1. Introduction
Protein detection is of great importance for medical research and diagnostic approaches. Until now, only antibody-based protein analysis has been available. Complementary to antibodies, aptamers could become advantageous recognition elements for protein analytics.
Aptamers are synthetically produced, single-stranded oligonucleotides consisting of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) and can be seen as an alternative to antibodies [
1]. These oligonucleotides are capable of binding proteins [
2], peptides [
3], small molecules [
4] or even whole cells [
5] with high affinity in the micro-to-picomolar range [
1]. The secondary and tertiary structures of aptamers, including hairpin loops, pseudo knots, and guanine (G)-quadruplexes, are of great importance for the formation of a three-dimensional (3D) structure and, thus, for target-binding [
6]. The major inter- and intra-molecular interactions of aptamers are based on hydrogen bonding, electrostatic interactions, and hydrophobic interactions [
6]. The 3D structure of an aptamer depends on the temperature, pH value, and ionic strength of the solution as well as on the sequence itself [
6] and can be determined by using nuclear magnetic resonance (NMR) spectroscopy or x-ray crystallography [
7]. Aptamers are selected against a specific target by a process termed the Systematic Evolution of Ligands by EXponential enrichment (SELEX) [
7]. During the SELEX process, chemically synthesized oligonucleotide libraries with randomized sequences are used and high affinity sequences are selected after binding the target [
7]. Furthermore, the specificity of aptamers can be high, since some of these oligonucleotides can even differ between enantiomers [
6]. Because of their high binding affinity and specificity, aptamers are applied in several technologies including sensor platforms [
4], affinity chromatography [
6,
8,
9], and microarrays [
10,
11]. One of the advantages of aptamers in comparison to antibodies is their cost-efficient production by chemical synthesis including their easy and directed modification with fluorophore, linker, quencher, or other functional groups [
11]. Moreover, aptamers can easily be immobilized on surfaces (e.g., microarrays), are stable at high temperatures, and do not denature as fast as proteins [
11]. In addition, immobilized aptamers are stable and can be stored for a long period of time [
11]. Since the target-binding of the aptamer is reversible, regeneration and multiple usage is possible [
12,
13,
14,
15].
The mentioned advantages of aptamers and sophisticated synthesis techniques make ready-to-use aptamer-microarrays a promising alternative to protein-microarrays [
11]. Printing technologies like fluid droplet dispensing allow for the high throughput generation of microarrays and photochemical methods like photochemical patterning or photolithography—which can also be combined with the in-situ synthesis of capture oligonucleotides—can be applied [
16]. The development of aptamer-microarrays requires the careful consideration and experimental investigation of suitable buffers, spacers, immobilization techniques, microarrays surfaces, etc. [
11,
15,
17]. Currently, a few commercial aptamer-microarray products are available for the detection of biomarkers by SomaLogic [
11]. Moreover, despite the current limitations in the application of aptamer microarrays, they represent a suitable platform technology to develop and optimize aptamer-based assays [
11].
Concerning the detection of the binding event, several principles can be used for aptamer microarrays. As in antibody microarrays, simple binding assays can be performed in the forward phase microarray format, which requires the labeling of the analyte, which is not easy to realize, especially when using complex samples. To avoid the labeling of the analyte, aptamer microarrays can also be set up in a sandwich format [
11]. In this sandwich format, one aptamer is immobilized on the microarray, and the bound target is detected by a second, labeled aptamer. These sandwich assays require the availability of two aptamers that are directed against different “aptatopes” of the target. In contrast, other aptamer-based assay principles are aptamer specific and cannot be realized using antibodies [
11]. These assays include the target-induced reassembly of aptamer fragments and the target-induced dissociation of complementary oligonucleotides [
4].
In this work, vascular endothelial growth factor (VEGF) was used as a model analyte. VEGF can take part in the development of different diseases like diabetes mellitus [
18,
19], age-related macular degeneration [
20], rheumatoid arthritis [
21,
22], and neurodegenerative diseases [
23] like Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis. In 2004, the first aptamer approved by the Food and Drug Administration (FDA) was a VEGF-targeting aptamer called pegaptanib [
20,
24]. Since then, different research groups have selected various aptamers against VEGF, such as the aptamers named V7t1 and Del5-1 [
2,
25,
26,
27]. The V7t1 aptamer contains a G-quadruplex structure [
13]. G-quadruplexes comprise G-rich regions which form more than one quadratic shape of the four guanines named G-quartet which are stapled vertically over each other [
28]. Moreover, various G-quadruplex structures are known to target different proteins like VEGF [
13], thrombin [
29], and nucleolin [
30]. Therefore, G-quadruplexes can be considered promising target binding sites in aptamers that can be useful for designing detection strategies based on the target-induced structural changes of the aptamer [
29].
Since most aptamers undergo structural changes upon target-binding, their target-induced conformational reorganization can be utilized for detection strategies, e.g., in aptamer-microarrays [
31] or other biosensing platforms [
21]. The nucleic acid-based sequence of aptamers allows for the design of oligonucleotides which are complementary to the target-binding site of the aptamer and can compete with the target in various assays [
31,
32]. The target-induced dissociation (TID) of complementary oligonucleotides allows for the detection of targets without labeling the analyte [
32]. The aptamer-based TID mechanism has been proven to allow for highly sensitive detection in several applications including lateral flow assays [
33], quantitative polymerase chain reaction (qPCR) [
34,
35], and other biosensing platforms [
19,
21]. The TID mode thereby uses oligonucleotides complementary to the aptamers sequence, which form a duplex with the aptamer via hybridization [
4]. In the presence of the aptamer target molecule, the complementary oligonucleotide dissociates from the aptamer while the aptamer forms a complex with the target [
4]. This dissociation can be optically detected by modifying the aptamer and/or the complementary oligonucleotide with fluorophores or other labels [
4].
Until now, TID mechanisms have been mainly applied to small molecule detection [
31,
34,
35]. Nonetheless, since TID assays do not require the labeling of the analyte (as required in conventional competitive assays) or the availability of two specific ligands binding to different binding sites of the analyte (as required for sandwich assays), the transfer of the TID mechanism to the detection of proteins could be advantageous. Therefore, efforts for TID-based protein detection have been already performed. Freeman et al. developed an optical aptasensor for VEGF detection using the TID method based on fluorescence resonance energy transfer with a detection limit of 1 nM [
19]. The VEGF-binding aptamer was used for the experiment, and the G-quadruplex of the aptamer was exploited for the detection [
19]. Therefore, an oligonucleotide complementary to the G-rich region of the aptamer was used to suppress quadruplex formation in absence of VEGF [
19].
In our research, we focused on the schematic development of a TID assay via thermophoresis and an aptamer-microarray. Using microarrays with immobilized aptamers excludes costly and unstable antibodies for VEGF detection and sustains the reusability of the aptamer. Therefore, we first investigated the affinity and specificity of different VEGF-binding aptamers by thermophoresis. Thereafter, an oligonucleotide complementary to the target-binding site of the chosen aptamer was designed for setting up a TID assay. Finally, the TID mechanism was transferred onto a microarray surface to prove the applicability of TID-based protein detection in the aptamer-microarray format. This systematic development of a TID assay for detecting the VEGF model protein can be transferred to various biosensing platforms for the detection of different biomarkers with other aptamers.
2. Materials and Methods
2.1. Chemicals
NaCl and EDTA (ethylenediaminetetraacetic acid) were purchased from Thermo Fisher Scientific, Waltham, Massachusetts, USA. Na2HPO4, KCl and KH2PO4 were obtained from Sigma-Aldrich, St. Louis, Missouri, USA. Ethanol (99.5%) and Tris were received from Carl Roth GmbH & Co. KG, Karlsruhe, Germany. Tween 20 was purchased from VWR, Radnor, PA, USA. SYBR Green I and II were obtained from Molecular Devices, San José, CA, USA. NaBH4 was received from Honeywell Riedel-de Haen AG, Seelze, Germany.
2.2. Buffers and Solutions
TBSET (Tris-buffered saline with EDTA and Tween 20) contained 10 mM of Tris, 100 mM of NaCl, 50 mM of KCl, 0.05 mM of EDTA and 0.05% Tween 20 (pH 7). The TBSET without KCl and 0.05% Tween 20 was used for the selection of the aptamers V7t1 and Del5-1, or at least their parent aptamer during the SELEX process [
2,
36]. Tween 20 was added to the buffer to reduce unspecific hydrophobic interactions. Marušič et al. investigated the increase of the stability of the G-quadruplex from V7t1 by adding potassium ions [
13]. Therefore, 50 mM of KCl was added to the buffer. For spotting and immobilizing aptamers onto the microarray (3D-aldehyde glass slides from PolyAn, Berlin, Germany) surface, a spotting solution consisting of 100 mM of NaCl, 50 mM of KCl and 0.05 mM of EDTA (pH 7) was prepared. The preparation of microarray slides required a blocking solution containing 0.1 g of NaBH
4 dissolved in 10 mL of 99.5% ethanol and 30 mL of PBS (phosphate-buffered saline), which was composed of 137 mM of NaCl, 2.7 mM of KCl, 4.3 mM of Na
2HPO
4, and 1.4 mM of KH
2PO
4 (pH 7.4). TBSETB (Tris-buffered saline with EDTA, Tween 20 and bovine serum albumin (BSA)) contained TBSET with 1% BSA. All solutions were prepared with deionized water (Arium661, Sartorius AG, Göttingen, Germany).
2.3. Oligonucleotides
Nonaka et al. developed the V7t1 aptamer specifically binding to VEGF with the sequence 5′-TGTGGGGGTGGACGGGCCGGGTAGA-3′ [
2]. The spacer sequence attached to the V7t1 aptamer is based on its maternal aptamer, termed Vap7 [
2]. During the selection of Vap7, this sequence was part of the primer region in the SELEX process [
2]. Lönne et al. investigated the performance of the immobilized V7t1 aptamer using different spacers [
8]. The binding affinity of the V7t1 aptamer to VEGF was highest when using the 14 nt spacer based on the primer sequence [
8]. The other VEGF-binding aptamer, Del5-1, was developed by Hasegawa et al. [
36]. As negative control, the Syl3C aptamer selected by Song et al. against the protein epithelial cell adhesion molecule (EpCAM) was used [
37]. The sequences of the used aptamers, as well as their corresponding complementary oligonucleotides and modifications are listed in
Table 1. Some oligonucleotides were modified with fluorophores cyanine 5 (Cy5) and fluorescein isothiocyanate (FITC), Dabcyl quencher or a terminal amino group (NH
2). All oligonucleotides were dissolved in deionized water (Arium661, Sartorius AG, Göttingen, Germany) at a concentration of 100 µM as stock solutions.
2.4. Proteins
The human, recombinant protein VEGF-A165 (product ID: GFH44, Cell Guidance Systems Ltd., Cambridge, UK) was produced in Escherichia coli. The negative control protein α-chymotrypsin was purchased from ApplyChem GmbH, Darmstadt, Germany. Lysozyme (as an additional negative control) and BSA were purchased from Sigma Aldrich, Germany. All proteins were dissolved in TBSET with a concentration of 15 µM as a stock solution and diluted for the experiments.
2.5. Preparation of Microscale Thermophoresis (MST) and Capillary Scan Experiments
Throughout the microscale thermophoresis (MST) experiments, the samples were protected from light including the incubation and the storage time as soon as fluorescent molecules were involved. To ensure that both fluorophores, Cy5 and FITC could be measured in the required device-specific fluorescence range (200–1500 fluorescence counts), a minimum concentration of 25 nM of the FITC-labeled aptamer was needed. To achieve good comparability, this concentration of labeled aptamer was used in all MST experiments.
2.5.1. Characterization of Aptamers via MST
MST experiments were performed to characterize different anti-VEGF aptamers. The advantage of the MST for the determination of K
D (dissociation constant) is that neither the aptamer nor the protein has to be immobilized. Therefore, the determination of binding affinities is facilitated without any influence of immobilization. Using this procedure thereby reduces the risk of excluding high affinity aptamers from further investigation, which failed due to non-optimal immobilization in other techniques, such as surface plasmon resonance (SPR) or microarrays. Therefore, the binding affinities towards recombinant human VEGF and the two negative controls lysozyme and α-chymotrypsin were investigated. Furthermore, an aptamer directed against EpCAM (named Syl3C) was used as a negative control. Both aptamers and proteins were dissolved and mixed together in TBSET. The aptamers were fluorescently labeled with FITC or Cy5 (
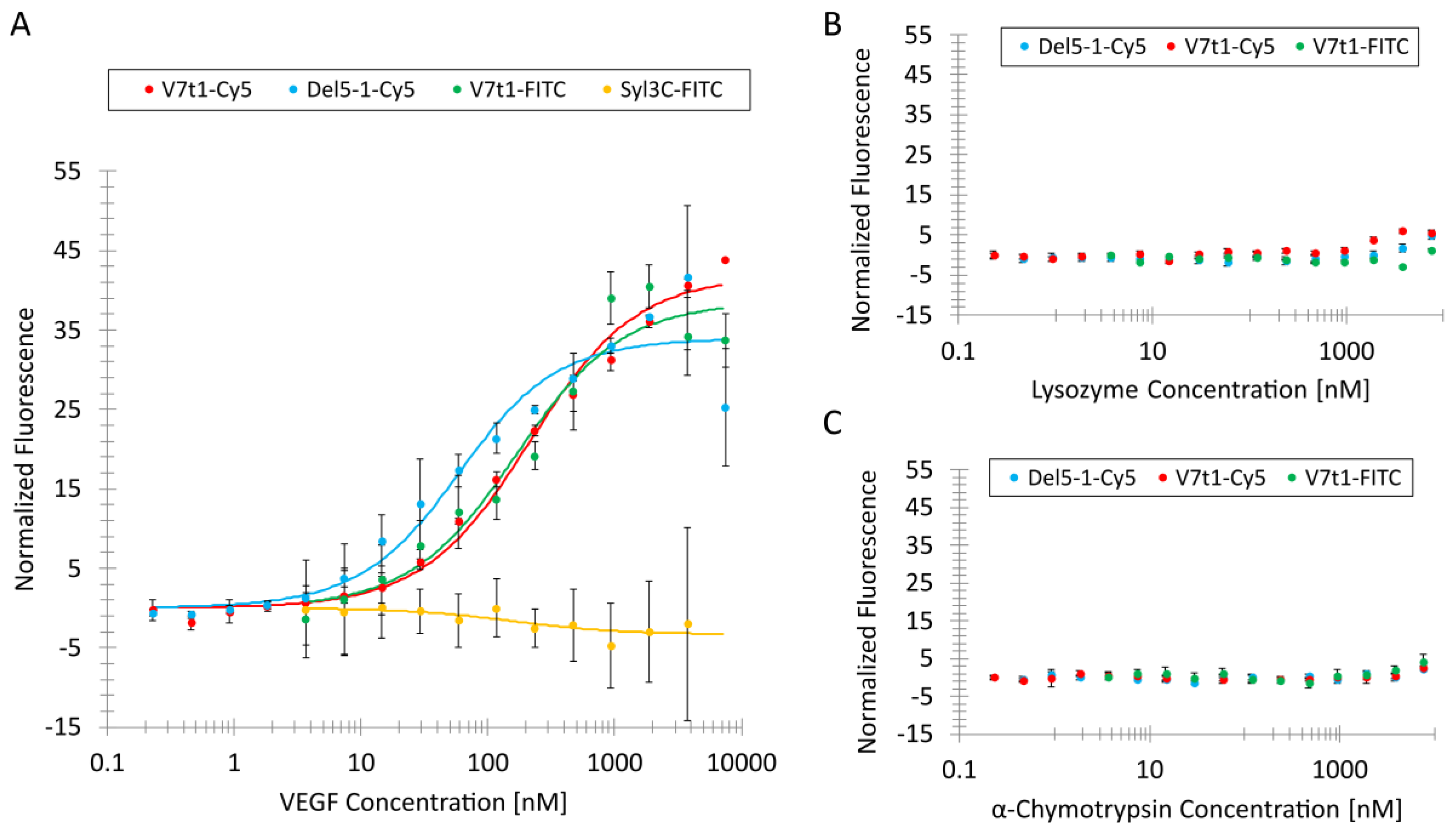
Table 1) and used in a constant concentration of 25 nM, and the protein concentration was varied from 7.5 µM to 3.7 nM (or 0.23 nM) by serial dilution. Before the MST measurements were started, the samples were stored for 20 min at 20 °C in an incubator IPP 30 (Memmert GmbH and Ko.KG, Schwabach, Germany).
2.5.2. Hybridization and Quenching of Complementary Oligonucleotides via Capillary Scan
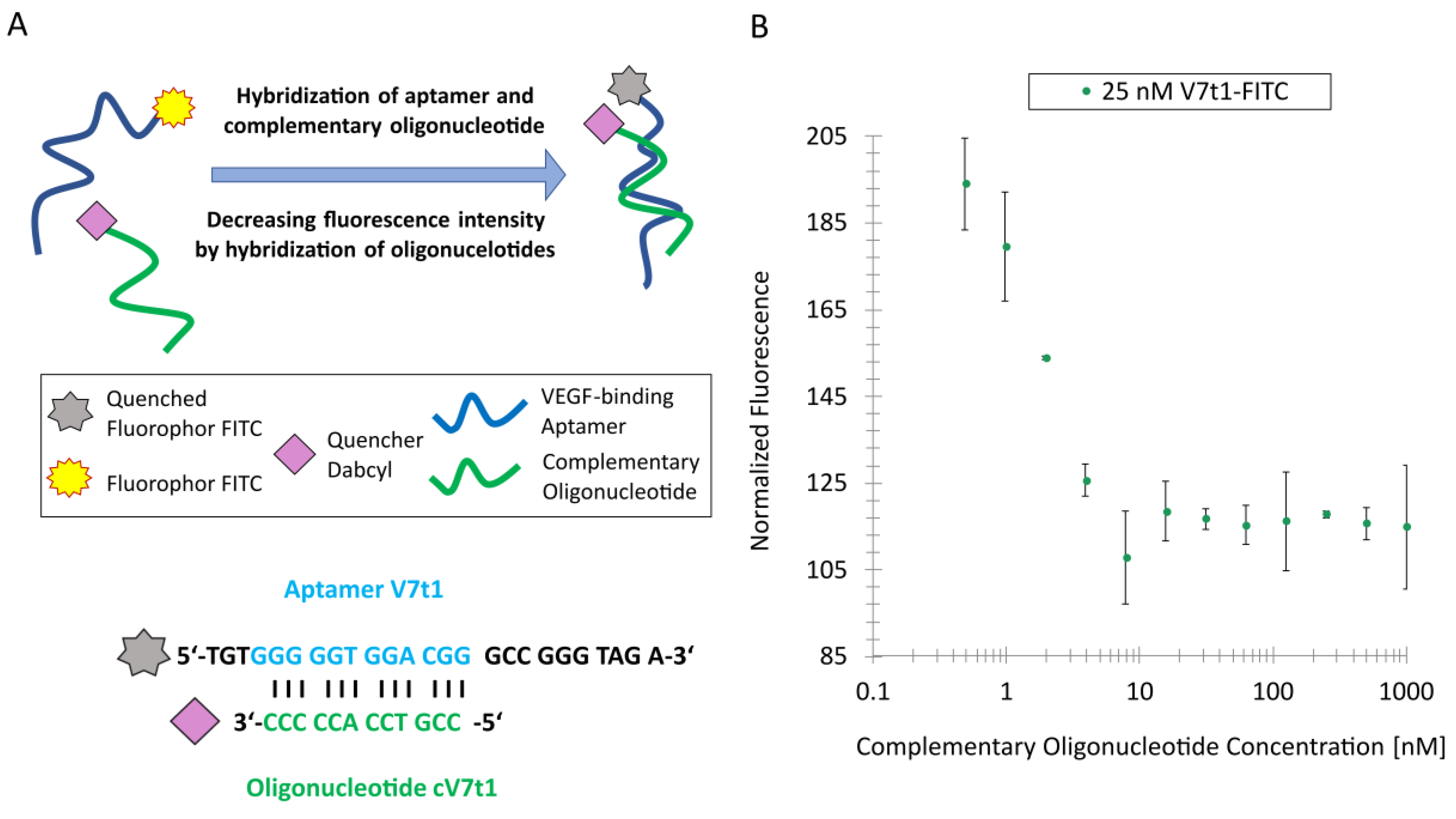
The quenching effect of hybridizing fluorescently labeled aptamers with complementary oligonucleotides modified with a quencher was investigated by using the fluorescence scanner of the MST Monolith NT.115. Therefore, the FITC-labeled aptamers were incubated with complementary oligonucleotides modified with a Dabcyl quencher in TBSET. While the aptamer concentration was kept constant at 25 nM, the concentration of the complementary oligonucleotide was varied from 1 µM to 0.49 nM by serial dilution. Before the fluorescence measurement via Monolith was started, the samples were heated for 10 min at 95 °C in a thermoblock VWR 732-1210-Doppio (VWR International GmbH, Darmstadt, Germany) to remove secondary structures in the oligonucleotides. Afterwards, the samples were cooled down to room temperature for 30 min for the hybridization of the complementary oligonucleotide strands.
2.5.3. TID of Complementary Oligonucleotides via Capillary Scan
For the TID experiment via the MST Monolith NT.115, an FITC-labeled aptamer and a complementary oligonucleotide modified with a Dabcyl quencher were used. Both the aptamer and the complementary oligonucleotide were diluted and mixed together in TBSET with a concentration of 50 and 14 nM, respectively. Afterwards, the mixture was heated to 95 °C for 10 min using a thermoblock. Then, the samples were cooled down for 30 min at room temperature for the hybridization of complementary oligonucleotide strands. While the oligonucleotides cooled down, the proteins were diluted in TBSET with a starting concentration of 15 µM. Then, the oligonucleotide solution was mixed with the protein solutions by ratio of 1:1. Consequently, the final concentrations of the aptamer and complementary oligonucleotide were 25 and 7 nM, respectively. The protein concentrations were varied starting from 7.5 µM. Before the MST measurements were started, the mixture of aptamers and oligonucleotides was incubated with the proteins. Therefore, the samples were stored for 20 min at 20 °C in an incubator to keep the temperature constant.
2.5.4. MST and Capillary Scan Settings
After incubation, the samples were filled into standard or premium capillaries (NanoTemper Technology GmbH, Munich, Germany). For samples containing VEGF, premium capillaries with low nonspecific binding (product number: MO-K025) were used to inhibit VEGF from sticking to the glass surface while all other samples were transferred into standard capillaries (product number: MO-K022). After that, the measurements were performed at room temperature with the Monolith NT.115 (NanoTemper Technology GmbH, Munich, Germany) using the NT-Control software Version 2.1.3 (NanoTemper Technology GmbH, Munich, Germany) to obtain capillary scans or analyze the samples by MST. Therefore, the MST power was set to 20%. Depending on the fluorophore, the light emitting diode (LED) power was set to 100% for the detection of FITC-labeled aptamers with the selection of the blue LED. Cy5-labeled aptamers were detected by setting the LED power at 20% and the selection of the red LED. Each experiment was performed three times. For the characterization of the aptamers via MST, the KD model of the NT-Analysis software Version 2.1.3 (NanoTemper Technology GmbH, Munich, Germany) was used to determine the KD values. The hybridization and quenching effects of the complementary oligonucleotides and the TID of complementary oligonucleotides were determined by analyzing the fluorescence data of the capillary scan using the NT-Analysis software Version 2.1.3.
2.6. Preparation of Microarray Slides
The slides used for the microarray experiments were 3D-aldehyde glass slides from PolyAn, Germany. The microarray slides were protected from light throughout the whole experiment including the incubation, the washing steps, and the storage time as soon as fluorescent molecules were involved. The following washing and incubation steps were carried out while the microarray slide was shaking on an MTS 4 shaker (IKA Werke, Staufen im Breisgau, Germany) at 300 rpm and at room temperature.
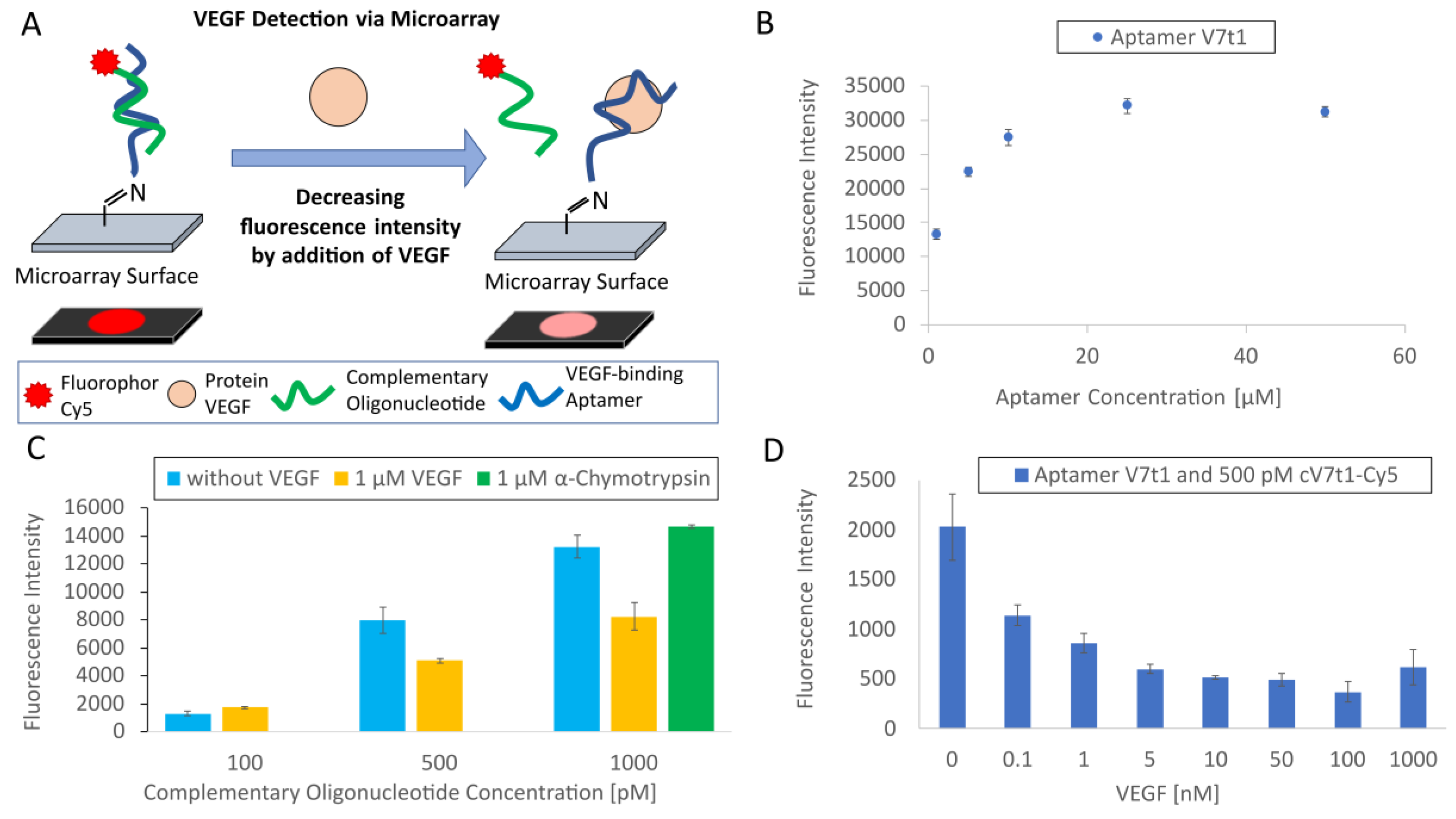
The spotting solution was prepared containing the amino-modified V7t1 aptamer diluted in concentrations of 50, 25, 10, 5 and 1 µM, and the resulting solutions were transferred into a PCR plate. For spotting aptamers onto the microarray slides, the Microarray Spotter NP 2.1 Nanoplotter (GeSiM, Großerkmannsdorf, Germany) was used. Five drops per spot were delivered with a volume of approximately 0.2 nL per drop for each aptamer concentration. The microarray was designed to contain 16 identical blocks. On each of the 16 blocks, every aptamer concentration was spotted in four replicates. Spotting was performed using a Nano-Tip A-J (GeSiM, Großerkmannsdorf, Germany). The amino-modification of the aptamer and the aldehyde group on the microarray surface reacted to an imine. A blocking solution containing NaBH4 was used for the reduction of the unstable imine to a stable amine before starting experiments. Therefore, the slides were incubated in a glass chamber with a blocking solution for 10 min.
2.6.1. SYBR Green Staining of Microarray Slides
By incubating the blocked microarray slides with a staining solution containing SYBR Green I and II, the immobilization of the amino-modified aptamer was investigated. SYBR Green I intercalates into double-stranded DNA, while SYBR Green II binds to single-stranded DNA. After incubating the microarray slide in the blocking solution, the microarray slide was transferred into a solution of SYBR Green I and SYBR Green II dissolved in deionized water in a ratio of 1:10,000. The slide was incubated in the SYBR Green staining solution for 5 min. Afterwards, the slide was washed two times with 100 mL deionized water for 1 min. Finally, the microarray slide was dried by compressed air before scanning and analysis.
2.6.2. Hybridization of Complementary Oligonucleotides on the Microarray
The blocked microarray slide was washed for 1 h with 100 mL TBSET in a glass chamber. Afterwards, the TBSET was removed, and the slide was mounted in a hybridization chamber (NEXTERION IC-16-incubation chamber, Schott, Jena, Germany). The 16 compartments were each loaded with 50 µL of the samples. Therefore, the complementary cV7t1 oligonucleotide labeled with Cy5 was diluted in different concentrations of 0.1, 0.5 and 1 nM in TBSETB and also in TBSETB containing 1 µM of VEGF. Furthermore, the complementary oligonucleotide was diluted in concentration of 1 nM in TBSETB containing 1 µM of α-chymotrypsin, with α-chymotrypsin used as a negative control. Afterwards, 50 µL of each sample were transferred in the wells of the hybridization chamber and incubated for 1 h. In microarray experiments, a longer incubation time was chosen compared to the MST experiments. Using the microarray format with immobilized aptamers results in solid-phase hybridization, which is known to be considerably slower than hybridization in the solution. Here, a high charge density and diffusion effects had to be considered, as they could have resulted in a longer hybridization time [
38]. After the incubation, each chamber was washed three times with 50 µL of TBSET for 5 min. Next, the microarray slide was taken out of the hybridization chamber and washed two times in a glass tray filled with 100 mL of TBSET. Then, the slide was dried with compressed air.
2.6.3. Microarray-Based TID Assay for Determination of Sensitivity
To further investigate the developed TID assay and to determine its sensitivity, the microarray slide was washed in TBSET for 1 h after blocking. Then, the microarray slide was fixed in the hybridization chamber. Furthermore, each well was filled with 50 µL of TBSETB and 0.5 nM of the complementary cV7t1 oligonucleotide labeled with the Cy5 fluorophore. The samples were incubated for 1 h. Afterwards, each well was washed three times with 50 µL of TBSET for 5 min. Then the wells were filled with 50 µL of TBSETB and incubated for 10 min. The buffer was removed and wells were incubated with 50 µL of TBSETB and VEGF in different concentrations for 1 h. The concentrations of VEGF prepared in TBSETB were 1000, 100, 50, 10, 5, 1, 0.1 and 0 nM. Afterwards, the wells were washed three times with 50 µL of TBSET for 5 min. Then, the slide was taken out of the hybridization chamber and washed two times in a glass chamber with 100 mL of TBSET for 5 min, and the microarray slide was dried with compressed air.
2.6.4. Scanning and Analyzing Microarray Slides
The microarray slides were scanned using a GenePix 4000B device (Molecular Devices, San José, CA, USA) at 100% laser power and a PhotoMultiplier Tube-Gain (PMT-Gain) between 450 and 650. The wave length was adjusted for each fluorophore. The Cy5 fluorophore was scanned at 635 nm and SYBR Green at 532 nm. Afterwards, the scanned slides were analyzed with the GenePix Pro 7 software (Molecular Devices, San José, CA, USA). The average and the standard deviation of the signal intensity of each concentration were determined.
4. Conclusions
In this work, we presented a strategy to systematically develop TID assays by using MST and fluorescence analysis to screen aptamers and complementary sequences for their sustainability and to prove the applicability of the developed system by transferring the TID method onto an aptamer-microarray.
The MST analysis showed that the VEGF-binding Del5-1 and V7t1 aptamers bind with high affinity and specificity to their target VEGF. The affinity of Del5-1-Cy5 towards VEGF (KD value 47 nM) was even higher than the affinity of V7t1-FITC, with a KD value of 155 nM. Nevertheless, V7t1 contains a G-quadruplex which is known to be important for the binding of VEGF and can be used for the development of a TID mechanism; however, the binding site of Del5-1 has yet to be revealed. Nonetheless, the proposed strategy is also applicable for the investigation of the aptamer binding site by screening different complementary oligonucleotides against a given aptamer, thereby identifying oligonucleotides that can be dissociated due to aptamer–target interaction.
In case of V7t1, a quencher-modified oligonucleotide complementary to the G-quadruplex motif of V7t1 was designed and hybridized to the V7t1-FITC aptamer. The maximal fluorescence quenching was obtained at a complementary oligonucleotide concentration of 7 nM. Afterwards, the TID mechanism was investigated by adding VEGF in different concentrations to the aptamer–oligonucleotide complex, thus revealing an increasing fluorescence signal with increasing VEGF concentration.
Finally, the method was transferred to an aptamer-microarray to prove the applicability of the developed TID assay. The incubation of microarrays with 500 pM of Cy5-labeled complementary oligonucleotides (cV7t1) competing with VEGF showed a significant lower fluorescence intensity than without VEGF. This leads to the conclusion that 500 pM of cV7t1-Cy5 was an appropriate concentration for hybridizing with the aptamer. Nonetheless, the concentration of the complementary oligonucleotide could be further optimized. Here, it has to be carefully considered that an increase of the oligonucleotide concentration will not only result in higher signal intensities, but the excess of oligonucleotide will also compete with VEGF for binding to the aptamer, which could impair the microarray sensitivity. At last, the sensitivity of the VEGF detection via TID mechanism on the developed aptamer-microarray was determined. Therefore, the immobilized aptamer (25 µM in the spotting solution) was incubated with its complementary oligonucleotide (500 pM) before adding VEGF. The difference between the fluorescence intensity at 0.1 nM of VEGF was 44% in contrast to the hybridized oligonucleotides without VEGF, suggesting that even lower VEGF concentrations might be detectable. Therefore, the sensitivity of the microarray could also be investigated with lower VEGF concentrations in future experiments.
An advantage of the TID assay is its high specificity, since not every binding results in a signal. In this study, an aptamer-based TID assay was schematically developed by MST and fluorescence analysis for the detection of the VEGF model protein. Furthermore, the TID assay was successfully transferred to a different application—an aptamer-microarray. All in all, we propose that the strategy to develop TID assays can be transferred to further aptamers, and the developed VEGF-sensitive TID assay could be transferred to different biosensing platforms for medical research and diagnostics in the future.