Unpuzzling Friunavirus-Host Interactions One Piece at a Time: Phage Recognizes Acinetobacter pittii via a New K38 Capsule Depolymerase
Abstract
:1. Introduction
2. Results
2.1. A. pittii Ap45 Has a OXA β-Lactamase and a K38 Capsular Type
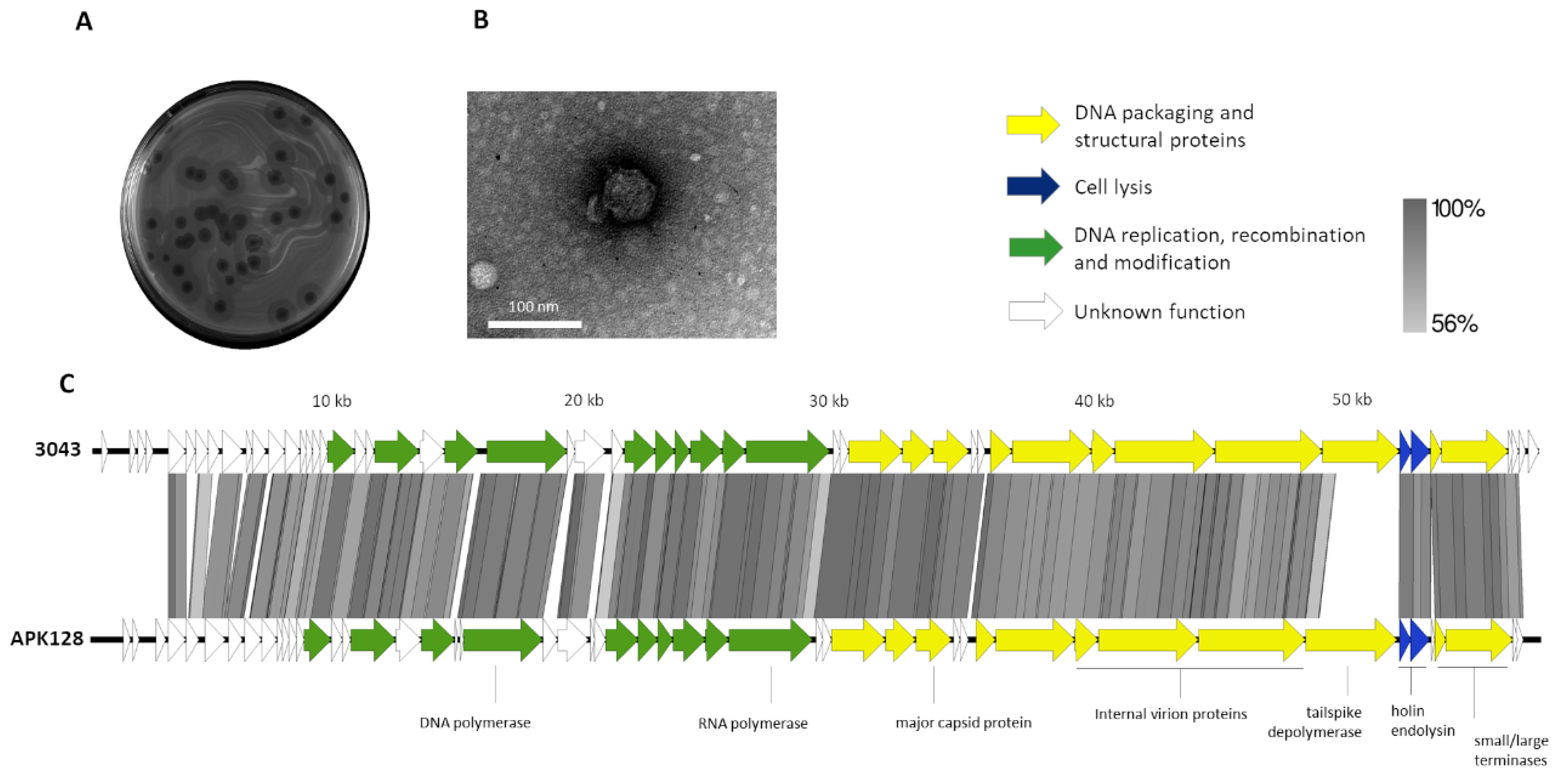
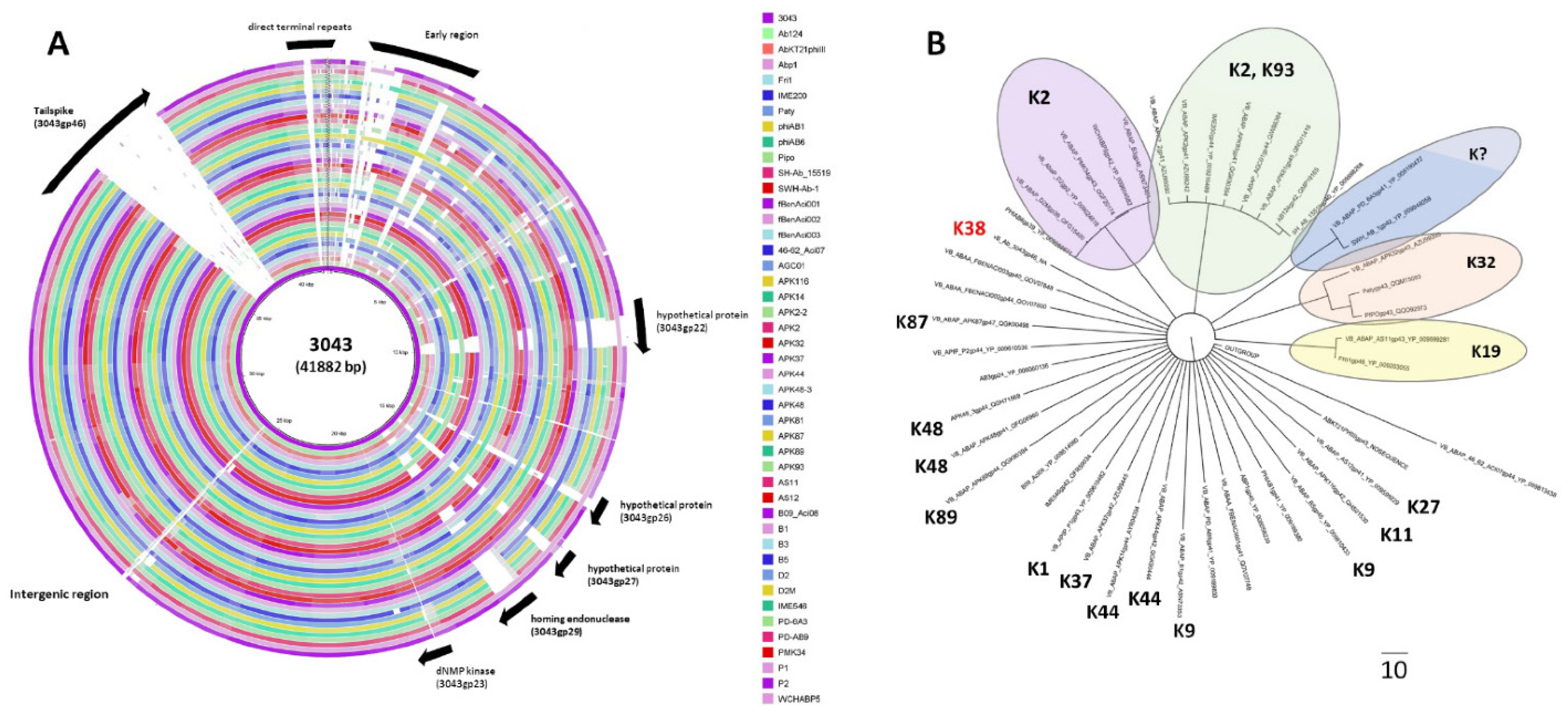
2.2. Phage 3043-K38 Is a New Fri1-like Virus Infecting the A. pittii K38 Capsule Type
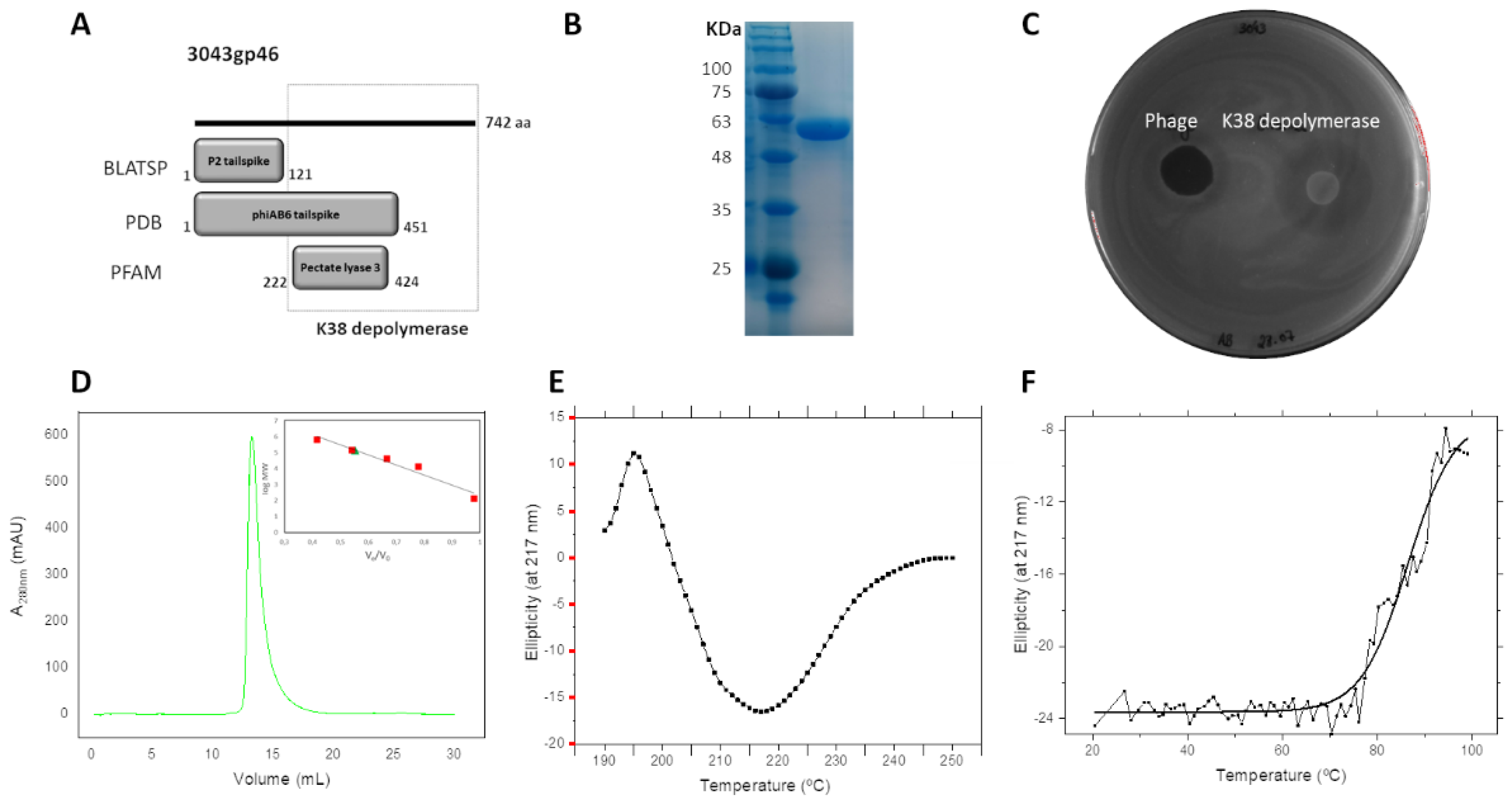
2.3. Tailspike Carries a Novel K38 Depolymerase Activity
2.4. K38 Depolymerase Is a Heat-Resistant Trimeric Protein
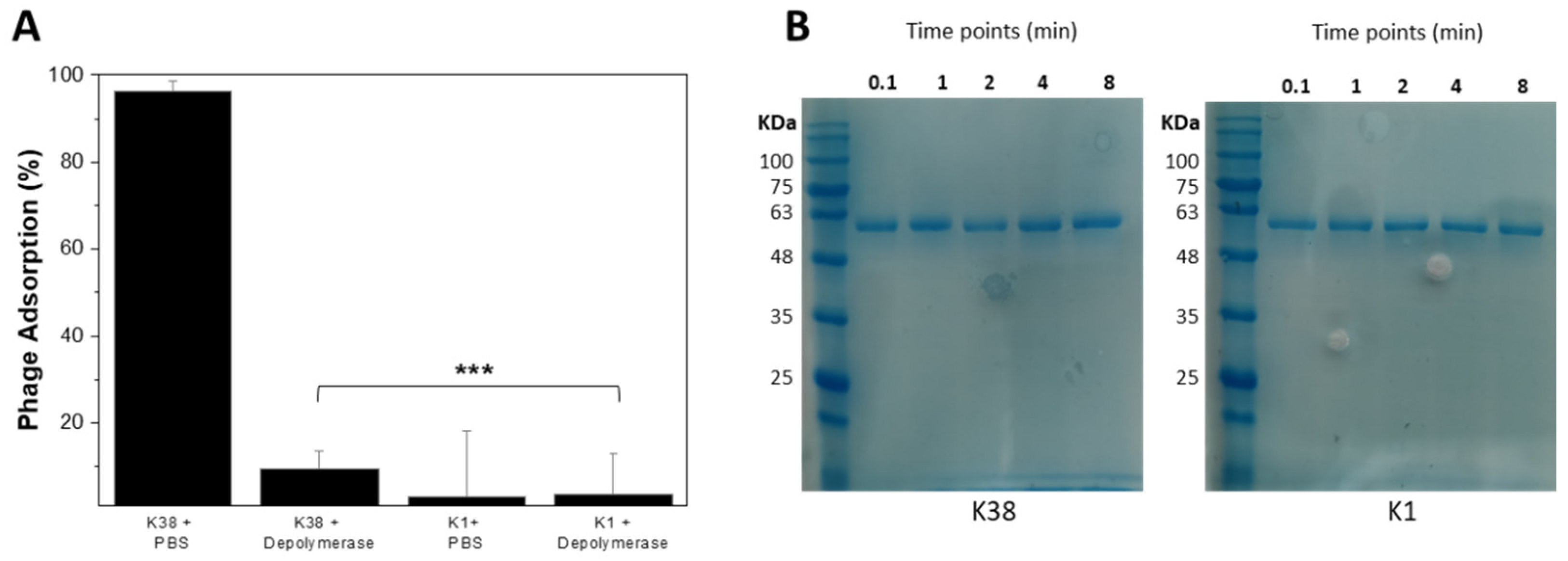
2.5. Recombinant K38 Depolymerase Quickly Removes Capsules, Making Bacterial Cells Immune to Phage
2.6. K38 Depolymerase Does Not Synergise with Antibiotics
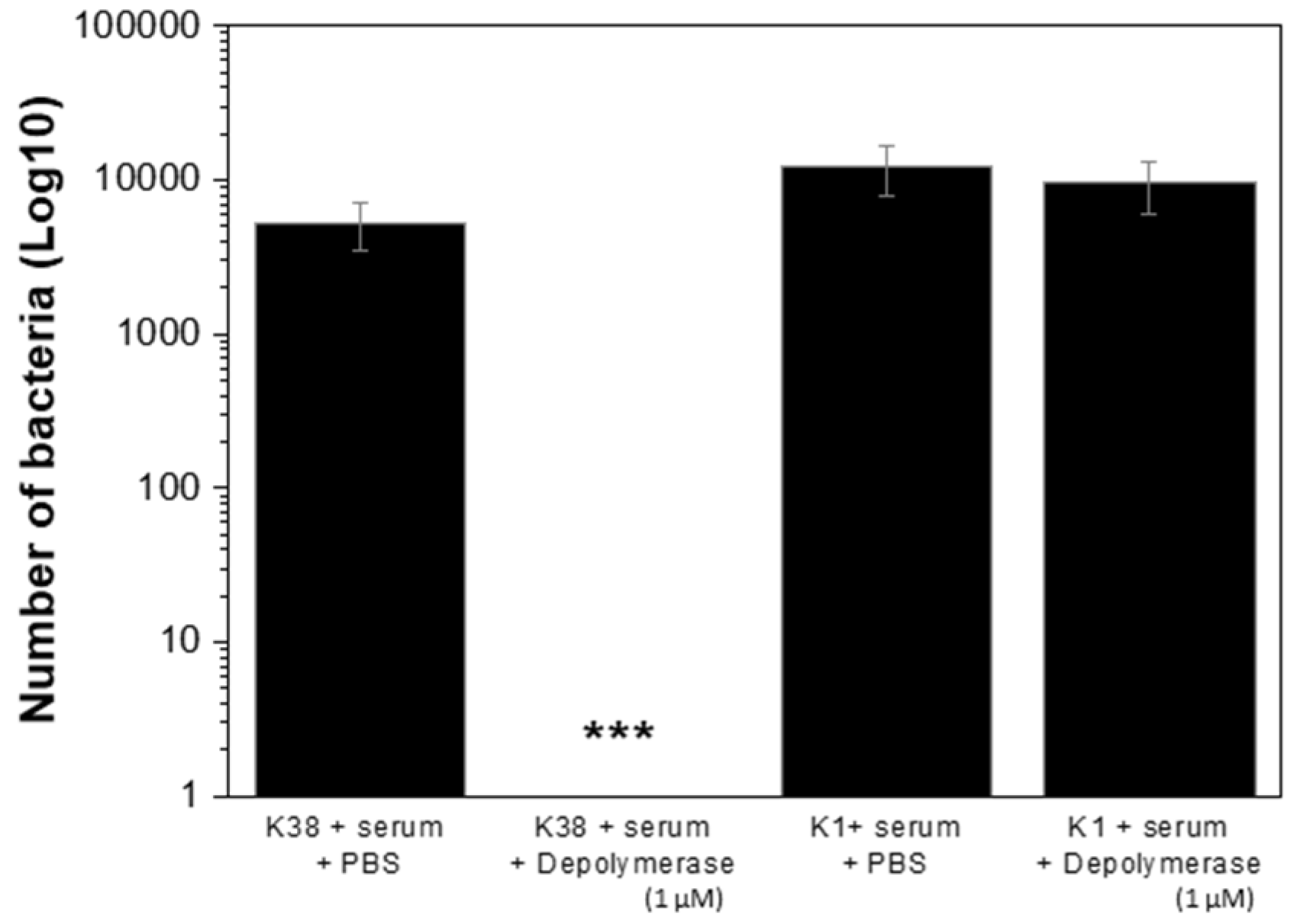
2.7. K38 Depolymerase Increased Susceptibility of K38 Cells to Host Immunity
3. Discussion
4. Material and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Bacterial Sequencing and Annotation
4.3. Phage Isolation and Propagation
4.4. Electron Microscopy
4.5. Phage Genome Sequencing and Annotation
4.6. Tailspike Cloning, Expression and Production
4.7. Phage Host Range and Depolymerase Activity Spectrum
4.8. Circular Dichroism Spectroscopy
4.9. Binding Assay
4.10. Adsorption Assay
4.11. MIC Determination and Antibiofilm Assays
4.12. Human Serum Assay
4.13. Nucleotide Sequence Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vrancianu, C.O.; Gheorghe, I.; Czobor, I.B.; Chifiriuc, M.C. Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter baumannii. Microorganisms 2020, 8, 935. [Google Scholar] [CrossRef]
- Pailhories, H.; Tiry, C.; Eveillard, M.; Kempf, M. Acinetobacter pittii isolated more frequently than Acinetobacter baumannii in blood cultures: The experience of a French hospital. J. Hosp. Infect. 2018, 99, 360–363. [Google Scholar] [CrossRef]
- Russo, T.A.; Luke, N.R.; Beanan, J.M.; Olson, R.; Sauberan, S.L.; MacDonald, U.; Schultz, L.W.; Umland, T.C.; Campagnari, A.A. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 2010, 78, 3993–4000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostowy, R.J.; Holt, K.E. Diversity-Generating Machines: Genetics of Bacterial Sugar-Coating. Trends Microbiol. 2018, 26, 1008–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbatsky, N.P.; Shneider, M.M.; Dmitrenok, A.S.; Popova, A.V.; Shagin, D.A.; Shelenkov, A.A.; Mikhailova, Y.V.; Edelstein, M.V.; Knirel, Y.A. Structure and gene cluster of the K125 capsular polysaccharide from Acinetobacter baumannii MAR13-1452. Int. J. Biol. Macromol. 2018, 117, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Costa, A.R.; Konstantinides, N.; Ferreira, A.; Akturk, E.; Sillankorva, S.; Nemec, A.; Shneider, M.; Dotsch, A.; Azeredo, J. Ability of phages to infect Acinetobacter calcoaceticus-Acinetobacter baumannii complex species through acquisition of different pectate lyase depolymerase domains. Environ. Microbiol. 2021, 23, 3334. [Google Scholar] [CrossRef]
- Oliveira, H.; Mendes, A.; Fraga, A.G.; Ferreira, A.; Pimenta, A.I.; Mil-Homens, D.; Fialho, A.M.; Pedrosa, J.; Azeredo, J. K2 Capsule Depolymerase Is Highly Stable, Is Refractory to Resistance, and Protects Larvae and Mice from Acinetobacter baumannii Sepsis. Appl. Environ. Microb. 2019, 85, e00934-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, A.V.; Lavysh, D.G.; Klimuk, E.I.; Edelstein, M.V.; Bogun, A.G.; Shneider, M.M.; Goncharov, A.E.; Leonov, S.V.; Severinov, K.V. Novel Fri1-like Viruses Infecting Acinetobacter baumannii-vB_AbaP_AS11 and vB_AbaP_AS12-Characterization, Comparative Genomic Analysis, and Host-Recognition Strategy. Viruses 2017, 9, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popova, A.V.; Shneider, M.M.; Arbatsky, N.P.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Dmitrenok, A.S.; Chizhov, A.O.; Mikhailova, Y.V.; Shagin, D.A.; et al. Specific Interaction of Novel Friunavirus Phages Encoding Tailspike Depolymerases with Corresponding Acinetobacter baumannii Capsular Types. J. Virol. 2021, 95, e01714-20. [Google Scholar] [CrossRef] [PubMed]
- Shchurova, A.S.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Skryabin, Y.P.; Mikhaylova, Y.V.; Sokolova, O.S.; Shelenkov, A.A.; Miroshnikov, K.A.; et al. Novel Acinetobacter baumannii Myovirus TaPaz Encoding Two Tailspike Depolymerases: Characterization and Host-Recognition Strategy. Viruses 2021, 13, 978. [Google Scholar] [CrossRef] [PubMed]
- Rossello-Mora, R.; Amann, R. Past and future species definitions for Bacteria and Archaea. Syst. Appl. Microbiol. 2015, 38, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Latka, A.; Leiman, P.G.; Drulis-Kawa, Z.; Briers, Y. Modeling the Architecture of Depolymerase-Containing Receptor Binding Proteins in Klebsiella Phages. Front. Microbiol. 2019, 10, 2649. [Google Scholar] [CrossRef] [PubMed]
- Leiman, P.G.; Molineux, I.J. Evolution of a new enzyme activity from the same motif fold. Mol. Microbiol. 2008, 69, 287–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarzer, D.; Stummeyer, K.; Gerardy-Schahn, R.; Muhlenhoff, M. Characterization of a novel intramolecular chaperone domain conserved in endosialidases and other bacteriophage tail spike and fiber proteins. J. Biol. Chem. 2007, 282, 2821–2831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plattner, M.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Nazarov, S.; Prokhorov, N.S.; Taylor, N.M.I.; Buth, S.A.; Gambino, M.; et al. Structure and Function of the Branched Receptor-Binding Complex of Bacteriophage CBA120. J. Mol. Biol. 2019, 431, 3718–3739. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Huang, J.; Yan, G.; Lei, L.; Wang, S.; Yu, L.; Zhou, L.; Gao, A.; Feng, X.; Han, W.; et al. Identification and Characterization of Dpo42, a Novel Depolymerase Derived from the Escherichia coli Phage vB_EcoM_ECOO78. Front. Microbiol. 2017, 8, 1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majkowska-Skrobek, G.; Latka, A.; Berisio, R.; Maciejewska, B.; Squeglia, F.; Romano, M.; Lavigne, R.; Struve, C.; Drulis-Kawa, Z. Capsule-Targeting Depolymerase, Derived from Klebsiella KP36 Phage, as a Tool for the Development of Anti-Virulent Strategy. Viruses 2016, 8, 324. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Costa, A.R.; Ferreira, A.; Konstantinides, N.; Santos, S.B.; Boon, M.; Noben, J.P.; Lavigne, R.; Azeredo, J. Functional Analysis and Antivirulence Properties of a New Depolymerase from a Myovirus That Infects Acinetobacter baumannii Capsule K45. J. Virol. 2019, 93, e01163-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Mi, Z.; Mi, L.; Huang, Y.; Li, P.; Liu, H.; Yuan, X.; Niu, W.; Jiang, N.; Bai, C.; et al. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 2019, 7, e6173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bansal, S.; Harjai, K.; Chhibber, S. Aeromonas punctata derived depolymerase improves susceptibility of Klebsiella pneumoniae biofilm to gentamicin. BMC Microbiol. 2015, 15, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020, 6, 000339. [Google Scholar] [CrossRef] [PubMed]
- Leshkasheli, L.; Kutateladze, M.; Balarjishvili, N.; Bolkvadze, D.; Save, J.; Oechslin, F.; Que, Y.A.; Resch, G. Efficacy of newly isolated and highly potent bacteriophages in a mouse model of extensively drug-resistant Acinetobacter baumannii bacteraemia. J. Glob. Antimicrob. Resist. 2019, 19, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Green, M.R. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor: New York, NY, USA, 2001. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domingues, R.; Barbosa, A.; Santos, S.B.; Pires, D.P.; Save, J.; Resch, G.; Azeredo, J.; Oliveira, H. Unpuzzling Friunavirus-Host Interactions One Piece at a Time: Phage Recognizes Acinetobacter pittii via a New K38 Capsule Depolymerase. Antibiotics 2021, 10, 1304. https://doi.org/10.3390/antibiotics10111304
Domingues R, Barbosa A, Santos SB, Pires DP, Save J, Resch G, Azeredo J, Oliveira H. Unpuzzling Friunavirus-Host Interactions One Piece at a Time: Phage Recognizes Acinetobacter pittii via a New K38 Capsule Depolymerase. Antibiotics. 2021; 10(11):1304. https://doi.org/10.3390/antibiotics10111304
Chicago/Turabian StyleDomingues, Rita, Ana Barbosa, Sílvio B. Santos, Diana Priscila Pires, Jonathan Save, Grégory Resch, Joana Azeredo, and Hugo Oliveira. 2021. "Unpuzzling Friunavirus-Host Interactions One Piece at a Time: Phage Recognizes Acinetobacter pittii via a New K38 Capsule Depolymerase" Antibiotics 10, no. 11: 1304. https://doi.org/10.3390/antibiotics10111304
APA StyleDomingues, R., Barbosa, A., Santos, S. B., Pires, D. P., Save, J., Resch, G., Azeredo, J., & Oliveira, H. (2021). Unpuzzling Friunavirus-Host Interactions One Piece at a Time: Phage Recognizes Acinetobacter pittii via a New K38 Capsule Depolymerase. Antibiotics, 10(11), 1304. https://doi.org/10.3390/antibiotics10111304







