Different Inhibitory Effects of Erythromycin and Chlortetracycline on Early Growth of Brassica campestris Seedlings
Abstract
:1. Introduction
2. Results
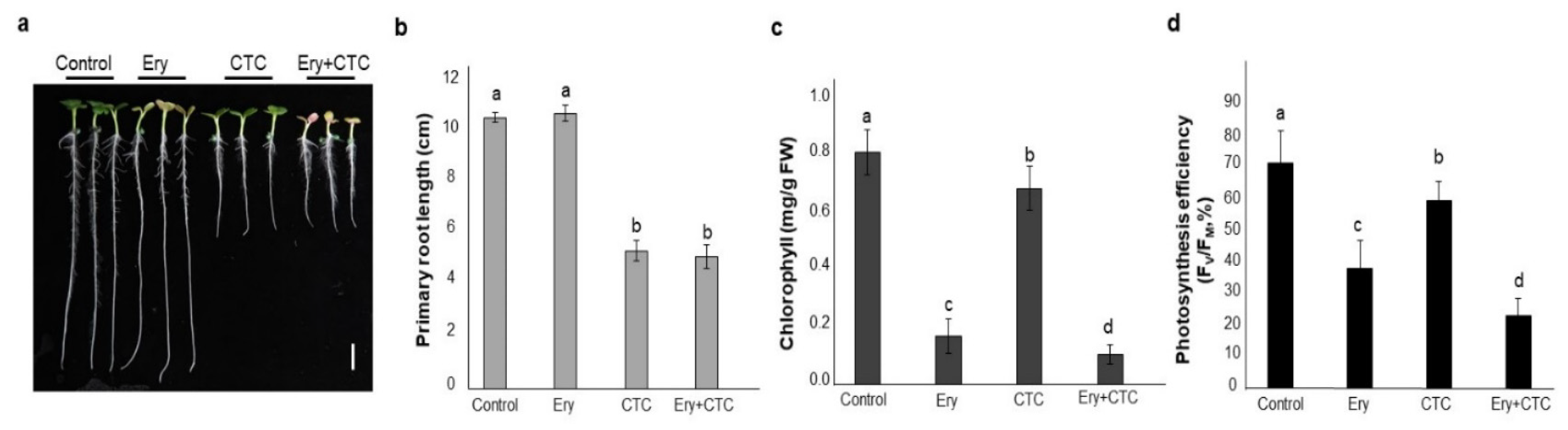
2.1. Erythromycin (Ery) and Chlortetracycline (CTC) Individually Affect Physiological Phenotypes
2.2. Ery and CTC Have Different Effects on Expression of Genes in a Tetrapyrrole Biosynthetic Pathway
2.3. Ery and CTC Affect Expression of the Photosynthetic Apparatus
2.4. Ery and CTC Inhibit Chloroplast Formation at the Ultrastructural Level
3. Discussion
4. Materials and Methods
4.1. Plant Growth for Chemical Treatments and the Measurement of Physiological Parameters
4.2. Total Protein Extraction and Western Blot Analysis
4.3. Total RNA Extraction and qRT-PCR
4.4. Transmission Electron Microscopy (TEM)
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Palme, K.J.; Jansen, M.A.K. Stress-induced morphogenic responses: Growing out of trouble? Trends Plant Sci. 2007, 12, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [Green Version]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, G.; Ding, J.; Zhang, Z.; Wang, Y. Tissue distribution, bioconcentration, metabolism, and effects of erythromycin in crucian carp (Carassius auratus). Sci. Total Environ. 2014, 490, 914–920. [Google Scholar] [CrossRef]
- Ezzariai, A.; Hafidi, M.; Khadra, A.; Aemig, Q.; El Fels, L.; Barret, M.; Merlina, G.; Patureau, D.; Pinelli, E. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. J. Hazard. Mater. 2018, 359, 465–481. [Google Scholar] [CrossRef]
- Kumar, K.; Gupta, S.C.; Baidoo, S.K.; Chander, Y.; Rosen, C.J. Antibiotic Uptake by Plants from Soil Fertilized with Animal Manure. J. Environ. Qual. 2005, 34, 2082–2085. [Google Scholar] [CrossRef] [Green Version]
- Kuppusamy, S.; Kakarla, D.; Venkateswarlu, K.; Megharaj, M.; Yoon, Y.E.; Lee, Y.B. Veterinary antibiotics (VAs) contamination as a global agro-ecological issue: A critical view. Agric. Ecosyst. Environ. 2018, 257, 47–59. [Google Scholar] [CrossRef]
- Bártíková, H.; Podlipná, R.; Skálová, L. Veterinary drugs in the environment and their toxicity to plants. Chemosphere 2016, 144, 2290–2301. [Google Scholar] [CrossRef]
- Pan, M.; Wong, C.K.C.; Chu, L.M. Distribution of antibiotics in wastewater-irrigated soils and their accumulation in vegetable crops in the Pearl River Delta, Southern China. J. Agric. Food Chem. 2014, 62, 11062–11069. [Google Scholar] [CrossRef]
- Bhatt, E.; Gauba, P. Impact of antibiotics on plants. Int. J. Pharm. Sci. Rev. Res. 2018, 52, 49–53. [Google Scholar]
- Zhang, C.; Xue, J.; Cheng, D.; Feng, Y.; Liu, Y.; Aly, H.M.; Li, Z. Uptake, translocation and distribution of three veterinary antibiotics in Zea mays L. Environ. Pollut. 2019, 250, 47–57. [Google Scholar] [CrossRef]
- Thompson, L.A.; Darwish, W.S. Environmental Chemical Contaminants in Food: Review of a Global Problem. J. Toxicol. 2019, 2019, 2345283. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.; Chu, L.M. Phytotoxicity of veterinary antibiotics to seed germination and root elongation of crops. Ecotoxicol. Environ. Saf. 2016, 126, 228–237. [Google Scholar] [CrossRef]
- Minden, V.; Deloy, A.; Volkert, A.M.; Leonhardt, S.D.; Pufal, G. Antibiotics impact plant traits, even at small concentrations. AoB Plants 2017, 9, plx010. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Chen, Q.; Sun, R.; Zhou, Q.; Liu, J. Eco-toxic effects of sulfadiazine sodium, sulfamonomethoxine sodium and enrofloxacin on wheat, Chinese cabbage and tomato. Ecotoxicology 2009, 18, 878–885. [Google Scholar] [CrossRef]
- Tripathy, B.C.; Pattanayak, G.K. Chlorophyll Biosynthesis in Higher Plants. In Photosynthesis; Springer: Dordrecht, The Netherlands, 2012; ISBN 9789400715790. [Google Scholar]
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The role of chloroplast gene expression in plant responses to environmental stress. Int. J. Mol. Sci. 2020, 21, 6082. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Goldschmidt-Clermont, M. Participation of nuclear genes in chloroplast gene expression. Biochimie 2000, 82, 559–572. [Google Scholar] [CrossRef] [Green Version]
- Sandelius, A.S.; Aronsson, H. The Chloroplast: Interactions with the Environment; Springer: Berlin/Heidelberg, Germany, 2009; Volume 13. [Google Scholar] [CrossRef]
- Leister, D.; Wang, X.; Haberer, G.; Mayer, K.F.X.; Kleine, T. Intracompartmental and intercompartmental transcriptional networks coordinate the expression of genes for organellar functions. Plant Physiol. 2011, 157, 386–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the Languages of the Chloroplast: Retrograde Signaling and beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53. [Google Scholar] [CrossRef]
- Paila, Y.D.; Richardson, L.G.L.; Schnell, D.J. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 2015, 427, 1038–1060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leister, D.; Wang, L.; Kleine, T. Organellar gene expression and acclimation of plants to environmental stress. Front. Plant Sci. 2017, 8, 387. [Google Scholar] [CrossRef] [Green Version]
- Mareš, J.; Strunecký, O.; Bučinská, L.; Wiedermannová, J. Evolutionary patterns of thylakoid architecture in Cyanobacteria. Front. Microbiol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.E.; Cho, H.M.; Bae, D.W.; Lee, S.J.; Choe, H.; Kim, M.C.; Cheong, M.S.; Lee, Y.B. Erythromycin treatment of Brassica campestris seedlings impacts the photosynthetic and protein synthesis pathways. Life 2020, 10, 311. [Google Scholar] [CrossRef]
- Cheong, M.S.; Yoon, Y.-E.; Kim, J.W.; Hong, Y.K.; Kim, S.C.; Lee, Y.B. Chlortetracycline inhibits seed germination and seedling growth in Brassica campestris by disrupting H2O2 signaling. Appl. Biol. Chem. 2020, 63, 1–8. [Google Scholar] [CrossRef]
- Wu, D.; Wright, D.A.; Wetzel, C.; Voytas, D.F.; Rodermel, S. The IMMUTANS variegation locus of arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 1999, 11, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [Green Version]
- Sakuraba, Y.; Park, S.Y.; Paek, N.C. The divergent roles of STAYGREEN (SGR) Homologs in chlorophyll degradation. Mol. Cells 2015, 38, 390–395. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, V.; Ingenfeld, A.; Apel, K. Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 2006, 60, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y. iTRAQ-based Proteomic Analysis of a Chlorophyll- De cient Mutant Caused by Single Base Change in RPS4 of Chinese Cabbage (Brassica Campestris L. ssp. Pekinensis). Res. Sq. 2020. preprint. [Google Scholar] [CrossRef]
- Lee, J. Leaf Proteome Analysis in Brassica rapa L. (Inbred line ‘Chiifu’) using Shotgun Proteome Approach. Plant Breed. Biotechnol. 2015, 3, 389–395. [Google Scholar] [CrossRef]
- Eberhard, S.; Finazzi, G.; Wollman, F.A. The dynamics of photosynthesis. Annu. Rev. Genet. 2008, 42, 463–515. [Google Scholar] [CrossRef] [Green Version]
- Boxall, A.B.A.; Fogg, L.A.; Blackwell, P.A.; Kay, P.; Pemberton, E.J.; Croxford, A. Veterinary medicines in the environment. Rev. Environ. Contam. Toxicol. 2004, 1–91. [Google Scholar] [CrossRef]
- Boxall, A.B.A.; Johnson, P.; Smith, E.J.; Sinclair, C.J.; Stutt, E.; Levy, L.S. Uptake of veterinary medicines from soils into plants. J. Agric. Food Chem. 2006, 54, 2288–2297. [Google Scholar] [CrossRef]
- Myouga, F.; Hosoda, C.; Umezawa, T.; Iizumi, H.; Kuromori, T.; Motohashi, R.; Shono, Y.; Nagata, N.; Ikeuchi, M.; Shinozaki, K. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in arabidopsis. Plant Cell 2008, 20, 3148–3162. [Google Scholar] [CrossRef] [Green Version]
- Schoefs, B. The Light-Dependent and Light-Independent Reduction of Protochlorophyllide a to Chlorophyllide a. Photosynthetica 2000, 36, 481–496. [Google Scholar] [CrossRef]
- Gaubier, P.; Wu, H.J.; Laudié, M.; Delseny, M.; Grellet, F. A chlorophyll synthetase gene from Arabidopsis thaliana. MGG Mol. Gen. Genet. 1995, 249, 58–64. [Google Scholar] [CrossRef]
- Woodson, J.D.; Perez-Ruiz, J.M.; Chory, J. Heme synthesis by plastid ferrochelatase i regulates nuclear gene expression in plants. Curr. Biol. 2011, 21, 897–903. [Google Scholar] [CrossRef] [Green Version]
- Roach, T.; Krieger-Liszkay, A. Regulation of Photosynthetic Electron Transport and Photoinhibition. Curr. Protein Pept. Sci. 2014, 15, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.S.; Berkowitz, G.A. Development and Use of Chlorotetracycline Fluorescence as a Measurement Assay of Chloroplast Envelope-Bound Mg2+. Plant Physiol. 1989, 89, 753–761. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Yu, J.W.; Park, J.S.; Li, J.; Yoo, S.C.; Lee, N.Y.; Lee, S.K.; Jeong, S.W.; Hak, S.S.; Koh, H.J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [Green Version]
- Rydzynski, D.; Piotrowicz-Cieslak, A.I.; Grajek, H.; Wasilewski, J. Investigation of chlorophyll degradation by tetracycline. Chemosphere 2019, 229, 409–417. [Google Scholar] [CrossRef]
- Hoober, J.K.; Eggink, L.L.; Chen, M. Chlorophylls, ligands and assembly of light-harvesting complexes in chloroplasts. Photosynth. Res. 2007, 94, 387–400. [Google Scholar] [CrossRef] [Green Version]
- Sadali, N.M.; Sowden, R.G.; Ling, Q.; Jarvis, R.P. Differentiation of chromoplasts and other plastids in plants. Plant Cell Rep. 2019, 38, 803–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wijk, K.J.; Kessler, F. Plastoglobuli: Plastid Microcompartments with Integrated Functions in Metabolism, Plastid Developmental Transitions, and Environmental Adaptation. Annu. Rev. Plant Biol. 2017, 68, 253–289. [Google Scholar] [CrossRef] [PubMed]
- Oosawa, N.; Masuda, T.; Awai, K.; Fusada, N.; Shimada, H.; Ohta, H.; Takamiya, K. ichiro Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett. 2000, 474, 133–136. [Google Scholar] [CrossRef] [Green Version]
- Barnes, S.A.; Nishizawa, N.K.; Quaggio, R.B.; Whitelam, G.C.; Chua, N.H. Far-red light blocks greening of arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 1996, 8, 601–615. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, R.; Kobayashi, K.; Masuda, T. Tetrapyrrole Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottet, S.; Besagni, C.; Kessler, F. The role of plastoglobules in thylakoid lipid remodeling during plant development. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 889–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowler, C.; Van Camp, W.; Van Montagu, M.; Inzé, D.; Asada, K. Superoxide Dismutase in Plants. CRC. Crit. Rev. Plant Sci. 1994, 13, 199–218. [Google Scholar] [CrossRef]
- Kuo, W.Y.; Huang, C.H.; Liu, A.C.; Cheng, C.P.; Li, S.H.; Chang, W.C.; Weiss, C.; Azem, A.; Jinn, T.L. CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol. 2013, 197, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Kroll, D.; Meierhoff, K.; Bechtold, N.; Kinoshita, M.; Westphal, S.; Vothknecht, U.C.; Soll, J.; Westhoff, P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 2001, 98, 4238–4242. [Google Scholar] [CrossRef] [Green Version]
- Babiychuk, E.; Bouvier-Nave, P.; Compagnon, V.; Suzuki, M.; Muranaka, T.; Van Montagu, M.; Kushnir, S.; Schaller, H. Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 3163–3168. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheong, M.S.; Choe, H.; Jeong, M.S.; Yoon, Y.-E.; Jung, H.S.; Lee, Y.B. Different Inhibitory Effects of Erythromycin and Chlortetracycline on Early Growth of Brassica campestris Seedlings. Antibiotics 2021, 10, 1273. https://doi.org/10.3390/antibiotics10101273
Cheong MS, Choe H, Jeong MS, Yoon Y-E, Jung HS, Lee YB. Different Inhibitory Effects of Erythromycin and Chlortetracycline on Early Growth of Brassica campestris Seedlings. Antibiotics. 2021; 10(10):1273. https://doi.org/10.3390/antibiotics10101273
Chicago/Turabian StyleCheong, Mi Sun, Hyeonji Choe, Myeong Seon Jeong, Young-Eun Yoon, Hyun Suk Jung, and Yong Bok Lee. 2021. "Different Inhibitory Effects of Erythromycin and Chlortetracycline on Early Growth of Brassica campestris Seedlings" Antibiotics 10, no. 10: 1273. https://doi.org/10.3390/antibiotics10101273
APA StyleCheong, M. S., Choe, H., Jeong, M. S., Yoon, Y.-E., Jung, H. S., & Lee, Y. B. (2021). Different Inhibitory Effects of Erythromycin and Chlortetracycline on Early Growth of Brassica campestris Seedlings. Antibiotics, 10(10), 1273. https://doi.org/10.3390/antibiotics10101273





