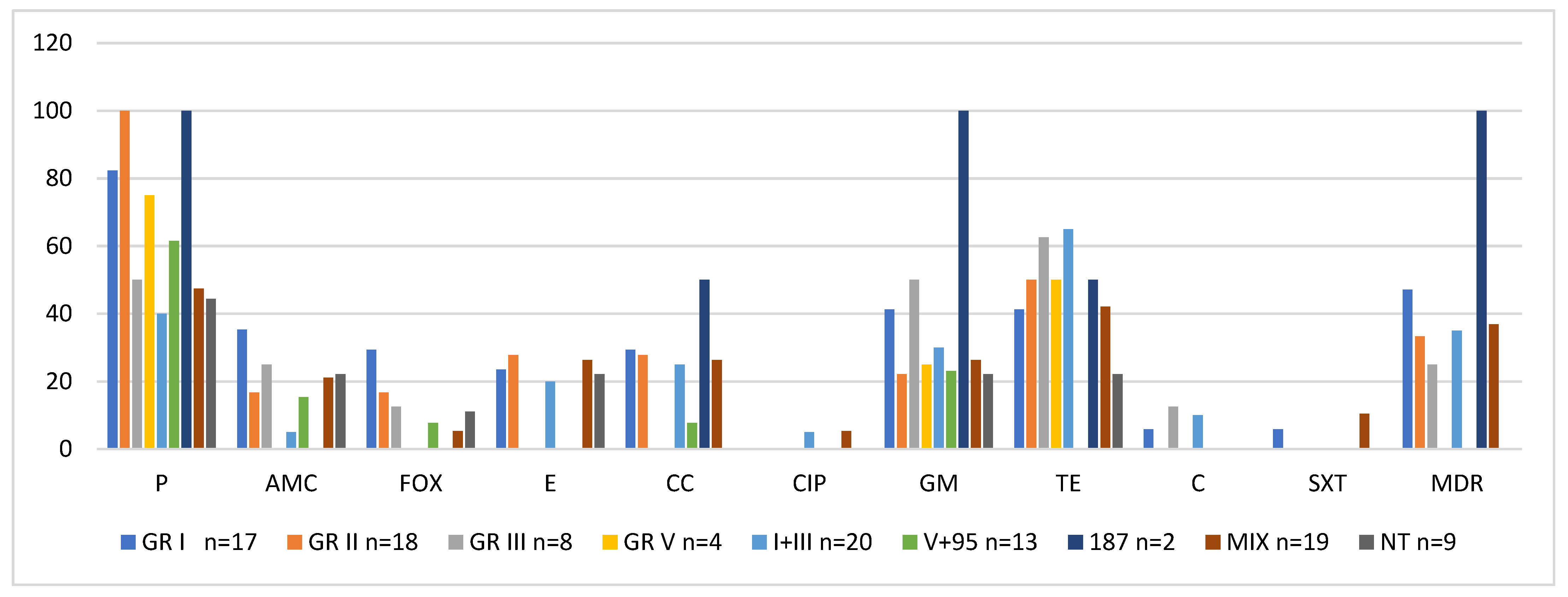
2.3. Activity of Phage Lytic Groups Versus Antibiotic Resistance
2.4. Carriage of Toxin Genes
2.5. Statistical Analysis
3. Discussion
4. Materials and Methods
4.1. Isolation of Oral S. Aureus Strains
4.2. Lytic Activity of Bacteriophages of Basic International Set
4.3. Antibiotic Resistance Testing
4.4. Methicillin-Resistance and Staphylococcal Cassette Chromosome Mec (SCCmec) Detection
4.5. Detection of Major Staphylococcal Toxin Genes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boswihi, S.S.; Udo, E.E. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pract. 2018, 8, 18–24. [Google Scholar] [CrossRef]
- Peng, H.; Liu, D.; Ma, Y.; Gao, W. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012–2017. Sci. Rep. 2018, 8, 17916. [Google Scholar] [CrossRef]
- Udo, E.E. Community-acquired methicillin-resistant Staphylococcus aureus: The new face of an old foe? Med. Princ. Pract. 2013, 22, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Kwapisz, E.; Garbacz, K.; Kosecka-Strojek, M.; Schubert, J.; Bania, J.; Międzobrodzki, J. Presence of egc-positive major clones ST 45, 30 and 22 among methicillin-resistant and methicillin-susceptible oral Staphylococcus aureus strains. Sci. Rep. 2020, 10, 18889. [Google Scholar] [CrossRef] [PubMed]
- Garvey, M. Bacteriophages and the one health approach to combat multidrug resistance: Is this the way? Antibiotics 2020, 9, 414. [Google Scholar] [CrossRef] [PubMed]
- Zalewska-Piątek, B.; Piątek, R. Bacteriophages as potential tools for use in antimicrobial therapy and vaccine development. Pharmaceuticals 2021, 14, 331. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Advances in bacteriophage therapy against relevant multidrug-resistant pathogens. Antibiotics 2021, 10, 672. [Google Scholar] [CrossRef] [PubMed]
- Racenis, K.; Kroica, J.; Rezevska, D.; Avotins, L.; Skuditis, E.; Popova, A.; Puide, I.; Kuzema, V.; Petersons, A.S. Aureus colonization, biofilm production, and phage susceptibility in peritoneal dialysis patients. Antibiotics 2020, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Simon, K.; Pier, W.; Krüttgen, A.; Horz, H.-P. Synergy between Phage Sb-1 and oxacillin against methicillin-resistant Staphylococcus aureus. Antibiotics 2021, 10, 849. [Google Scholar] [CrossRef]
- Tian, F.; Li, J.; Nazir, A.; Tong, Y. Bacteriophage-a promising alternative measure for bacterial biofilm control. Infect Drug Resist. 2021, 14, 205–217. [Google Scholar] [CrossRef]
- Alvarez, A.; Fernandez, L.; Gutierrez, D.; Iglesias, B.; Rodriguez, A.; Garcia, P. Methicillin-resistant Staphylococcus aureus in hospitals: Latest trends and treatments based on bacteriophages. J. Clin. Microbiol. 2019, 57, e01006-19. [Google Scholar] [CrossRef]
- Piechowicz, L.; Garbacz, K. Poultry-Like pA+ Biotype of Staphylococcus aureus CC346/084 clone in human population. Curr. Microbiol. 2016, 73, 124–131. [Google Scholar] [CrossRef]
- Garbacz, K.; Piechowicz, L.; Galiński, J. Presence of enterotoxin C and toxic shock syndrome toxin--1 (TSST-1) genes in population of Staphylococcus aureus phage type 187. Med. Doświadczalna 2006, 58, 191–198. [Google Scholar]
- Garbacz, K.; Piechowicz, L.; Mroczkowska, A. Distribution of toxin genes among different spa types and phage types of animal Staphylococcus aureus. Arch. Microbiol. 2015, 197, 935–940. [Google Scholar] [CrossRef][Green Version]
- Garbacz, K.; Piechowicz, L.; Haras, K.; Wiśniewska, K. Clone of Staphylococcus aureus phage type 187 isolated from people. Med. Doświadczalna 2007, 59, 195–200. [Google Scholar]
- Garbacz, K.; Piechowicz, L. Phage type 187 as a separate subunit MboI restriction site within the Staphylococcus aureus species. Curr. Microbiol. 2013, 66, 578–581. [Google Scholar] [CrossRef]
- Meinen, A.; Reuss, A.; Willrich, N.; Feig, M.; Noll, I.; Eckmanns, T.; Al-Nawas, B.; Markwart, R. Antimicrobial resistance and the spectrum of pathogens in dental and oral-maxillofacial infections in hospitals and dental practices in Germany. Front. Microbiol. 2021, 12, 676108. [Google Scholar] [CrossRef]
- Garbacz, K.; Jarzembowski, T.; Kwapisz, E.; Daca, A.; Witkowski, J. Do the oral Staphylococcus aureus strains from denture wearers have a greater pathogenicity potential? J. Oral. Microbiol. 2018, 11, 1536193. [Google Scholar] [CrossRef]
- Garbacz, K.; Wierzbowska, M.; Kwapisz, E.; Kosecka-Strojek, M.; Bronk, M.; Saki, M.; Międzobrodzki, J. Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J. Oral Microbiol. 2021, 13, 1983322. [Google Scholar] [CrossRef]
- Garbacz, K.; Kwapisz, E.; Wierzbowska, M. Denture stomatitis associated with small-colony variants of Staphylococcus aureus: A case report. BMC Oral Health 2019, 19, 219. [Google Scholar] [CrossRef]
- Marples, R.R.; Rosdahl, V.T. International quality control of phage typing of Staphylococcus aureus. International Union of Microbial Societies Subcommittee. J. Med. Microbiol. 1997, 46, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Vazhavandal, G.; Uma, A. Prevalence of Staphylococcus aureus phage types and their correlation to antimicrobial resistance in a tertiaty care hospital. Int. J. Pharm. Sci. Rev. 2017, 42, 201–204. [Google Scholar]
- Al-Khulaifi Manal, M.; Amin Aref Nagwa, M.; Al Salamah, A.A. Phage typing, PCR amplification for mecA gene, and antibiotic resistance patterns as epidemiologic markers in nosocomial outbreaks of methicillin resistant Staphylococcus aureus. Saudi J. Biol. Sci. 2009, 16, 37–49. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mehndiratta, P.L.; Gur, R.; Saini, S.; Bhalla, P. Staphylococcus aureus phage types and their correlation to antibiotic resistance. Indian J. Pathol. Microbiol. 2010, 53, 738–741. [Google Scholar] [CrossRef]
- Wiśniewska, K.; Szewczyk, A.; Piechowicz, L.; Bronk, M.; Samet, A.; Świeć, K. The use of spa and phage typing for characterization of clinical isolates of methicillin-resistant Staphylococcus aureus in the University Clinical Center in Gdańsk, Poland. Folia Microbiol. 2012, 57, 243–249. [Google Scholar] [CrossRef][Green Version]
- Kareiviene, V.; Pavilonis, A.; Sinkute, G.; Liegiūte, S.; Gailiene, G. Staphylococcus aureus resistance to antibiotics and spread of phage types. Medicina 2006, 42, 332–339. [Google Scholar]
- Abarna, V.; Jayavarthinni, M.; Sindhanai, V.; Bhaskaran, K.; Sethumadhavan, K. Bacteriophage typing of methicillin resistant Staphylococcus aureus and changing trend in their antibiotic profile. Ann. Int. Med. Den. Res. 2017, 3, MB01–MB05. [Google Scholar]
- DeLeo, F.R.; Kennedy, A.D.; Chen, L.; Bubeck Wardenburg, J.; Kobayashi, S.D.; Mathema, B.; Braughton, K.R.; Whitney, A.R.; Villaruz, A.E.; Martens, C.A.; et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2011, 108, 18091–18096. [Google Scholar] [CrossRef] [PubMed]
- Piechowicz, L.; Garbacz, K.; Budzyńska, A.; Dąbrowska-Szponar, M. Outbreak of bullous impetigo caused by Staphylococcus aureus strains of phage type 3C/71 in a maternity ward linked to nasal carriage of a healthcare worker. Eur. J. Dermatol. 2012, 22, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Vintov, J.; Aarestrup, F.M.; Zinn, C.E.; Olsen, J.E. Association between phage types and antimicrobial resistance among bovine Staphylococcus aureus from 10 countries. Vet. Microbiol. 2003, 95, 133–147. [Google Scholar] [CrossRef]
- McCarthy, A.J.; Witney, A.A.; Lindsay, J.A. Staphylococcus aureus temperate bacteriophage: Carriage and horizontal gene transfer is lineage associated. Front. Cell Infect Microbiol. 2012, 2, 6. [Google Scholar] [CrossRef]
- Xia, G.; Wolz, C. Phages of Staphylococcus aureus and their impact on host evolution. Infect. Genet. Evol. 2014, 21, 593–601. [Google Scholar] [CrossRef]
- Whittard, E.; Redfern, J.; Xia, G.; Millard, A.; Ragupathy, R.; Malic, S.; Enright, M.C. Phenotypic and genotypic characterization of novel polyvalent bacteriophages with potent in vitro activity against an international collection of genetically diverse Staphylococcus aureus. Front. Cell Infect Microbiol. 2021, 11, 698909. [Google Scholar] [CrossRef]
- Ladhani, S.; Evans, R.W. Staphylococcal scalded skin syndrome. Arch. Dis. Child. 1998, 78, 85–88. [Google Scholar] [CrossRef]
- Lina, G.; Piemont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef]
- Blair, J.E.; Williams, R.E. Phage typing of staphylococci. Bull World Health Organ. 1961, 24, 771–784. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Available online: http://www.eucast.org/ (accessed on 30 September 2021).
- Khairalla, A.; Wash, R.; Ashour, H.M. Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care presonnel, patients, and environment. Sci. Rep. 2017, 7, 7390. [Google Scholar] [CrossRef]
- Stegger, M.; Andersen, P.S.; Kearns, A.; Pichon, B.; Holmes, M.A.; Edwards, G.; Laurent, F.; Teale, C.; Skov, R.; Larsen, A.R. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 2012, 18, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef]
- Milheiriço, C.; Oliveira, D.C.; de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007, 51, 3374–3377. [Google Scholar] [CrossRef]
- Becker, K.; Roth, R.; Peters, G. Rapid and specific detection of toxigenic staphylococcus aureus: Use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcoal enterotoxin genes, exfoliatine toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 1998, 36, 2548–2553. [Google Scholar] [CrossRef]
- Bania, J.; Dabrowska, A.; Bystron, J.; Korzekwa, K.; Chrzanowska, J.; Molenda, J. Distribution of newly described enterotoxin-like genes in Staphylococcus aureus from food. Int. J. Food Microbiol. 2006, 108, 36–41. [Google Scholar] [CrossRef]
| Phage Group | Phage Susceptible | Antibiotic Resistance | mecA/mecC Genes | Type SCCmec | Toxin Genes |
|---|---|---|---|---|---|
| I | 81 ++ | P, AMC, FOX, E, CC | mecA | V | lukS-PV/lukF-PV seb, sek |
| I | 80 ++ | P, AMC, FOX, TE, SXT | mecA | V | lukS-PV/lukF-PV seb, sek |
| I | 52A ++ | P, AMC, FOX, | mecA | IV | sec |
| I | 81 ++ | P, AMC, FOX | mecA | IV | none |
| I | 81 ++ | P, AMC, FOX, TE, GM | mecA | IV | egc |
| II | 3C ++ | P, AMC, FOX, | mecA | IV | none |
| II | 3C ++, 71 ++ | P, AMC, FOX, E, CC, TE | mecA | IV | egc, eta |
| II | 3C ++, 71 ++ | P, AMC, FOX, E, CC, TE | mecA | IV | egc |
| III | 75 + | P, AMC, FOX, TE | mecA | V | tst, egc |
| V and 95 | 95 ++, 96 ++ | P, AMC, FOX, | mecA | IV | egc |
| I, III, V and 95 | 29 ++, 52 ++, 52A ++, 79 ++, 80 ++,, 81 ++, 6 ++, 42E ++, 53 ++, 54 ++, 75 ++, 77 ++, 83A ++, 85 ++, 88 ++, 89 ++, 95 ++, 96 ++ | P, AMC, FOX, GM | mecA | IV | sec |
| NT | NT | P, AMC, FOX | mecA | V | sec |
| Phage Lytic Groups | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I Group II Group III Group V | |||||||||||||||||||||||||||
| Specific phages | |||||||||||||||||||||||||||
| 29 | 52 | 52A | 79 | 80 | 81 | 3A | 3C | 55 | 71 | 6 | 42E | 47 | 53 | 54 | 75 | 77 | 83A | 84 | 85 | 94 | 96 | 95 | 187 | 88 | 89 | ||
| Ant. Number of phage-sensitive MSSA strains Total (%) | |||||||||||||||||||||||||||
| 6 | 7 | 30 | 36 | 17 | 6 | 5 | 7 | 17 | 2 | 17 | 9 | 4 | 2 | 7 | 7 | 0 | 3 | 13 | 14 | 27 | 23 | 24 | 2 | 5 | 17 | 307 (100) | |
| P | 5 | 4 | 10 | 17 | 10 | 3 | 5 | 7 | 17 | 2 | 5 | 4 | 2 | 2 | 4 | 3 | 2 | 3 | 6 | 16 | 12 | 12 | 2 | 3 | 4 | 160 (52.1) | |
| AMC | 1 | 1 | 1 | 4 | 3 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 2 | 4 | 2 | 2 | 1 | 32 (10.4) | |||||||
| FOX | |||||||||||||||||||||||||||
| E | 1 | 1 | 10 | 3 | 3 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 4 | 3 | 4 | 2 | 3 | 1 | 4 | 52 (16.9) | ||||||
| CC | 1 | 2 | 9 | 6 | 1 | 1 | 1 | 2 | 3 | 1 | 2 | 2 | 5 | 2 | 4 | 1 | 1 | 4 | 48 (15.6) | ||||||||
| CIP | 1 | 1 | 1 | 1 | 1 | 1 | 6 (2.0) | ||||||||||||||||||||
| TE | 2 | 4 | 13 | 17 | 11 | 5 | 3 | 3 | 8 | 1 | 9 | 8 | 1 | 5 | 3 | 3 | 7 | 5 | 11 | 4 | 5 | 1 | 2 | 9 | 140 (45.6) | ||
| GM | 1 | 2 | 9 | 8 | 7 | 4 | 1 | 5 | 5 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 9 | 6 | 6 | 2 | 1 | 5 | 84 (27.4) | ||||
| C | 2 | 1 | 2 | 1 | 1 | 1 | 2 | 10 (3.3) | |||||||||||||||||||
| SXT | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 8 (2.6) | |||||||||||||||||||
| Phage lytic groups | |||||||||||||||||||||||||||
| Group I Group II Group III Group V | |||||||||||||||||||||||||||
| Specific phages | |||||||||||||||||||||||||||
| 29 | 52 | 52A | 79 | 80 | 81 | 3A | 3C | 55 | 71 | 6 | 42E | 47 | 53 | 54 | 75 | 77 | 83A | 84 | 85 | 94 | 96 | 95 | 187 | 88 | 89 | ||
| Ant. Number of phage-sensitive MRSA strains Total (%) | |||||||||||||||||||||||||||
| 1 | 1 | 2 | 1 | 2 | 4 | 0 | 3 | 0 | 2 | 1 | 1 | 0 | 1 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | 2 | 2 | 0 | 1 | 1 | 31 (100) | |
| P | 1 | 1 | 2 | 1 | 2 | 4 | 3 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 31 (100) | ||||||
| AMC | 1 | 1 | 2 | 1 | 2 | 4 | 3 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 31 (100) | ||||||
| FOX | 1 | 1 | 2 | 1 | 2 | 4 | 3 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 31 (100) | ||||||
| E | 1 | 2 | 2 | 5 (16.1) | |||||||||||||||||||||||
| CC | 1 | 2 | 2 | 5 (16.1) | |||||||||||||||||||||||
| CIP | |||||||||||||||||||||||||||
| TE | 1 | 1 | 2 | 2 | 1 | 7 (22.3) | |||||||||||||||||||||
| GM | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 19 (61.3) | ||||||||
| C | |||||||||||||||||||||||||||
| SXT | 1 | 1 (3.2) | |||||||||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garbacz, K.; Kwapisz, E.; Piechowicz, L.; Wierzbowska, M. Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance. Antibiotics 2021, 10, 1329. https://doi.org/10.3390/antibiotics10111329
Garbacz K, Kwapisz E, Piechowicz L, Wierzbowska M. Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance. Antibiotics. 2021; 10(11):1329. https://doi.org/10.3390/antibiotics10111329
Chicago/Turabian StyleGarbacz, Katarzyna, Ewa Kwapisz, Lidia Piechowicz, and Maria Wierzbowska. 2021. "Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance" Antibiotics 10, no. 11: 1329. https://doi.org/10.3390/antibiotics10111329
APA StyleGarbacz, K., Kwapisz, E., Piechowicz, L., & Wierzbowska, M. (2021). Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance. Antibiotics, 10(11), 1329. https://doi.org/10.3390/antibiotics10111329






