Abstract
A human mission to Mars can be viewed as the apex of human technological achievement. However, to make this dream a reality several obstacles need to be overcome. One is devising practical ways to safeguard the crew health during the mission through the development of easy operable and compact sensors. Lately, several smartphone-based sensing devices (SBDs) with the purpose to enable the immediate sensitive detection of chemicals, proteins or pathogens in remote settings have emerged. In this critical review, the potential to piggyback these systems for in situ analysis in space has been investigated on application of a systematic keyword search whereby the most relevant articles were examined comprehensively and existing SBDs were divided into 4 relevant groups for the monitoring of crew health during space missions. Recently developed recognition elements (REs), which could offer the enhanced ability to tolerate those harsh conditions in space, have been reviewed with recommendations offered. In addition, the potential use of cell free synthetic biology to obtain long-term shelf-stable reagents was reviewed. Finally, a synopsis of the possibilities of combining novel SBD, RE and nanomaterials to create a compact sensor-platform ensuring adequate crew health monitoring has been provided.
1. Introduction
1.1. Need for Miniaturized Sensors for Future Space Missions
Scott Kelly and Mikhail Korniyenko spent 342 days in orbit on the international space station (ISS). Their achievement shows that long-term space flights are feasible and brings humanity one step closer to one of the biggest scientific challenges of this century: Human settlement on other worlds, with the most ambitious endeavor being a human mission to Mars. Significant resources are invested in the technological advancement of rocket science in order to make this dream a reality. However, some other important facets of the challenge should not be forgotten. One of these is a better understanding of the effects of a prolonged stay in space to one’s health. For instance, it has been found that microgravity can lead to muscle atrophy after only a few weeks in space [1]. Moreover, decreased oxygen consumption during space flight can lead to a decrease in exercise capacity and might affect performance upon arrival [2]. In addition, a significant increased risk for renal stones has been reported [3] and is mainly subscribed to increased turnover of bone minerals due to bone atrophy [4]. Another possible threat to crew health is the emergence of infectious diseases. Indeed, it should not be forgotten that where humans go, microbes go. A study on the presence of microbes on a space shuttle has shown that the amount of colony forming units (CFUs) in the shuttle air increased by 300% within 12 days [5]. Moreover, it has been shown that microgravity can actually increase the growth rate and secondary metabolite production of microbes [6] and that the susceptibility of opportunistic pathogens to antibiotics as well as their virulence may change aboard the space ship [7,8]. Next to this, it has been reported that space travel can reduce the efficiency of the immune system, increase cytokine blood plasma levels, and cause reactivation of Herpes virus (HV) [9], including Latent Epstein–Barr Virus, which can lead to infectious mononucleosis [10]. Moreover, increased exposure to radiation and stress can lead to a higher risk of cancer. Indeed, all the health risks mentioned above are included in the NASA human research roadmap (https://humanresearchroadmap.nasa.gov/) and will need to be closely monitored during any long term space mission [4]. Evidently, strict monitoring of the physical and biochemical parameters as indicators of the crew health during a mission or simulated events by use of portable analytical tools could improve the understanding of these biological phenomena and enable early diagnosis, treatment and intervention measures aboard the spacecraft. Similarly, continuous environmental monitoring of both inorganic and organic compounds present in the air, water and other surfaces as well as other systems such as waste/feces and biological life support of a spacecraft can help prevent the growth of pathogenic microbes on board. Requirements for such devices, as stated by NASA in the Human Research Program Requirements Document HRP 47052 Revision E, are especially low mass, volume and power consumption. Moreover, the devices should be reliable and durable, whilst avoiding laborious analysis with bulky instruments in microgravity. Proof of concept studies for the accurate detection of microbes in space using miniature devices have already been reported [11,12]. These include the use of a miniaturized PCR system [12] and a portable DNA sequencing device based on nucleotide recognition through conformational changes in the protein-based pore (Nanopore technology) [13]. The latter was used successfully for rapid microorganism identification and possible disease diagnostics through DNA sequencing was suggested. Miniaturized Gram positive/negative bacterial and fungal detection aboard the ISS using the Lab-On-a-Chip Application Development Portable Test System (LOCAD-PTS) has also been reported as a successful system for the quantification of microorganisms aboard a spacecraft [14]. Other such miniaturized devices have been developed to perform screening tests for the detection of extra-terrestrial life. Some examples are: an automated microfluidic device using capillary electrophoresis and laser induced fluorescence for amino acid detection [15], an antibody microarray for biomolecule detection [16] and an in situ DNA sequencing device based on Nanopore technology [17]. Although these sensors do not deliver pertinent conclusions regarding the discovery of extra-terrestrial life, they could filter out which samples are potentially interesting for more detailed sophisticated analysis back on earth (https://ntrs.nasa.gov/search.jsp?R=20180002121).
1.2. Smartphone Based Devices for Facile Crew Health Monitoring during Deep Space Missions
The development of portable lab on a chip (LOC) or point of care (POC) bio-sensing devices is currently experiencing a major boost in many sectors, including environmental science [18], animal and human health monitoring, disease diagnosis, and food safety monitoring [19,20]. An interesting development is the use of smartphone-hyphenated biosensors that use the phone’s built in sensors and hardware to directly analyze the sample in situ. Phones nowadays are equipped with a plethora of sensors including cameras, with ever increasing resolution, and ever more sophisticated processing units, memory storage capabilities and connectivity, all in a highly compacted design. So why not utilize this? In other words, why not benefit from the already optimized miniaturization of smartphones for the further development of sophisticated sensing devices. As previously mentioned, similar systems (in terms of compactness and simplicity), are already being developed by NASA such as the mentioned LOCAD system [14,21] and water monitoring systems for in-flight microbial contamination [22]. Such systems are being developed to enable more in-flight analysis instead of relying on analysis on the ground using bulky bench-top instruments, which is often still the case [22]. However, these systems are not capable to detect between species making it impossible to distinguish pathogens from rather harmless species. Moreover, more elegant solutions using SBDs, which might be able to replace current bench-top instruments for direct inflight analysis, might already exist. In fact, one could bring this question even one step further i.e., why not piggyback already existing smartphone based sensing devices developed for biochemical sensing on Earth, for deep space missions? Indeed, such devices are often developed for use as robust and simple point of care devices in remote locations and, as such, inherently meet miniaturization and reduced power requirements. Moreover, additional costs implicated in the development of novel sensors complete with processing and memory units can be avoided by adopting currently developed systems. Thus, research in this direction seems a logical choice, if adequate robustness and sensitivity can be reached. Studies have shown that these devices can be quite sensitive. Long et al., for instance, compared the performance of a SBD spectrometer against conventional bench-top analyzers for quantifying analyte concentrations using two commercial assays based on transmission/reflection measurements (ELISA), or fluorescence intensity measurements [23]. This SBD uses either the flashlight or an integrated green laser diode for illumination of the sample which is held in a microfluidic chamber. Emerging light is then piped to the rear camera with fiber optic cable. The camera is covered by a diffraction grating which generates spectra when images are collected. The authors found that the SBD was able to predict analyte concentrations as accurately as bench-top analyzers and, in some cases, even outperform the latter. Moreover, the finding that SBDs can perform equally to bench-top instruments is not limited to a single study. Ludwig et al., tested a fluorescent protein micro array SBD for the detection of recombinant bovine somatotropin (rbST) in milk [24]. Briefly, UV light from LEDs embedded in a 3D-printed smartphone attachment was used for excitation of quantum dot fluorescent labels used to visualize the amount of rbST. The images, collected with the rear camera, were then corrected by a developed Android-based software on the SBD and used to estimate rbST concentration. The system was compared with a flow cytometry reference method and obtained excellent agreement. The usefulness of such devices has equally been demonstrated for the detection of infectious diseases. Laksanasopin et al., has developed and compared a SBD for the simultaneous detection of syphilis and HIV within 15 min [25]. In this system microfluidic channels are coated with antigen recognized by marker antibodies present in whole blood samples of HIV and/or Syphilis patients. Whole blood samples are flown through the channels (using a hand driven vacuum pump), followed by Gold labeled IgM antibodies held in a chamber in the microfluidic cassette. Then, silver reagent is added to amplify the signal and the optical density is measured. This device was compared to laboratory-based tests in a small (n = 96) clinical trial in Rwanda and obtained excellent results. Finally, Priye et al., has recently developed a multiplex SBD for the detection of zika, chikungunya and dengue viruses. The authors use an isothermal PCR technique (reverse-transcription loop-mediated isothermal amplification) for fluorescent detection of the RNA viruses. The system is integrated in a small 3D printed box fitted with an excitation source and emission filters and powered with a 5V USB power source. Spectra are taken with the smartphone rear camera and analyzed using a developed smartphone app equipped with an algorithm to analyze the fluorescent signal. Again, it was shown that the SBD was capable to detect the targets in crude matrixes (blood, urine and saliva) with similar performance to bench-top devices [26]. Such techniques, embedded within a simple compact device that does not require extensive training before use, could potentially be used to scan for signs of stress, detect opportunistic microbes, keep track of an astronaut’s metabolism and perform preliminary in situ scans for extra-terrestrial life. Moreover, hyphenated biosensors, combined with immunosorbent assays, have been proven to work both under microgravity and Martian gravity [27].
1.3. Obstacles to Overcome to Enable SBD Use in Space
However, most SBDs have been developed for implementation on earth. In order to optimize a smartphone based device (SBD) for use in space travel special considerations are required with regards to the construction of the device such as the principle of detection, the biorecognition element and the sample type to be applied. One particular concern can be the degradation of protein-based recognition elements (antibodies/enzymes) used for bio-recognition. Thus, long-term stability to ensure functionality of the sensor throughout the trip is vital. Another possible obstacle for piggybacking SBDs for deep space analysis is the lack of protection of the electronics of the SBD from galactic cosmic rays (GCR) and solar particle events (SPE). Luckily, the protective shielding integrated in a crewed spacecraft might mitigate the need for additional electronic shielding. Indeed several commercial off the shelve (COTS) devices are being tested and used on the ISS with interesting results [28], including a COTS device using smartphone software (mobiPV) [29]. However, GCR and SPE outside of the earth’s magnetic field might still cause damage to such devices in deep space. Thus more research in the use of novel shielding materials, like doping polymeric casing with carbon nanotubes [30,31], or nanometals [32] would be useful to ensure the safe use of COTS devices in those settings. Another issue might be impaired functioning of REs such as proteins and DNA due to radiation damage. However, recent work has shown that proteins [33], including antibodies in protein arrays [34,35], as well as DNA based aptamers [36] show little to no loss of function at radiation levels which are orders of magnitude higher than the levels measured by the Curiosity rover on its mission to Mars [37]. Nonetheless antibody storage conditions remain stringent and long term storage can lead to reduced activity of antibodies [33]. Moreover, a study on radiation resistance of gDNA and primers showed PCR function inactivation at 180 Gy exposure to proton radiation at SPE energy levels [38]. This suggests higher radiation sensitivity for nucleic acid based systems as observed in [36]. Thus the use of more stable, synthetic REs might further increase the shelf-life of such devices and enable long term storage in less stringent conditions as needed for SBDs with protein based components. The aim of this review is to provide a guide to the presently developed SBDs that could prove utile, after limited adaptations, for their implementation in long-term space missions. Firstly, the possibilities to monitor human health with these devices will be discussed with an emphasis on the screening for infectious diseases, viruses, and the monitoring of biomarkers indicating the state of a person’s health, e.g., stress and immune response levels, as well as options to monitor the external environment for the presence of pathogens. In each of these sections effort is made to identify which are the most pressing risks that could be addressed using, or adapting, an existing SBD and which requirements such a device should meet to be utile for deep space missions by using information gathered from the human research roadmap (https://humanresearchroadmap.nasa.gov/Risks/). Secondly, the possible weak points of these biosensors for in situ analysis in space will be critiqued including the choice of the integrated recognition element (RE) and the use of synthetic biology to obtain shelf-stable reagents. Finally, a short synopsis will focus on the predicted further development of such devices for this purpose.
2. Existing SBDs Useful for Space Missions
2.1. General Overview of Available SBDs to Monitor the Crew’s Health
The tasks that an SBD could perform during deep space missions are numerous and include monitoring the crew’s health, checking the health of any biological elements of life support systems, investigating microbiome health and soil health, and screening for signs of life upon landing. In this paper the main focus is to investigate the possibility to piggyback on existing SBDs for crew health monitoring during space missions. This specific application was chosen for two main reasons: (i) By measure of time priority. If SBDs are to be used for any application at the final destination they must first undertake the journey. Thus advantageously they might firstly be utilized to monitor crew health during that journey. (ii) Use of SBDs on a manned spacecraft instead of an unmanned craft, send ahead for life detection for example, implies protection of both the electronics and RE elements of the SBD from radiation. This limits the threat of malfunctioning and allows faster actual implementation of such instruments for use in space. This however, does not mean that the systems discussed here below cannot be useful for the other bespoken applications simply by changing the RE. Possible use of SBDs to monitor crew health was further classified into four groups: (i) Monitoring the general health of the crew (cancer, reduced immunity and signs of stress); (ii) screening food for freshness and contamination; (iii) monitoring the air and water quality on-board and (iv) diagnosis of infectious diseases. In order to classify the SBD reported in the scientific literature a keyword search in Scopus was conducted. The general query was: “(smartphone OR cell phone) AND (portable OR mobile) AND (instrument OR sensor OR device OR platform) AND (sensing OR testing OR analysis OR detection OR measurement OR monitoring OR diagnostics)”. The following terms were added to that query for the individual groups: AND (biomarker OR protein OR cancer OR stress) for group (i). AND (food OR foodstuff OR milk OR fruit OR cereal OR meal AND NOT allergen OR allergy) for group (ii). AND (air OR water OR environment OR volatile OR inorganic) for group (iii). AND (pathogen OR bacteria OR virus OR infection) for group (iv). This search yielded 155 original research articles for group (i), 78 for group (ii), 579 for group (iii) and 118 for group (iv). In group (iii) all articles related to computer science mainly discussing advances in app development and algorithm improvement were then excluded in order to focus on articles related to advances in mobile biochemical analysis which led to a reduction in that group to 199 documents. Overall, articles were considered in scope if the focus was on mobile biochemical analysis and if at least one of the possible group-specific application-related keywords was mentioned in the article as a detection target. After deletion of duplicates (5) a total of 550 articles remained for analysis. Based on the abstract content this number was condensed to 186 articles. Finally, after full article analyses and cross referencing, the following number of articles were deemed to enter into the scope of this review: 51 for group (i), 22 for group (ii), 27 for group (iii) and 25 for group (iv). Another 23 articles were deemed relevant but difficult to classify into one of these groups and were denoted “other”. Thus a total of 148 articles were reviewed and further classified into subgroups (Figure 1). There are approximately twice as many SBDs reported for general health monitoring compared to the other groups as perhaps more funding is available for research targeting all these issues including cancer (25% of the articles reported in the group) and cardiovascular and stress related problems (± 30% of the group). Moreover, the SBDs proposed to monitor cardiovascular and stress related disease often analyze the heart rate, using a variety of measurements such as pulse to pulse intervals and ECG, using a smartphone for the determination of stress levels [39,40,41,42,43,44,45,46], which is easier to accomplish than the specific detection of a pathogen. Some articles report the use of SBDs to monitor infectious diseases or to diagnose diseases that are unlikely to present during a space mission. However, these systems may still prove useful since they can be used to detect other targets by simply changing the bio-recognition element for detection of a specific contaminant. Interestingly, only one article was reported for the detection of fungi (Fusarium) using an SBD [47] indicating that more research in this direction is needed since fungi are major players in food spoilage [48] and several Fusarium species can produce toxic compounds [49]. The polyvalent group “Other” (16%) reports SBDs that use REs which require modification to become useful such as a printed paper assay for the quantification of streptavidin [50], or articles difficult to classify into a group as applied to diverse applications.
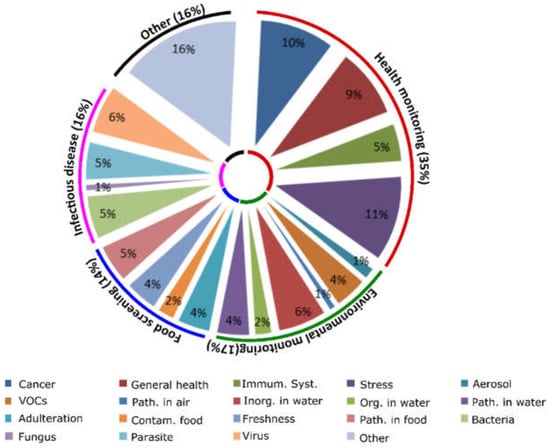
Figure 1.
Classification of relevant SBD related literature. A pie chart showing the classification of the 148 articles kept for thorough analyses after the keyword searches. Groups (outer circle) are divided into subgroups (inner pie chart). Percentages indicate percentage of articles presenting the group or subgroup in relation to the total amount of articles kept for analyses.
2.2. SBDs for Health Monitoring in Space
General health monitoring can be performed by observing physical and biochemical parameters. Key examples are highlighted of SBDs for their current use in detecting stress, reduced immune response and general health monitoring and in cancer diagnosis of importance for prolonged space travel.
2.2.1. Detecting Stress
The human research program integrated research plan identifies stress as a very real problem which can jeopardize the mission due to several factors (e.g., high workload, circadian desynchrony, elevated CO2 levels, radiation and diet and nutrition), and stresses the need for early detection mechanisms to monitor the mental and cognitive health of crew members [4]. Moreover, risk of elevated cardiac rhythm problems during space flights was identified as a red zone in the likelihood consequence (LxC) rating in the human research plan and it is planned to conduct more detailed in-flight heart rate measurements to better predict risks for environmentally induced cardiovascular disease and determine the causes of this (https://go.nasa.gov/2OEifZN). Evidently, stress, equally listed as a risk factor in the human research plan, can be a contributor to this risk factor. On the other hand the multimodal nature of stress makes early detection using one single parameter virtually unattainable [51]. In fact, information regarding cortisol levels, heart rate (mainly ECG and heart rate variability (HRV)) and behavior should be integrated [51,52]. Unfortunately, most SBDs use only the heart rate to monitor stress levels although there are some exceptions where additional data from motion activity, posture [53] and communication data [54] are being used. This development increases the prediction accuracy [54]. However, it does not absolve the need for proper psychological assessment. Extended psychoanalyses and treatment can however be tedious during a mission to Mars since it requires intensive dialog which is complicated due to the extended one way light time (approx. 14 min) between Earth and Mars. Thus, the use of mobile devices that use a multimodal approach for early stress diagnosis and at the same time offer self-help solutions can be a welcome complementary approach. The U.S. military has recently developed an SBD that uses such a complementary approach. The SBD uses multiple sensors to detect stress and other psychological health problems and is equipped with an option for self-help via an app [55]. Another interesting paper reports the development of an application designed to deliver breathing awareness meditation to reduce stress levels [56]. Although data are preliminary, such systems could potentially not only allow diagnosis but also help reduce stress on a prolonged space mission. Apart from these developments other devices for monitoring an array of health related parameters including ECG measurements, blood oxygen levels, body temperature, and sleep quality are already commercially available and their potential usefulness for a mission to Mars was discussed in a recent perspective [57]. Although the paper might overestimate the ease with which the futuristic suggestions could be implicated and does not critically compare the functioning of the mentioned devices, it does provide an extensive list of commercial devices that are potentially interesting to test. In addition, other commercial devices like Google Glass, albeit in a slightly adapted format, have been tested and found interesting to use during space missions as a mobile procedure viewer assisting astronauts during various operations while enabling full two way video communication [29,58]. Moreover, commercially available wearable devices like smart-watches are already used in space and potentially integrate most measurements mentioned here. The interest of NASA for such devices is showcased by the crowd-sourced astronaut app competition that was recently held by NASA and won by I. Calvo and J. Richard for the design of such a system (https://bit.ly/2N1MDNy). In addition, major interest in tricorder like personal health monitoring devices became apparent during the Tricorder X-prize competition (https://tricorder.xprize.org/).
2.2.2. Detecting Reduced Immune Response and General Health Monitoring
The production of naïve T-cells, which form an important protection mechanism against opportunistic viral and fungal pathogens as well as latent viruses, has been shown to decrease in astronauts after space travel [59]. Moreover, this reduction in thymopoiesis was linked to increased amounts of glucocorticoids in plasma and urine. Interestingly, Geiger et al., recently suggested that cortisol levels increased by stress exposure can indeed negatively affect the immune response to pathogens [60]. Indeed, reduced immune response during space missions is a concern included in the human research program integrated research plan [4] and the need for in-flight evidence regarding this is identified as a gap in the program (https://go.nasa.gov/2Mctbkt). More research regarding stress and immune response interplay might reveal the cause of the observed reduced immune response during space travel. One interesting avenue to investigate the link between stress and immune response during space missions is monitoring the cortisol levels in saliva and to link this to CD4+ cell count. Recently, a competitive lateral flow immunoassay (LFIA) and horse radish peroxidase (HRP) conjugated cortisol was developed to detect and quantify chemo luminescence [52]. A 3D printed device shields the strip from background noise and operator variation thus creating a robust test that can be operated by the layman yet allows cortisol quantification directly in saliva with a LOQ of 0.3 ng/mL and a linear range between 0.3 and 60 ng/L. Cortisol levels in saliva vary depending the time of day but typically remain between 0.6 and 10 ng/L [52]. Thus the developed assay (which takes about 30 min) can reveal clinically relevant information. Interestingly, the performance of a variation of this device has very recently been tested aboard the ISS station (mission 52/53) using saliva samples from an Italian astronaut, further underlining the interest in such rapid tests for in situ monitoring of crew health, and the results are pending (https://go.nasa.gov/2Kb854L). Such a device, in combination with an SBD that can determine specific T-cell densities in blood, could then be used to further investigate this phenomenon directly in space while keeping tabs on stress levels and other major markers indicating reduced immunity or infection like reduced CD4+ levels. Indeed SBD based cell counting devices exist already including an SBD using fluorescent imaging cytometry [61] and another using magnetic bead ELISA [62]. The latter may be more fit for the purpose of rapid on-site cell counting since the device uses highly specific monoclonal antibodies and does not require extensive treatment of the blood sample prior to analysis, in contrast to the former. Moreover, since it has been shown in several studies using different detection mechanisms that SBDs can have a similar performance as bench-top reference methods [23,24,25,26], and since the discussed magnetic bead ELISA assay shows a CD4+ T-cell count accuracy of 97% at levels below normal (350 cells/µL instead of ~1000 cells/µL which is the normal level), it can be considered feasible to do such precise measurements with an SBD. Vitamin D (VD) is another important biomarker as levels have been reported to decrease during space missions whereby supplements are required to limit bone loss [63] and minimize effects on both the innate and adaptive immune response [64]. Recently an SBD has been developed that allows quantification of VD [65]. For this purpose VD was aminopropylated and immobilized on a glass substrate whereby antibody coated gold nanoparticles (Ab-GNPs) were allowed to bind to this substrate in a competitive assay (fewer Ab-GNPs will bind if free VD is present in the sample). Finally silver ion reduction on the gold surface of the Ab-GNPs bound to the immobilized VD allows for sensitive colorimetric detection by using a mobile application that applies the hue saturation brightness model to quantify VD at nanomolar concentrations. However, the method does hold some weak points such as a required lengthy 6-h incubation. Finally some interesting work has been reported on mobile devices to detect glaucoma i.e., a 3D printed retinal imager [66] and a pressure sensitive microchannel [67] to measure blood pressure behind the eye.
2.2.3. Detecting Cancer
The detection of certain cancers can be performed by SBD through the cellular image analysis or the detection of biomarker indicators. An exact risk assessment for cancer due to GCR is difficult due to the lack of sufficient data [68]. However, compelling indications that ionizing radiation can increase the risk for melanoma [69] exist and that exposure to GCR and SPE during a Mars mission can increase the risk of skin cancer in astronauts by >1% [70]. Evidently, this does not mean that astronauts have a high risk of developing cancer during the mission but rather that the life-time cancer risk will be elevated due to the stay in deep space. This being said, the use of facile screening methods for melanoma and other cancers can still be considered desirable on long-term missions to ensure optimum crew health and allow early detection. Indeed developing technologies for risk mitigation and monitoring are mentioned as desirable in the human research roadmap (https://go.nasa.gov/2KSjIt8). Several interesting SBDs targeting skin cancer came to surface in the cancer subgroup detecting malignancies via image analysis [71,72,73,74]. Of these the most complete system uses deep convolutional neural networks (CNN) to diagnose keratinocyte carcinomas versus benign seborrheic keratosis and malignant melanomas versus benign nevi in a binary classification system [72]. The system was trained using a substantial dataset (129,450 clinical images representing over 2000 diseases), tested and found as proficient in its diagnostic capabilities as skilled dermatologists. However, such a system does require the mobile device to be fitted with CNN which remains difficult since CNN requires considerable computing power. However, this situation might change in the future since graphical processing units (GPUs) are becoming more common in mobile devices. These GPUs have parallel processing capabilities which can be exploited to accelerate CNN computations on mobile devices. Moreover, an open source, GPU accelerated, library has recently become available on github [75]. Apart from this there is also a neural compute stick (Movidius) available on the market which shows promising results for the use of some CNN on low power devices [76]. This being said, mobile GPUs remain constrained for the use of very deep CNN networks and much work remains to be done on the development of mobile devices for the use of such sophisticated machine learning techniques [77]. An alternative to computed image analysis is visual microscopic analysis via a SBD microscope [78]. Here sensitivity and specificity of a SBD microscope was compared to a conventional light microscope for the expert analyses of dermatopathologic samples. It was found that the SBD had a similar performance as the conventional microscope except for the diagnosis of malignant melanoma where the sensitivity was only 60% but with good specificity (99.9%). However effective, this method still requires expert knowledge for diagnosis and thus requires the images to be sent back to earth. In theory this is not a problem since one-way light time (around 14 min) is not limiting for sending such data between earth and Mars although this delay can quickly add up to hours if active guidance of an expert is required while acquiring images. Thus, it may be more interesting to limit sending microscopic images that have popped up interesting during a preliminary screening test to limit unnecessary data analysis by experts. To this end it might be a more fruitful approach to use the microscope to gather images that could be further processed using image analysis. In this manner more direct analysis with less background noise and variables could also reduce the need for algorithms that require excessive computing power. Another way to reduce background noise might be to use a SBD spectrometer [79]. Such a system could potentially prove useful if enough data is collected to build a solid database for chemometric analysis. Indeed NIR spectroscopy with commercial smartphones is already coming available making such an endeavor more feasible (https://bit.ly/2Kong71). An SBD to detect ovarian cancer using a microchip ELISA to detect human epididymis protein (HE4) in urine [80,81], as well as prostate cancer using a microchip ELISA targeting prostate specific antigen (PSA) [82]. The latter SBD uses magnetic nanoparticles and a magnet rather than pumps thus simplifying the design. Moreover, the surface to volume ratio is thus increased which has reportedly reduced the analysis time to 30 min in contrast to 5 h for the assay. In addition, a SBD using a spectrometer to detect Interleukin 6 (IL-6), a biomarker for several cancers, using conventional ELISA [83], and a SBD using microfluidic dielectrophoresis combined with image analyses on a smartphone camera to count MCF-7 breast cancer cells in culture media [84] have been reported. Apart from these systems other intriguing SBDs exist that could be used for monitoring the crew health. The target, detection method and pros and cons of these systems are illustrated in Table 1.

Table 1.
A list of SBDs developed to monitor general health features relevant for space missions.
2.3. Environmental Monitoring
Environmental monitoring including both water and air quality both whilst on board the spacecraft and on arrival in Mars will be an important element of consideration for a SBD to be valuable.
2.3.1. Inorganic and Organic Compounds in Water
The storage of drinking water on a prolonged space mission must be economized to avoid adding unnecessary weight to the spacecraft. However, recycling water from urine, air humidity and hygiene water can lead to higher amounts of toxic metals due to leaching from metal coatings and filter resin failure and cause hazardous levels of multiple toxic metals in the water supply calling for sensitive detection of these metals at the ppb level [95]. SBD sensors have been reported for the detection of lead(II) ions [96,97] with LODs around 20 µg/L which is 2 fold below the requirements for regenerated potable water aboard the ISS (http://emits.sso.esa.int/emits-doc/RD5-ITT-1-5247.pdf). Unfortunately one of the methods is based on gravitational force [96]. Three SBDs are reported for Hg2+ quantification [98,99,100] and one for fluoride quantification [101]. Of these, one is especially [98] interesting since the LOD reported is almost 10 fold lower than bespoken requirements for potable water on the ISS (0.28 µg/L in [98] versus 2 µg/L aboard the ISS). Moreover, the testing time is limited to 20 min, and the sensor size is under 5 cm thus responding to the goal of reducing human systems resource requirements stipulated in the Human Research Plan-47052 revision E. However, none of these systems were used for the multiplex detection of toxic metals. Wang et al., used a paper fluidics device that uses stacked paper layers fitted with channels bordered by hydrophobic walls and adhesive tape to construct a device allowing the detection of 4 metals in 16 zones [102]. The system was tested for Cd, Ni, Cu and Cr using selective chromogenic reagents for a colorimetric readout that was then quantified using a smartphone camera and application. The authors found that quantification in the low ppm level was possible using this setup. Although sensing at ppb was not achieved the sensitivity could be increased by using other techniques for the detection like the immune detection of metals using HRP, GNP or quantum dots (QD) for enhancement while keeping the design of the device. As for the detection of organic compounds in water three SBDs were described including one that uses electrochemical detection of nitrate in water at the ppm level [103], one using colorimetric detection of catechols in river water [104] and finally one using an acetylcholinesterase inhibition assay to detect organophosphate pesticides in natural water resources [105]. Of these, one is especially [103] of interest since its LOD is 5 fold lower than ISS requirements (0.2 µg/L versus 10 µg/L respectively), while keeping the sensor mass at ~65 g and analysis time around 1 min.
2.3.2. Aerosols, Pathogens and Volatile Organic Compounds (VOCs) in Air
Three existing SBDs have been described for the detection of small particles in air [106,107]. One focuses on the detection of particles on a miniaturized aerosol filter via subsequent image analysis of the observed color change and is effective to measure particle (mainly black carbon) concentrations but not particle size [106], an important parameter to determine particle carcinogenicity [108]. Another SBD, developed by the Ozcan group and termed c-Air [107], uses computational lens free imaging and machine learning to calculate particle size and distribution. In brief, the camera registers the holograms produced by the captured particles on a sticky surface. An iterative particle-peeling algorithm (which takes into account the generated twin image artifact and corrects for it) is then used to reconstruct the particle size from the interference patterns. Finally machine learning is used to further avoid the measurement of false positives. The system, which has a cut-off at 1.4 µm, was tested and found proficient when compared with a conventional device (BAM-1020, Met One Instruments, Inc., Grants Pass, OR, USA). Moreover, c-Air works at a ±15 times higher debit (amount of air analyzed per time unit) as other portable devices [107]. Moreover, the system would fit requirements (particle size ≤ 10 µM) described in NASA-STD-3001, VOLUME 2, REVISION A and adheres to the requirements for such in flight analysis devices (reduced mass, analysis time and power use) mentioned in the Human Research Plan-47052 revision E. As for the measurement of VOCs several SBDs have been developed. Chen et al., [109] developed a system that uses a porous graphitized carbo-pack fitted with a tungsten heating wire that enables pre-concentration of VOCs followed by sudden release at 300 °C. A small GC-column (varying from 4 m to 19 m depending on sample complexity) is used for separation. Detection is achieved via quartz mechanical resonators fitted with molecular imprinted polymers (MIPs) leading to the selective detection of a number of mono-aromatic and alkyl hydrocarbons at the ppb level. The entire procedure (from pre-concentration to detection and flushing the system) only takes a few minutes and has proven efficient in real-life situations making this an attractive portable method to detect VOCs. Finally, one SBD has been reported that enables the detection of pathogens in air (influenza; H3N2) [110]. The detection of this pathogen is especially interesting since the majority of infectious disease incidents reported among approximately 742 crew members in 106 space missions were fever/chills and flu-like illnesses (11 out of 29) [111]. The system uses antibody functionalized silicon nanowires (Ab-Si-NW) in microfluidic channels to detect conductance changes created by the binding event. Information regarding air quality is then displayed on a smartphone through wireless connectivity [110]. Such a system however only works in a conductive medium such as water and not in air. Thus the authors used an electrostatic air sampling system that allowed transferring aerosols into hydrosols which could then be transported to the Ab-Si-NW via microfluidic channels. This innovative system is a prime example of opening up the real-time sensitive water-world of Ab-based label free sensing to the detection of pathogens in air whereby it may be made multiplex by splitting up the microfluidic channel before detection. Such capability would potentially be very facilitating for space missions and could be remotely monitored on a computer. Moreover, the aerodynamic reach of such a system could be increased significantly by using a Venturi system for aerosol sampling [112]. In such a set-up the inlet tube used for air sampling could be reduced in diameter (from 16 mm in the original setup to about 5 mm) thus allowing the reduction of the water volume used for hydrosol formation while improving vapor collection which could lead to a higher concentration of hydrosols in the microfluidic channels, improve the detection limit and limit water use. Figure 2 shows a schematic of this futuristic device. Evidently, the device depicted can transfer information to a SBD for data processing while stationary, a system which was used by the authors for data processing, or could even be used as a portable device due to the reduced need of water for hydrosol formation.
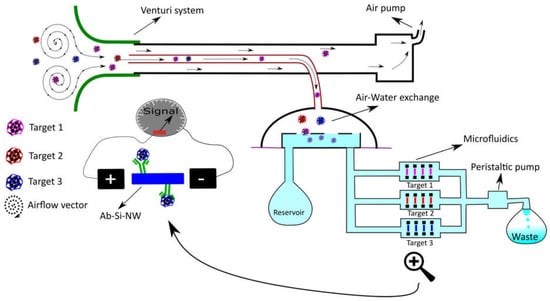
Figure 2.
Schematic representation of a multiplex system for pathogen detection in air. The system depicted here follows the same principle as suggested in [100]. To this principle we added a presentation of enhanced aerodynamic reach as developed by [102] in combination with a simple microfluidic system to reach multiplex detection of several targets (as presented by pink, red and blue color particles).
2.4. Food Screening
Testing for microbial food contamination currently requires sample return and identification on Earth using culture-based methodology. Utilizing such a system in-flight on long missions is unfit for purpose due to the limited shelf life and mass of the consumables and other methods should be considered (https://go.nasa.gov/2P5XdUK). A recent review has already discussed the subject of SBDs for the screening of food quite thoroughly [19]. The most relevant articles on the testing of food freshness and screening for pathogens in food by SBDs are highlighted. This selection was chosen, excluding allergens and chemicals since the detection of these could be done prior to the mission whilst problems due to contamination with pathogens can emerge during the mission. Regarding food freshness most SBDs are used to determine the quality of fresh fruits using portable spectroscopy [113,114,115]. However, one interesting article focuses on the discrimination and semi-quantification of volatile amines emitted by microorganisms indicating rot [116]. In this article cellulose acetate membranes were spotted with 5 pH indicators. The membrane was then exposed to the amines that represent typical metabolic products from protein degradation by microorganisms. Red green blue (RGB) values were extracted and used to generate scores for a principal component analysis (PCA). The first 2 components of the PCA managed to explain over 72% of the variance (n = 4) indicating good separation of these VOCs. In a separate experiment the authors also managed to explain 85% of the variance between several biogenic amines (tyramine, putrescine, cadaverine) in a proof of principle test using analytical standards. Thus this simple test might prove useful to check the quality of rehydrated, lyophilized food after long term storage. Apart from this work there have been several reports on the detection of E. coli [117,118,119,120] and Salmonella [121] individually and E. coli and Salmonella together [122] in various fresh food products. Of these, three articles reported a detection limit at 10 CFUs or lower in real sample matrix (milk, yoghurt or egg) [117,119,121]. Thus these sensors show promise to be used on long space missions since they approach the limit of 0 CFU per food sample set in NASA-STD-3001, VOLUME 2, REVISION A.
2.5. Infectious Disease Detection
As for infectious diseases, many reports focused on the detection of infectious diseases unlikely to occur during any space mission (e.g., malaria [123,124,125], HIV [25,126,127] schistosomiasis [128,129,130], tuberculosis [131,132], and leprosies [133]). Nonetheless, these systems could be adapted for the detection of other infectious diseases. However, some reports of SBDs focused on the detection of HV [134,135,136]. Of these systems one uses fluorescent imaging as a detection method and thus there is a requirement to first label the virus particles for detection [136]. A second system is based on the detection of virus DNA by measuring the changes in optical density in a DNA-GNP solution specific for HV DNA upon HV addition. The system is promising but remains at a proof of principle stage for the moment [135]. The final system however, developed by the Ozcan group, has been thoroughly tested utilizing real clinical samples and proven to attain over 98% accuracy [134]. This system uses standard ELISA tests, in a 96 well plate. The wells are illuminated by LEDs and light is transported from each well to the smartphone camera via optical fibers. The data is then remotely interpreted using a machine-learning algorithm. Although this system is portable and well beyond the proof of principle stage, it would need further simplification to make it suitable for non-expert use upon a spacecraft. Overall, the majority of SBDs currently developed for the detection of infectious diseases focus on diseases unlikely to develop during space missions (malaria, schistosomiasis, HIV, etc.). For space missions however, the focus should be on microbial infections known to occur during space missions such as HV, urine tract infection and subcutaneous skin infections https://ntrs.nasa.gov/search.jsp?R=20140002769. In fact, a list of recommended specific infectious disease targets to screen for during deep space missions can be found in [111] and include meningococcus, pneumococcus, typhoid and several fungi.
2.6. Other, Unclassified SBDs
Many interesting technical papers describing the development of novel smartphone sensors fall within this group. Two describe the development of mobile spectrometers [137,138]. Another describes the development of an SBD with image resolution beyond the pixel size using lens free microscopy [138]. Fluorescent microscopy is evaluated also with the invention of a QD based Förster resonant energy transfer (FRET) SBD [139]. Another technique [140] describes the integration of an optical sensor into the touchscreen of smartphones by fabricating an optical waveguide just below the screen surface. The system is interesting because it allows the measurement of changes in the refractive index of liquids directly from the screen surface opening up the door to direct surface plasmon resonance (SPR) without the use of additional add-on devices. Although these SBDs can be promising in the future, none of them have been tested on real-life examples. Other SBDs, some of which show remarkable detection sensitivity, have been tested on targets from several groups. The targets and detection methods of these SBDs are illustrated in Table 2 together with their strengths and limitations.

Table 2.
A list of the SBDs that were not classified in any group since they have targets from various groups. Targets, detection methods, advantages and limitations of each SBD are highlighted.
3. Limitations of the Smartphone for In Situ Analysis in Space
3.1. Novel Recognition Elements
The RE used in a biosensor has a great influence on the price as well as the shelf life, selectivity and possibility of reuse of that sensor. The latter is especially of importance if quantitative, more expensive, SBDs are required for use during a space mission. Thus an informed choice regarding the RE to use for a certain type of SBD is paramount. The development, possible improvements and lurking pitfalls for effective sensor development using a variety of REs including antibodies [147,148], aptamers [149,150], MIPs [151,152], enzymes (divided in detection via substrate conversion [153,154] and inhibition of this conversion [155], riboswitches [156], affibodies [157] and cell-based biosensors (CBBs) [158] were recently reviewed. Table 3 lists each of these REs together with a summary of the findings of these reviews regarding the pros and cons of each RE. However, upcoming REs that can potentially entail major advancements in the development of rugged portable sensors with a longer shelf life and with expectations to be more capable to withstand harsh conditions, have not as yet been critically reviewed elsewhere and are described herein.

Table 3.
A description of REs which illustrates the advantages and limitations of each.
3.2. MIP-Aptamer Hybrids
Aptamers have been used for the construction of biosensors since SELEX was invented in the 1990s [160]. Although, the development of Aptasensors is promising, there are a few drawbacks to the system such as degradation of the aptamer by nucleases and lower affinity compared to antibodies [150]. One interesting sensor that was recently built to overcome these limits is a novel aptasensor that uses an aptamer covered with a layer of MIPs (Apta-MIPs). This RE has been developed for the electrochemical (EC) detection of PSA [161]. Briefly, a gold surface was functionalized with a PSA specific aptamer after which PSA was added to allow the aptamer-target complex to be formed. Then electro polymerization of dopamine was initiated around the complex. Finally PSA was washed away leaving the aptamer in a stable polymer layer which protects against degradation. The group showed that the LOD (1 pg/mL) was three times better than the LOD of the aptamer alone. Shortly after a similar technique was used by another group for the detection of enrofloxacin, (a fluoroquinolone antibiotic) via up-conversion fluorescence [162]. Again a very low LOD (0.04 ng/mL) was achieved as well as a good quantitation limit (0.12 ng/mL) with a relative standard deviation of 1–5%. These sensors are sensitive, stable and need less template as MIP sensors (a common obstacle for MIP production) [151], thus combining the best of 2 worlds. Finally another group showed that the polymerization of the fragments of an adenosine specific aptamer can rescue the binding of these fragments for adenosine, which was virtually absent for the free individual fragments [163]. This work opens the doors to new MIP fabrication using nucleic acids as functional monomers; the development of which is much needed to further boost MIP development for more diverse targets [151]. The basic production mechanism for Apta-MIPs is shown in Figure 3A.
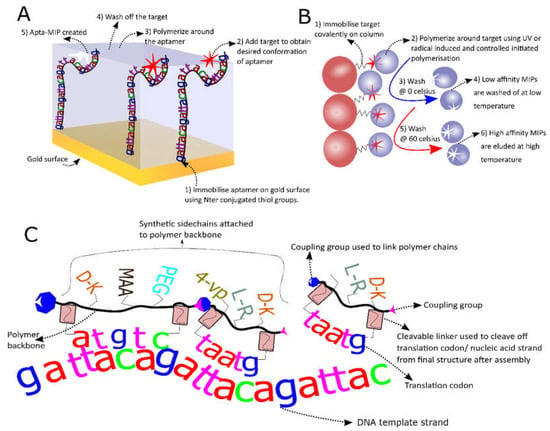
Figure 3.
Novel recognition elements (REs). (A) This Schematic shows Apta-MIP hybrid fabrication steps where the aptamer is first immobilized on a gold surface after which the target is added. Next electropolymerization is followed by a washing step to produce the final hybrid; (B) Solid phase MIP production. The principle of solid phase MIP production starts with the immobilization of the target and ends with stringent washing to eluate the high affinity MIPs after whereby step 2 can be repeated several times; (C) A schematic showing the principle of enzyme free translation of polymers. The system that can be used to achieve the enzyme free translation of polymers carrying a pool of various side chains (here methacrylic acid (MAA) polyethylenglycol (PEG), 4-vinylpyridine (4-VP) and some amino acids with varying chirality are shown but others can be used).
3.3. Solid Phase MIPs
The use of MIPs is summarized in Table 3 and more information can be found in reviews [151,152]. However, one particular type of MIP, which is believed to be especially interesting for SBD development (Solid phase MIPs or MIP nanoparticles (MIP-NPs)), was not treated in any identified review. Thus, these will be reviewed here in more detail. NP-MIPs are produced by covalently fixing the template molecule to a substrate (glass beads) after which initiated polymerization occurs around this fixed template. These “plastic antibodies” were first developed by Poma et al., for melamine, vancomycin and a model octapeptide [164]. MIP-NPs are promising because they show high affinity (Kd in the nM range) at a lower production cost, improved shelf life and stability compared to antibodies [165]. MIP-NPs can be further functionalized with fluorescent groups, thiol groups, electro active groups or molecules with antifouling properties for further biosensor development [166]. Figure 3B shows the basic steps for MIP-NP construction. The system was also used for the construction of MIP-NPs targeting histamine [165,167], vacomycin [168,169] and fumonisin B2 as a first MIP-NP targeting a toxin [165]. Furthermore, the performance of the latter was compared with monoclonal antibody (mAb) performance in an ELISA assay and showed a 3X lower LOD (6.1 pM for the MIP-NPs in comparison with the mAb at 25 pM) as well as an improved linearity range [170]. Moreover, the system can be automated both for MIP synthesis in solvents [164] and in water [171]. The latter is preferable if proteins or other biomolecules are used as a template since buffers can be used that conserve the natural steric conformation of the templates [170,171]. Production in organic solvents is better for small molecules where H-bridge formation between template and functional monomer is important [151]. A protocol for the development of these MIP-NPs in both media [172] as well as a protocol describing how to perform enthalpy calculations of the monomer-template complex [173] were recently published. Unfortunately, the latter is based on software (Sybyl) that is no longer available. Although these developments are very interesting it must be stated that MIP-NPs generally require more template than aptamers or antibody production. Although this problem might be overcome by the use of rational design and multiple reuse of the covalently linked template in solid phase production, laborious and costly optimization might be needed to attain this goal. Moreover, non-covalent free radical or UV initiated polymerization used for MIP-NPs can attack double bonds in the template molecule, thus covalently linking template and MIP-NP. This can potentially make it very hard to remove the template molecules containing alkene groups.
3.4. In Vitro Selection of More Diverse Polymers
In order to block nucleases and increase structural diversity, thus increasing the chance of better target recognition, aptamers are often modified with non-natural nucleic acids or peptide chains [150,174]. Recently some intriguing advances in this area have been made like the use of peptide nucleic acid (PNA), a nucleic acid sequence with a peptide backbone that cannot be degraded by nucleases nor proteases with lower salt sensitivity and greater affinity to its target nucleic acids as its DNA counter sequence [175], directly for the in vitro selection process using PNA transcription enzyme [176]. A similar technique was used by the Chaput team, which used enzymatic transcription of threose nucleic acid (TNA) for the in vitro selection of a TNA aptamer targeting thrombin [177]. The limitation of these techniques however is the enzymatic transcription step which ultimately limits the amount of possible polymers that can be used for in vitro selection. Recently, a novel system that uses enzyme free translation of non-nucleic acid polymers (NNAP) was developed [178]. This system uses a quintuplet codon, much like a T-RNA, to proximate monomers to each other using a random ss-DNA pool for template (Figure 3C). After in vitro selection the sequence of the polymer can be recuperated via a fluorescent PAGE assay and ESI-MS [178]. Although this system has not been used directly to develop an SBD yet it does hold great potential to develop more diverse polymers targeting a structurally highly diverse group of compounds more efficiently.
3.5. Cell Free Synthetic Biology, the Answer to Long-Term Storage?
Some of the issues for deep space missions and extra-terrestrial settlement are the cost and impracticality of shuttling goods to the destination. Indeed it was estimated that the transport of 1 unit of goods requires 99 additional units of mass in fuel to get the product into space [179]. Moreover, the cost is approximately $10,000 per pound payload [180]. Thus, solutions have been sought to limit payload including self-sustaining life support systems [181] and bio-printing for onsite food production [180]. To this end use of synthetic biology has often been suggested as a viable solution [179,180,182]. An interesting use of synthetic biology is cell free protein production. Here, reagents e.g., DNA, transcription and translation machinery, are mixed to produce metabolic products and proteins in a cell free reaction chamber. The main advantages of such a system are the high production rate, yield, product pureness and ease of gene editing. Post translational modified (PTM) protein production however, such as disulphide bridge formation and protein folding (paramount for antibody production), remains difficult [183]. Luckily, different strategies to surmount this problem exist, such as changing the redox potential using glutamate [184,185], or adding chaperone proteins to the reaction mix [186]. A detailed protocol for cell free protein production of reporter proteins is freely available [187]. Another big advantage of such a system, which is especially valid for deep space missions, is the possibility to transport the reagents in lyophilized form at room temperature (RT). This technique was recently used to manufacture a low cost (less than 1 $ per test), user friendly, paper based colorimetric test for the detection of Zika virus [188] as well as Ebola virus [189] from Escherichia coli based extracts. It was shown that these systems can remain stable over 1 year at RT which greatly improves storage facility. Other work from the same group also demonstrated the production of antimicrobial peptides, cancer biomarkers (HER2, CEA5), fluorescent protein (mCherry), cytokines, small molecules, Clostridium difficile exotoxin (TcdA) and several antibodies using freeze-dried cell free E. coli based extracts [185]. However, these systems remain quite novel and not all reports show similar performance. A report from Smith et al., for instance, shows that protein activity can indeed be partially preserved at sub-optimal temperatures (4 °C) although it does decrease drastically after only 90 days of storage at RT even when sucrose is added as a lyoprotectant [190]. Nonetheless the potential advantages that these techniques have for long space missions justify further development in this intriguing field.
4. Conclusions and Outlook
A plethora of SBDs for human health related purposes have been developed in recent years. These compact and novel sensors stretch from simple sensors that allow the detection of stress through heart rate measurements using photoplethysmography to sophisticated biosensors using microfluidics and isothermal nucleic acid amplification methods with single copy DNA detection limits. In order to view and improve those currently available, the cornucopia of sensors were divided into four groups based on their usefulness for the monitoring of the crew’s health and further divided into subgroups as the most promising sensors for use aboard a spacecraft on a mission to Mars. Using this classification system and building on previous work mapping out LOC use in environmental monitoring [18] and food analyses [19,20], it became apparent that several SBDs for the monitoring of stress levels, skin cancer diagnostics, water screening for toxic metals, and infectious disease monitoring have been developed well beyond the proof of principle concept. However, different targets, more fit to the needs for microbial detection in space, would be necessary for the latter as pointed out in [111]. Moreover, the development of other groups such as testing for food spoilage (other than fruit) and scanning for airborne pathogens with SBDs is clearly lagging behind while demand for miniaturized devices for astronaut crew health monitoring is rising [191]. Thus more effort should be invested in these areas. Another observation is the prevalence of antibody based SBDs, in particular LFIAs with a smartphone readout that are often mentioned. Interestingly, LFIAs were previously identified as ideal systems for use in space [191] and thus LFIA SBDs might be the logical step forward. However, other more complex formats using unconventional REs have also been described herein as interesting alternatives. In particular MIP-NPs, MIP-Aptas and NNAPs have been identified as promising REs for space missions due to their increased stability, shelf life and protection against degradation. In summary, it is believed that several of the currently available SBDs could in theory be adapted for the use in space and aid in the monitoring of the crew’s health on a mission to Mars in a user-friendly fashion as light portable devices. When peering into the future, one could imagine an ideal SBD adapted for a prolonged space mission. This “galactic” SBD, would have to be equipped with a maximum number of sensors that are able to withstand harsh environments, without losing sensitivity, for a prolonged time as a primary requirement. Moreover, such sensors would ideally be integrated in the phone to maximize space, use shelf-stable components, have a user friendly “one step only” approach and work in (near) real-time to facilitate rapid and easy use by non-experts. A good sensor for the galactic, that potentially meets these requirements, is an integrated optical waveguide in the smartphone screen [140]. Such a system potentially allows direct multiplex SPR measurements on the screen. Covalently attached MIP-NPs could be used to enable sensitive sensing without degradation caused by proteases, temperature and pH variations and mechanical stress associated with touch screen use. This would in theory enable label free detection of multiple harmless analytes such as stress or cancer related biomarkers, by simply depositing a drop of sample on the indicated area of the screen. A limit for this technique would be the molecular weight of the target, which is correlated with the generated refractive index change upon binding. Moreover, it would be unfit to screen for pathogens. For the direct detection of smaller analytes an aptasensor, with electrochemical detection, could be used. Such a system is advantageous for small compounds since its sensing capabilities are based on the conformational change of the aptamer making the weight/electric resistance of the analyte less important. Moreover, a rapid one step approach can be achieved if a signal enhancer like methylene blue is attached to the aptamer [192]. Again, a stable synthetic RE like an NNAP would avoid degradation by proteases and nucleases and might enable direct analyses at the surface. An add-on device could be used for this purpose and could further be equipped with a VIS-NIR spectrometer/camera to allow spectrum analyses over a larger spectrum range and allow for the detection of more abundant compounds [115]. Moreover, the camera could be used for optical detection of a cornucopia of microfluidic assays such as fluorescent imaging cytometry or microfluidic dielectrophoresis for cell counting, magnetic bead ELISA for the detection of cancer or infectious disease markers or pathogens and an aerosol/hydrosol exchange system for the detection of pathogens in air. Microfluidic cassettes could be designed using a capillary plug-in so that multiple tests can be performed on the galactic by simply changing the cassette. This way the galactic could be made ready to perform even more tasks needed upon the arrival at Mars such as performing preliminary scans for extra-terrestrial life, or detecting the presence of essential nutrients or contaminants to realize Martian crop growth. Figure 4 shows an illustration of this futuristic sensing device indicating some possibilities for multiplex sensing. The envisaged compactness of such a system, combined with the vast arsenal of possible types of analyses that could be conducted by such a device, could greatly reduce the amount of space needed aboard. Moreover, analysis time, and thus workload (an ever persisting pressure on the crew), could be greatly reduced due to the multiplex nature of the “galactic” combined with the one-step only approaches mentioned. Keeping tabs on the concentration of multiple biomarkers in-flight might also improve our understanding of the effects of space travel on several biological phenomena and be beneficial for future missions. Thus such a system potentially provides interwoven benefits for the actual crew travelling to the faraway destination, and for those who will follow. Evidently, this ideal galactic sensing system remains fictional and may seem farfetched. However, technological advancement in the biotechnology sector is moving incredibly fast and the foundations for such a device have already been laid. This means that it might be more a question of perseverance and the will to dream and dare to turn this idea into a reality than anything else.
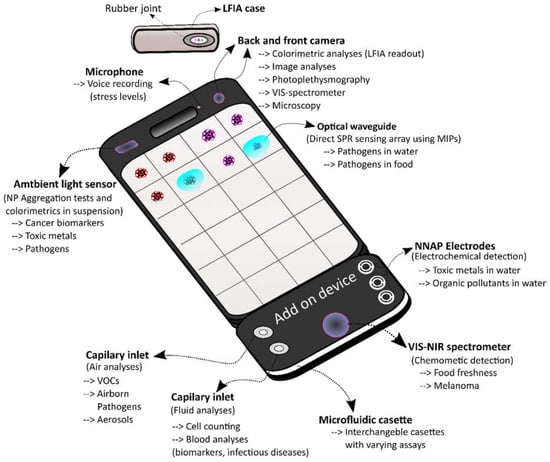
Figure 4.
The galactic SBD. This figure shows a futuristic SBD that incorporates novel sensing technology with synthetic REs directly into the smartphone as well as using an add-on device equipped with interchangeable microfluidic cassettes. Possible uses and basic functioning mechanisms of these sensors are indicated.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 720325.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hawkey, A. Physiological and biomechanical considerations for a human Mars mission. J. Br. Interplanet. Soc. 2005, 58, 117–130. [Google Scholar]
- Ade, C.J.; Broxterman, R.M.; Moore, A.D.; Barstow, T.J. Decreases in maximal oxygen uptake following long-duration spaceflight: Role of convective and diffusive O2 transport mechanisms. J. Appl. Physiol. 2017, 122, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Drinnan, N.R.; de Juniac, A.B. The effects of microgravity on the urological system: A review. J. Clin. Urol. 2013, 6, 391–394. [Google Scholar] [CrossRef]
- Human Research Program Integrated Research Plan HRP 47065. Available online: https://www.nasa.gov/pdf/651214main_hrp47065_revc_IRP.pdf (accessed on 28 June 2011).
- Pierson, D.L. Microbial contamination of spacecraft. Graviatational Space Biol. Bull. 2001, 14, 1–6. [Google Scholar]
- Horneck, G.; Klaus, D.M.; Mancinelli, R.L. Space Microbiology. Microbiol. Mol. Biol. Rev. 2010, 74, 121–156. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.W. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect. Drug Resist. 2015, 8, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, C.A.; Ott, C.M.; Wilson, J.W.; Ramamurthy, R.; Pierson, D.L. Microbial Responses to Microgravity and Other Low-Shear Environments. Microbiol. Mol. Biol. Rev. 2004, 68, 345–361. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.K.; Laudenslager, M.L.; Stowe, R.P.; Crucian, B.E.; Feiveson, A.H.; Sams, C.F.; Pierson, D.L. Latent virus reactivation in astronauts on the international space station. Npj Microgr. 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.K.; Crucian, B.E.; Stowe, R.P.; Simpson, R.J.; Ott, C.M.; Sams, C.F.; Pierson, D.L. Reactivation of latent viruses is associated with increased plasma cytokines in astronauts. Cytokine 2013, 61, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Roberts, M.; Castro, S.; Oubre, C.; Makimura, K.; Leys, N.; Grohmann, E.; Sugita, T.; Ichijo, T.; Nasu, M. Microbial Monitoring of Crewed Habitats in Space—Current Status and Future Perspectives. Microbes Environ. 2014, 29, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Boguraev, A.-S.; Christensen, H.C.; Bonneau, A.R.; Pezza, J.A.; Nichols, N.M.; Giraldez, A.J.; Gray, M.M.; Wagner, B.M.; Aken, J.T.; Foley, K.D.; et al. Successful amplification of DNA aboard the International Space Station. Npj Microgr. 2017, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Castro-Wallace, S.L.; Chiu, C.Y.; John, K.K.; Stahl, S.E.; Rubins, K.H.; McIntyre, A.B.R.; Dworkin, J.P.; Lupisella, M.L.; Smith, D.J.; Botkin, D.J.; et al. Nanopore DNA Sequencing and Genome Assembly on the International Space Station. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.C.; Damon, M.; Maule, J.; Monaco, L.A.; Wainwright, N. Rapid Culture-Independent Microbial Analysis Aboard the International Space Station (ISS) Stage Two: Quantifying Three Microbial Biomarkers. Astrobiology 2012, 12, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Mora, M.F.; Greer, F.; Stockton, A.M.; Bryant, S.; Willis, P.A. Toward Total Automation of Microfluidics for Extraterrestrial in Situ Analysis. Anal. Chem. 2011, 83, 8636. [Google Scholar] [CrossRef] [PubMed]
- Parro, V.; de Diego-Castilla, G.; Rodríguez-Manfredi, J.A.; Rivas, L.A.; Blanco-López, Y.; Sebastián, E.; Romeral, J.; Compostizo, C.; Herrero, P.L.; García-Marín, A.; et al. SOLID3: A Multiplex Antibody Microarray-Based Optical Sensor Instrument for In Situ Life Detection in Planetary Exploration. Astrobiology 2011, 11, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Carr, C.E.; Mojarro, A.; Hachey, J.; Saboda, K.; Tani, J.; Bhattaru, S.A.; Smith, A.; Pontefract, A.; Zuber, M.T.; Doebler, R.; et al. Towards in situ sequencing for life detection. In Proceedings of the 2017 IEEE Aerospace Conference, Big Sky, MT, USA, 4–11 March 2017. [Google Scholar] [CrossRef]
- Pol, R.; Céspedes, F.; Gabriel, D.; Baeza, M. Microfluidic lab-on-a-chip platforms for environmental monitoring. TrAC Trends Anal. Chem. 2017, 95, 62–68. [Google Scholar] [CrossRef]
- Rateni, G.; Dario, P.; Cavallo, F. Smartphone-based food diagnostic technologies: A review. Sensors (Switzerland) 2017, 17, 1453. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.Y.; Kim, B. Lab-on-a-chip pathogen sensors for food safety. Sensors (Switzerland) 2012, 12, 10713–10741. [Google Scholar] [CrossRef] [PubMed]
- Maule, J.; Wainwright, N.; Steele, A.; Monaco, L.; Morris, H.; Gunter, D.; Flroes, G.; Effinger, M.; Damon, M.; Wells, M.; et al. LOCAD-PTS: Operation of a new system for microbial monitoring aboard the International Space Station (ISS). In Proceedings of the AIAA SPACE 2008 Conference & Exposition, San Diego, CA, USA, 9–11 September 2008; pp. 1–9. [Google Scholar] [CrossRef]
- Van Houdt, R.; Mijnendonckx, K.; Leys, N. Microbial contamination monitoring and control during human space missions. Planet. Space Sci. 2012, 60, 115–120. [Google Scholar] [CrossRef]
- Long, K.D.; Woodburn, E.V.; Le, H.M.; Shah, U.K.; Lumetta, S.S.; Cunningham, B.T. Multimode smartphone biosensing: The transmission, reflection, and intensity spectral (TRI)-analyzer. Lab Chip 2017, 17, 3246–3257. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, S.K.J.; Tokarski, C.; Lang, S.N.; Van Ginkel, L.A.; Zhu, H.; Ozcan, A.; Nielen, M.W.F. Calling biomarkers in milk using a protein microarray on your smartphone. PLoS ONE 2015, 10, e134360. [Google Scholar] [CrossRef] [PubMed]
- Laksanasopin, T.; Guo, T.W.; Nayak, S.; Sridhara, A.A.; Xie, S.; Olowookere, O.O.; Cadinu, P.; Meng, F.; Chee, N.H.; Kim, J.; et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci. Transl. Med. 2015, 7, 273re1. [Google Scholar] [CrossRef] [PubMed]
- Priye, A.; Bird, S.W.; Light, Y.K.; Ball, C.S.; Negrete, O.A.; Meagher, R.J. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Maule, J.; Fogel, M.; Steele, A.; Wainwright, N.; Pierson, D.L.; McKay, D.S. Antibody binding in altered gravity: Implications for immunosorbent assay during space flight. J. Gravit. Physiol. 2003, 10, 47–55. [Google Scholar] [PubMed]
- Korth, D.W. Exercise Countermeasure Hardware Evolution on ISS: The First Decade. Aerosp. Med. Hum. Perform. 2015, 86, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.; Fortunato, A.; Wolff, M.; Oliveira, D.M. mobiPV: A new, wearable real-time collaboration software for Astronauts using mobile computing solutions. In Proceedings of the SpaceOps 2016 Conference, Daejeon, Korea, 16–20 May 2016; pp. 1–10. [Google Scholar] [CrossRef]
- Li, Z.; Nambiar, S.; Zheng, W.; Yeow, J.T.W. PDMS/single-walled carbon nanotube composite for proton radiation shielding in space applications. Mater. Lett. 2013, 108, 79–83. [Google Scholar] [CrossRef]
- Li, Z.; Chen, S.; Nambiar, S.; Sun, Y.; Zhang, M.; Zheng, W.; Yeow, J.T.W. PMMA/MWCNT nanocomposite for proton radiation shielding applications. Nanotechnology 2016, 27, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Atxaga, G.; Marcos, J.; Jurado, M.; Carapelle, A.; Orava, R. Radiation Shielding of Composite Space Enclosures. Available online: https://orbi.uliege.be/bitstream/2268/132394/1/IAC-12%2CC2%2C6%2C6%2Cx13735.pdf (accessed on 1 June 2012).
- de Diego-Castilla, G.; Cruz-Gil, P.; Mateo-Martí, E.; Fernández-Calvo, P.; Rivas, L.A.; Parro, V. Assessing Antibody Microarrays for Space Missions: Effect of Long-Term Storage, Gamma Radiation, and Temperature Shifts on Printed and Fluorescently Labeled Antibodies. Astrobiology 2011, 11, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, S.; Berlenbach, P.; Langenfelder, S.; Hörl, D.; Lehn, N.; Hiller, K.A.; Schmalz, G.; Durchschlag, H. Integrity of proteins in human saliva after sterilization by gamma irradiation. Appl. Environ. Microbiol. 2011, 77, 749–755. [Google Scholar] [CrossRef] [PubMed]
- Coussot, G.; Moreau, T.; Faye, C.; Vigier, F.; Baqué, M.; Le Postollec, A.; Incerti, S.; Dobrijevic, M.; Vandenabeele-Trambouze, O. Biochip-based instruments development for space exploration: Influence of the antibody immobilization process on the biochip resistance to freeze-drying, temperature shifts and cosmic radiations. Int. J. Astrobiol. 2016, 16, 190–199. [Google Scholar] [CrossRef]
- Baqué, M.; Le Postollec, A.; Ravelet, C.; Peyrin, E.; Coussot, G.; Desvignes, I.; Incerti, S.; Moretto, P.; Dobrijevic, M.; Vandenabeele-Trambouze, O. Investigation of Low-Energy Proton Effects on Aptamer Performance for Astrobiological Applications. Astrobiology 2011, 11, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, C.; Hassler, D.; Cucinotta, F.A.; Ehresmann, B.; Wimmer-Schweingruber, R.F.; Brinza, D.E.; Kang, S. Measurements of Energetic Particle Radiatino in Transit to Mars on the Mars Science Laboratory. Am. Assoc. Adv. Sci. 2013. [Google Scholar] [CrossRef]
- Carr, C.E.; Rowedder, H.; Vafadari, C.; Lui, C.S.; Cascio, E.; Zuber, M.T.; Ruvkun, G. Radiation Resistance of Biological Reagents for In Situ Life Detection. Astrobiology 2013, 13, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Choo, K.-Y.; Ling, H.-C.; Lo, Y.-C.; Yap, Z.-H.; Pua, J.-S.; Phan, R.C.-W.; Goh, V.-T. Android based self-diagnostic electrocardiogram system for mobile healthcare. Technol. Heal. Care 2015, 23, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, P.; Garde, A.; Karlen, W.; Petersen, C.L.; Wensley, D.; Dumont, G.A.; Mark Ansermino, J. Evaluation of cardiac modulation in children in response to apnea/hypopnea using the Phone OximeterTM. Physiol. Meas. 2016, 37, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kwon, S.; Yoo, C.; Seo, S.; Park, K.; Song, J.; Lee, Y. Sinabro: Opportunistic and unobtrusive mobile electrocardiogram monitoring system. Assoc. Comput. Mach. 2014, 15, 1–6. [Google Scholar] [CrossRef]
- Kennedy, A.P.; Epstein, D.H.; Jobes, M.L.; Agage, D.; Tyburski, M.; Phillips, K.A.; Ali, A.A.; Bari, R.; Hossain, S.M.; Hovsepian, K.; et al. Continuous in-the-field measurement of heart rate: Correlates of drug use, craving, stress, and mood in polydrug users. Drug Alcohol Depend. 2015, 151, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Muhlestein, J.B.; Le, V.; Albert, D.; Moreno, F.L.; Anderson, J.L.; Yanowitz, F.; Vranian, R.B.; Barsness, G.W.; Bethea, C.F.; Severance, H.W.; et al. Smartphone ECG for evaluation of STEMI: Results of the ST LEUIS Pilot Study. J. Electrocardiol. 2015, 48, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Pierleoni, P.; Pernini, L.; Belli, A.; Palma, L. An android-based heart monitoring system for the elderly and for patients with heart disease. Int. J. Telemed. Appl. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rachim, V.P.; Chung, W.-Y. Wearable Noncontact Armband for Mobile ECG Monitoring System. IEEE Trans. Biomed. Circuits Syst. 2016, 10, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Sinddhuja, A.K.; Mounika, M.; Dass, P. A heartbeat and temperature measuring system for remote health monitoring using gsm technology. Int. J. Pharm. Technol. 2016, 8, 20847–20855. [Google Scholar] [CrossRef]
- Agarwal, T.; Bandivadekar, P.; Satpathy, G.; Sharma, N.; Titiyal, J.S. Detection of fungal hyphae using smartphone and pocket magnifier: Going cellular. Cornea 2015, 34, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, L.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Microbial Spoilage of Foods: Fundamentals. Microbiol. Qual. Food Foodborne Spoilers 2016, 1–21. [Google Scholar] [CrossRef]
- Escrivá, L.; Font, G.; Manyes, L. In vivo toxicity studies of fusarium mycotoxins in the last decade: A review. Food Chem. Toxicol. 2015, 78, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.X.H.; Liu, F.S.F.; Yu, H.-Z. Mobile app-based quantitative scanometric analysis. Anal. Chem. 2014, 86, 11966–11971. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aztiria, A.; Basarab, A. Towards an automatic early stress recognition system for office environments based on multimodal measurements: A review. J. Biomed. Inform. 2016, 59, 49–75. [Google Scholar] [CrossRef] [PubMed]
- Zangheri, M.; Cevenini, L.; Anfossi, L.; Baggiani, C.; Simoni, P.; Di Nardo, F.; Roda, A. A simple and compact smartphone accessory for quantitative chemiluminescence-based lateral flow immunoassay for salivary cortisol detection. Biosens. Bioelectron. 2015, 64, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, A.; Pioggia, G.; Tartarisco, G.; Baldus, G.; Ferro, M.; Cipresso, P.; Serino, S.; Popleteev, A.; Gabrielli, S.; Maimone, R.; et al. A system for automatic detection of momentary stress in naturalistic settings. Annu. Rev. CyberTherapy Telemed. 2012, 10, 182–186. [Google Scholar] [CrossRef]
- Muaremi, A.; Arnrich, B.; Tröster, G. Towards Measuring Stress with Smartphones and Wearable Devices During Workday and Sleep. Bionanoscience 2013, 3, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Kizakevich, P.N.; Hubal, R.; Brown, J.; Lyden, J.; Spira, J.; Eckhoff, R.; Zhang, Y.; Bryant, S.; Munoz, G. PHIT for duty, a mobile approach for psychological health intervention. Annu. Rev. CyberTherapy Telemed. 2012, 10, 268–272. [Google Scholar] [CrossRef]
- Gregoski, M.J.; Vertegel, A.; Shaporev, A.; Treiber, F.A. Tension tamer: Delivering meditation with objective heart rate acquisition for adherence monitoring using a smart phone platform. J. Altern. Complement. Med. 2013, 19, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Doule, O.; Poulet, L. Ergonomy of Head Mounted Displays Inside Analog. In Proceedings of the AIAA SPACE 2014 Conference and Exposition, San Diego, CA, USA, 4–7 August 2014; pp. 1–26. [Google Scholar] [CrossRef]
- Chintamani, K.; Lierde, B.V.; Maloney, S.; Kiernan, P. Wearable crew support technology on the International Space Station: The mobile Procedure Viewer (mobiPV). HFES Eur. 2014, 4959, 1–11. [Google Scholar]
- Benjamin, C.L.; Stowe, R.P.; St. John, L.; Sams, C.F.; Mehta, S.K.; Crucian, B.E.; Pierson, D.L.; Komanduri, K.V. Decreases in thymopoiesis of astronauts returning from space flight. JCI Insight 2016, 1, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Geiger, A.M.; Pitts, K.P.; Feldkamp, J.; Kirschbaum, C.; Wolf, J.M. Cortisol-dependent stress effects on cell distribution in healthy individuals and individuals suffering from chronic adrenal insufficiency. Brain. Behav. Immun. 2015, 50, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Mavandadi, S.; Coskun, A.F.; Yaglidere, O.; Ozcan, A. Optofluidic fluorescent imaging cytometry on a cell phone. Anal. Chem. 2011, 83, 6641–6647. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tasoglu, S.; Chen, P.Z.; Chen, M.; Akbas, R.; Wach, S.; Ozdemir, C.I.; Gurkan, U.A.; Giguel, F.F.; Kuritzkes, D.R.; et al. Micro-a-fluidics ELISA for rapid CD4 cell count at the point-of-care. Sci. Rep. 2014, 4, 3796. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; McCoy, T.; Gazda, D.; Morgan, J.L.L.; Heer, M.; Zwart, S.R. Space flight calcium: Implications for astronaut health, spacecraft operations, and Earth. Nutrients 2012, 4, 2047–2068. [Google Scholar] [CrossRef] [PubMed]
- Manuscript, A.; Blood, W.; Count, C. Vitamin D and the Immune System. J. Investig. Med. 2009, 49, 1841–1850. [Google Scholar] [CrossRef]
- Lee, S.; Oncescu, V.; Mancuso, M.; Mehta, S.; Erickson, D. A smartphone platform for the quantification of vitamin D levels. Lab Chip Miniaturisation Chem. Biol. 2014, 14, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.C. 3D printable retinal imaging adapter for smartphones could go global. Graefe’s Arch. Clin. Exp. Ophthalmol. 2015, 253, 1831–1833. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.E.; Koh, J.; Lin, J.; Di Carlo, D. Research highlights: Translating chips. Lab Chip 2015, 15, 1984–1988. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Schimmerling, W.; Wilson, J.W.; Peterson, L.E.; Badhwar, G.D.; Saganti, P.B.; Dicello, J.F. Space radiation cancer risks and uncertainties for Mars missions. Radiat. Res. 2001, 156, 682–688. [Google Scholar] [CrossRef]
- Fink, C.A.; Bates, M.N.; Fink, C.A.; Bates, M.N. Melanoma and Ionizing Radiation: Is There a Causal Relationship? Radiat. Res. 2005, 164, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.Y.; George, K.A.; Cucinotta, F.A. Evaluation of skin cancer risk for lunar and Mars missions. Adv. Space Res. 2006, 37, 1798–1803. [Google Scholar] [CrossRef]
- Ramlakhan, K.; Shang, Y. A mobile automated skin lesion classification system. In Proceedings of the International Conference on Tools with Artificial Intelligence (ICTAI), Boca Raton, FL, USA, 7–9 November 2011; Volume 23, pp. 138–141. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Zouridakis, G.; Wadhawan, T.; Situ, N.; Hu, R.; Yuan, X.; Lancaster, K.; Queen, C.M. Melanoma and other skin lesion detection using smart handheld devices. Methods Mol. Biol. 2015, 1256, 459–496. [Google Scholar] [CrossRef] [PubMed]
- Wadhawan, T.; Situ, N.; Lancaster, K.; Yuan, X.; Zouridakis, G. SkinScan©: A portable library for melanoma detection on handheld devices. In Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Chicago, IL, USA, 30 Match–2 April 2011; Volume 11, pp. 133–136. [Google Scholar] [CrossRef]
- Oskouei, S.S.L.; Golestani, H.; Hashemi, M.; Ghiasi, S. CNNdroid: GPU-Accelerated Execution of Trained Deep Convolutional Neural Networks on Android. AC. ISBM 2015, 1201–1205. [Google Scholar] [CrossRef]
- Gregg, D.; Ionica, M.H. The Movidius Myriad Archetecture’s Potential for Scientific Computing. IEEE Micro 2009, 6–14. [Google Scholar] [CrossRef]
- Rallapalli, S.; Qiu, H.; Bency, A.J.; Karthikeyan, S.; Govindan, R.; Urgaonkar, R. Are Very Deep Neural Networks Feasible on Mobile Devices? In Usc Conference Proceedings; USC University of Southern California: Oakland, CA, USA, 2015. [Google Scholar]
- Jahan-Tigh, R.R.; Chinn, G.M.; Rapini, R.P. A comparative study between smartphone-based microscopy and conventional light microscopy in 1021 dermatopathology specimens. Arch. Pathol. Lab. Med. 2016, 140, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Swedish, T.; Wahi, A.; Moufarrej, M.; Noland, M.; Gurry, T.; Aranda-Michel, E.; Aksel, D.; Wagh, S.; Sadashivaiah, V.; et al. Mobile phone based mini-spectrometer for rapid screening of skin cancer. Proc. SPIE 2015, 9482, 1–5. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, X.; Khimji, I.; Akbas, R.; Qiu, W.; Edwards, D.; Cramer, D.W.; Ye, B.; Demirci, U. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip 2011, 11, 3411–3418. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Akbas, R.; Demirci, U. Microchip ELISA coupled with cell phone to detect ovarian cancer HE4 biomarker in urine. Methods Mol. Biol. 2015, 1256, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Adel Ahmed, H.; Azzazy, H.M.E. Power-free chip enzyme immunoassay for detection of prostate specific antigen (PSA) in serum. Biosens. Bioelectron. 2013, 49, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Long, K.D.; Yu, H.; Cunningham, B.T. Smartphone instrument for portable enzymelinked immunosorbent assays. Biomed. Opt. Express 2014, 5, 3792–3806. [Google Scholar] [CrossRef] [PubMed]
- Aslan, M.K.; Kulah, H. Android based portable cell counting system for label free quantification of dep manipulated cancer cells. In Proceedings of the 19th International Conference Solid-State Sensors, Kaohsiung, Taiwan, 19–22 June 2017; Volume 17, pp. 556–559. [Google Scholar] [CrossRef]
- Archibong, E.; Konnaiyan, K.R.; Kaplan, H.; Pyayt, A. A mobile phone-based approach to detection of hemolysis. Biosens. Bioelectron. 2017, 88, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Felton, E.J.; Velasquez, A.; Lu, S.; Murphy, R.O.; Elkhal, A.; Mazor, O.; Gorelik, P.; Sharda, A.; Ghiran, I.C. Detection and quantification of subtle changes in red blood cell density using a cell phone. Lab Chip 2016, 16, 3286–3295. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Gu, C.; Li, C.; Lin, J. Doppler radar noncontact vital sign monitoring. Neural Comput. Neural Devices Neural Prosthes. 2014, 41–62. [Google Scholar] [CrossRef]
- Reyes, B.A.; Reljin, N.; Kong, Y.; Nam, Y.; Ha, S.; Chon, K.H. Employing an incentive spirometer to calibrate tidal volumes estimated from a smartphone camera. Sensors (Switzerland) 2016, 16, 397. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wu, J.; Peretz-Soroka, H.; Zhu, L.; Li, Z.; Sang, Y.; Hipolito, J.; Zhang, M.; Santos, S.; Hillier, C.; et al. Mkit: A cell migration assay based on microfluidic device and smartphone. Biosens. Bioelectron. 2018, 99, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, M.; Nemade, H.B.; Bandyopadhyay, D. Nano-enabled paper humidity sensor for mobile based point-of-care lung function monitoring. Biosens. Bioelectron. 2017, 94, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.L.; Ouyang, Y.; Li, J.; Krauss, S.T.; Shukla, N.; Kessel, B.G.; Haverstick, D.M.; Garner, G.T.; Landers, J.P. Protein quantitation from whole blood on polyester-toner laser-printed microfluidic discs with cell phone image analysis. In Proceedings of the 18th International Conference Miniaturized Systems for Chemistry & Life Science MicroTAS, San Antonio, TX, USA, 26–30 October 2014; pp. 1434–1436. [Google Scholar]
- Harder, R.; Diedrich, A.; Whitfield, J.S.; Buchowski, M.S.; Pietsch, J.B.; Baudenbacher, F.J. Smart Multi-Frequency Bioelectrical Impedance Spectrometer for BIA and BIVA Applications. IEEE Trans. Biomed. Circuits Syst. 2016, 10, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Patnaik, R.; Kuhlmann, K.; Rai, A.J.; Sia, S.K. Smartphone dongle for simultaneous measurement of hemoglobin concentration and detection of HIV antibodies. Lab Chip 2015, 15, 3514–3520. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, H.; Dong, T. Smartphone-Based Rapid Screening of Urinary Biomarkers. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Dweik, B.; Argun, A.; Tempelman, L.; Mackenzie, N.; Forchione, J.; Hamdan, M. Portable Sensor for Rapid Measurement of Trace Toxic Metals in Water. Techport NASA 2015, 1–4. [Google Scholar]
- Sun, H.; Li, W.; Dong, Z.-Z.; Hu, C.; Leung, C.-H.; Ma, D.-L.; Ren, K. A suspending-droplet mode paper-based microfluidic platform for low-cost, rapid, and convenient detection of lead(II) ions in liquid solution. Biosens. Bioelectron. 2018, 99, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-H.; Tseng, W.-L. Ultrasensitive detection of target analyte-induced aggregation of gold nanoparticles using laser-induced nanoparticle Rayleigh scattering. Talanta 2015, 132, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Xiao, M.; Fu, Q.; Yu, S.; Shen, H.; Bian, H.; Tang, Y. A portable smart-phone readout device for the detection of mercury contamination based on an aptamer-assay nanosensor. Sensors (Switzerland) 2016, 16, 1871. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-H.; Chen, W.-Y.; Yen, Y.-C.; Wang, C.-W.; Chang, H.-T.; Chen, C.-F. Detection of mercury(II) ions using colorimetric gold nanoparticles on paper-based analytical devices. Anal. Chem. 2014, 86, 6843–6849. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, B.; Xu, F.; Shi, X.; Feng, D.; Wei, D.; Li, Y.; Feng, Y.; Wang, Y.; Jia, D.; et al. High-yield synthesis of strong photoluminescent N-doped carbon nanodots derived from hydrosoluble chitosan for mercury ion sensing via smartphone APP. Biosens. Bioelectron. 2016, 79, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Levin, S.; Krishnan, S.; Rajkumar, S.; Halery, N.; Balkunde, P. Monitoring of fluoride in water samples using a smartphone. Sci. Total Environ. 2016, 551–552, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.-J.; Wei, J.-F.; Xu, J.-R.; Wang, Y.-H.; Zheng, G.-X. Paper-based three-dimensional microfluidic device for monitoring of heavy metals with a camera cell phone. Anal. Bioanal. Chem. 2014, 406, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gartia, M.R.; Jiang, J.; Chang, T.-W.; Qian, J.; Liu, Y.; Liu, X.; Liu, G.L. Audio jack based miniaturized mobile phone electrochemical sensing platform. Sens. Actuators B Chem. 2015, 209, 677–685. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Bao, X.; Han, J.; Xia, J.; Tian, X.; Ni, L. A smartphone-based colorimetric reader coupled with a remote server for rapid on-site catechols analysis. Talanta 2016, 160, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Sicard, C.; Glen, C.; Aubie, B.; Wallace, D.; Jahanshahi-Anbuhi, S.; Pennings, K.; Daigger, G.T.; Pelton, R.; Brennan, J.D.; Filipe, C.D.M. Tools for water quality monitoring and mapping using paper-based sensors and cell phones. Water Res. 2015, 70, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, N.; Lukac, M.; Ahmed, T.; Kar, A.; Praveen, P.S.; Honles, T.; Leong, I.; Rehman, I.H.; Schauer, J.J.; Ramanathan, V. A cellphone based system for large-scale monitoring of black carbon. Atmos. Environ. 2011, 45, 4481–4487. [Google Scholar] [CrossRef]
- Wu, Y.-C.; Shiledar, A.; Li, Y.-C.; Wong, J.; Feng, S.; Chen, X.; Chen, C.; Jin, K.; Janamian, S.; Yang, Z.; et al. Air quality monitoring using mobile microscopy and machine learning. Light Sci. Appl. 2017, 6, e17046. [Google Scholar] [CrossRef] [PubMed]
- Mirowsky, J.; Hickey, C.; Horton, L.; Blaustein, M.; Galdanes, K.; Peltier, R.E.; Chillrud, S.; Chen, L.C.; Ross, J.; Nadas, A.; et al. The effect of particle size, location and season on the toxicity of urban and rural particulate matter. Inhal. Toxicol. 2013, 25, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tsow, F.; Campbell, K.D.; Iglesias, R.; Forzani, E.; Tao, N. A wireless hybrid chemical sensor for detection of environmental volatile organic compounds. IEEE Sens. J. 2013, 13, 1748–1755. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Tan, M.; Wang, Z.; Yao, M.; Xu, Z.; Wu, Y.; Wang, J.; Guo, X.; Zhu, T. Integrating silicon nanowire field effect transistor, microfluidics and air sampling techniques for real-time monitoring biological aerosols. Environ. Sci. Technol. 2011, 45, 7473–7480. [Google Scholar] [CrossRef] [PubMed]
- Mermel, L.A. Infection prevention and control during prolonged human space travel. Clin. Infect. Dis. 2013, 56, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Forbes, T.P.; Staymates, M. Enhanced aerodynamic reach of vapor and aerosol sampling for real-time mass spectrometric detection using Venturi-assisted entrainment and ionization. Anal. Chim. Acta 2017, 957, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Das, A.J.; Wahi, A.; Kothari, I.; Raskar, R. Ultra-portable, wireless smartphone spectrometer for rapid, non-destructive testing of fruit ripeness. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Intaravanne, Y.; Sumriddetchkajorn, S.; Nukeaw, J. Cell phone-based two-dimensional spectral analysis for banana ripeness estimation. Sens. Actuators B Chem. 2012, 168, 390–394. [Google Scholar] [CrossRef]
- Yu, X.; Lu, Q.; Gao, H.; Ding, H. Development of a handheld spectrometer based on a linear variable filter and a complementary metal-oxide-semiconductor detector for measuring the internal quality of fruit. J. Near Infrared Spectrosc. 2016, 24, 69–76. [Google Scholar] [CrossRef]
- Bueno, L.; Meloni, G.N.; Reddy, S.M.; Paixão, T.R.L.C. Use of plastic-based analytical device, smartphone and chemometric tools to discriminate amines. RSC Adv. 2015, 5, 20148–20154. [Google Scholar] [CrossRef]
- Zeinhom, M.M.A.; Wang, Y.; Song, Y.; Zhu, M.-J.; Lin, Y.; Du, D. A portable smart-phone device for rapid and sensitive detection of E. coli O157:H7 in Yoghurt and Egg. Biosens. Bioelectron. 2018, 99, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Borysiak, M.D.; Kimura, K.W.; Posner, J.D. NAIL: Nucleic Acid detection using Isotachophoresis and Loop-mediated isothermal amplification. Lab Chip 2015, 15, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Sikora, U.; Ozcan, A. Quantum dot enabled detection of Escherichia coli using a cell-phone. Analyst 2012, 137, 2541. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.-S.; Park, T.S.; Yoon, J.-Y. Rapid and reagentless detection of microbial contamination within meat utilizing a smartphone-based biosensor. Sci. Rep. 2014, 4, 5953. [Google Scholar] [CrossRef] [PubMed]
- Park, T.S.; Li, W.; McCracken, K.E.; Yoon, J.-Y. Smartphone quantifies Salmonella from paper microfluidics. Lab Chip 2013, 13, 4832–4840. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, V.K.; Bakthavathsalam, P.; Jaffar Ali, B.M. Smartphone based bacterial detection using biofunctionalized fluorescent nanoparticles. Microchim. Acta 2014, 181, 1815–1821. [Google Scholar] [CrossRef]
- Dallet, C.; Kareem, S.; Kale, I. Real time blood image processing application for malaria diagnosis using mobile phones. In Proceedings of the IEEE International Symposium on Circuits & Systems (ISCAS), Melbourne, Australia, 1–5 June 2014; pp. 2405–2408. [Google Scholar] [CrossRef]
- Lillehoj, P.B.; Huang, M.-C.; Ho, C.-M. A handheld, cell phone-based electrochemical biodetector. In Proceedings of the 26th IEEE International Conference on Micro Electro Mechanical Systems, Taipei, Taiwan, 20–24 January 2013; pp. 53–56. [Google Scholar] [CrossRef]
- Stemple, C.C.; Angus, S.V.; Park, T.S.; Yoon, J.-Y. Smartphone-Based Optofluidic Lab-on-a-Chip for Detecting Pathogens from Blood. J. Lab. Autom. 2014, 19, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Mauk, M.G.; Liu, C.; Sadik, M.; Bau, H.H. Microfluidic devices for nucleic acid (NA) isolation, isothermal NA amplification, and real-time detection. Methods Mol. Biol. 2015, 1256, 15–40. [Google Scholar] [CrossRef] [PubMed]
- Sandoz, P.A.; Coskun, A.F.; Chung, A.J.; Weaver, W.M.; Adeyiga, O.; Khodadadi, D.; Ozcan, A.; Di Carlo, D. Digital readout platform for water-in-oil droplet immunoassays running on a cell-phone for point of care viral load sensing. In Proceedings of the The 16th International Conference on Microsystems for Chemistry and Life Sciences MicroTAS, Okinawa, Japan, 28 October–1 November 2012; pp. 338–340. [Google Scholar]
- Coulibaly, J.T.; Ouattara, M.; D’Ambrosio, M.V.; Fletcher, D.A.; Keiser, J.; Utzinger, J.; N’Goran, E.K.; Andrews, J.R.; Bogoch, I.I. Accuracy of Mobile Phone and Handheld Light Microscopy for the Diagnosis of Schistosomiasis and Intestinal Protozoa Infections in Côte d’Ivoire. PLoS Negl. Trop. Dis. 2016, 10, e0005550. [Google Scholar] [CrossRef] [PubMed]
- Ephraim, R.K.D.; Cybulski, J.S.; Duah, E.; Prakash, M.; D’Ambrosio, M.V.; Fletcher, D.A.; Keiser, J.; Andrews, J.R.; Bogoch, I.I. Diagnosis of Schistosoma haematobium infection with a mobile phone-mounted Foldscope and a reversed-lens CellScope in Ghana. Am. J. Trop. Med. Hyg. 2015, 92, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Holmen, S.D.; Kjetland, E.F.; Taylor, M.; Kleppa, E.; Lillebø, K.; Gundersen, S.G.; Onsrud, M.; Albregtsen, F. Colourimetric image analysis as a diagnostic tool in female genital schistosomiasis. Med. Eng. Phys. 2015, 37, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Jacob, J.M.; Costa, M.N.; Santos, D.S.; Viveiros, M.; Inácio, J.; Martins, R.; Barquinha, P.; Fortunato, E.; Baptista, P.V. Gold on paper-paper platform for Au-nanoprobe TB detection. Lab Chip 2012, 12, 4802–4808. [Google Scholar] [CrossRef] [PubMed]
- Veigas, B.; Fortunato, E.; Baptista, P.V. Mobile based gold nanoprobe TB diagnostics for point-of-need. Methods Mol. Biol. 2015, 1256, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Duthie, M.S.; Balagon, M.F.; Maghanoy, A.; Orcullo, F.M.; Cang, M.; Dias, R.F.; Collovati, M.; Reed, S.G. Rapid quantitative serological test for detection of infection with Mycobacterium leprae, the causative agent of leprosy. J. Clin. Microbiol. 2014, 52, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Berg, B.; Cortazar, B.; Tseng, D.; Ozkan, H.; Feng, S.; Wei, Q.; Chan, R.Y.-L.; Burbano, J.; Farooqui, Q.; Lewinski, M.; et al. Cellphone-Based Hand-Held Microplate Reader for Point-of-Care Testing of Enzyme-Linked Immunosorbent Assays. ACS Nano 2015, 9, 7857–7866. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Cesarman, E.; Erickson, D. Detection of Kaposi’s sarcoma associated herpesvirus nucleic acids using a smartphone accessory. Lab Chip Miniaturisation Chem. Biol. 2014, 14, 3809–3816. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Qi, H.; Luo, W.; Tseng, D.; Ki, S.J.; Wan, Z.; Göröcs, Z.; Bentolila, L.A.; Wu, T.-T.; Sun, R.; et al. Fluorescent imaging of single nanoparticles and viruses on a smart phone. ACS Nano 2013, 7, 9147–9155. [Google Scholar] [CrossRef] [PubMed]
- Gallegos, D.; Long, K.D.; Yu, H.; Clark, P.P.; Lin, Y.; George, S.; Nath, P.; Cunningham, B.T. Label-free biodetection using a smartphone. Lab Chip 2013, 13, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Yang, C. A smartphone-based chip-scale microscope using ambient illumination. Lab Chip Miniaturisation Chem. Biol. 2014, 14, 3056–3063. [Google Scholar] [CrossRef] [PubMed]
- Petryayeva, E.; Algar, W.R. Multiplexed homogeneous assays of proteolytic activity using a smartphone and quantum dots. Anal. Chem. 2014, 86, 3195–3202. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, J.; Parent, F.; De Lima Filho, E.S.; Loranger, S.; Kashyap, R. Toward the integration of optical sensors in smartphone screens using femtosecond laser writing. Opt. Lett. 2015, 40, 5654–5657. [Google Scholar] [CrossRef] [PubMed]
- DuVall, J.A.; Borba, J.C.; Shafagati, N.; Luzader, D.; Shukla, N.; Li, J.; Kehn-Hall, K.; Kendall, M.M.; Feldman, S.H.; Landers, J.P. Optical imaging of paramagnetic bead-DNA aggregation inhibition allows for low copy number detection of infectious pathogens. PLoS ONE 2015, 10, e0129830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Ozcan, A. Wide-field fluorescent microscopy and fluorescent imaging flow cytometry on a cell-phone. J. Vis. Exp. 2013, e50451. [Google Scholar] [CrossRef] [PubMed]
- Gantelius, J.; Bass, T.; Sjöberg, R.; Nilsson, P.; Andersson-Svahn, H. A lateral flow protein microarray for rapid and sensitive antibody assays. Int. J. Mol. Sci. 2011, 12, 7748–7759. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; van Oordt, T.; Schneider, E.M.; Zengerle, R.; von Stetten, F.; Luong, J.H.T. A smartphone-based colorimetric reader for bioanalytical applications using the screen-based bottom illumination provided by gadgets. Biosens. Bioelectron. 2015, 67, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wu, Z.; Xu, F.; Li, X.; Yao, C.; Xu, M.; Sheng, L.; Yu, S.; Tang, Y. A portable smart phone-based plasmonic nanosensor readout platform that measures transmitted light intensities of nanosubstrates using an ambient light sensor. Lab Chip 2016, 16, 1927–1933. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.J.; Chu, K.; Wachsmann-Hogiu, S. Nanometer-Scale Sizing Accuracy of Particle Suspensions on an Unmodified Cell Phone Using Elastic Light Scattering. PLoS ONE 2012, 7, e46030. [Google Scholar] [CrossRef] [PubMed]
- Byrne, B.; Stack, E.; Gilmartin, N.; O’Kennedy, R. Antibody-based sensors: Principles, problems and potential for detection of pathogens and associated toxins. Sensors (Switzerland) 2009, 9, 4407–4445. [Google Scholar] [CrossRef] [PubMed]
- Welch, N.G.; Scoble, J.A.; Muir, B.W.; Pigram, P.J. Orientation and characterization of immobilized antibodies for improved immunoassays (Review). Biointerphases 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, F.; Mayer, G. Selection and Biosensor Application of Aptamers for Small Molecules. Front. Chem. 2016, 4, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Ruscito, A.; DeRosa, M.C. Small-Molecule Binding Aptamers: Selection Strategies, Characterization, and Applications. Front. Chem. 2016, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, X.; Lu, W.; Wu, X.; Li, J. Molecular imprinting: Perspectives and applications. Chem. Soc. Rev. 2016, 45, 2137–2211. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Griffete, N.; Lamouri, A.; Felidj, N.; Chehimi, M.M.; Mangeney, C. Nanocomposites of Gold Nanoparticles@Molecularly Imprinted Polymers: Chemistry, Processing, and Applications in Sensors. Chem. Mater. 2015, 27, 5464–5478. [Google Scholar] [CrossRef]
- Sharma, S.K.; Leblanc, R.M. Biosensors based on β-galactosidase enzyme: Recent advances and perspectives. Anal. Biochem. 2017, 535, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rocchitta, G.; Spanu, A.; Babudieri, S.; Latte, G.; Madeddu, G.; Galleri, G.; Nuvoli, S.; Bagella, P.; Demartis, M.I.; Fiore, V.; et al. Enzyme biosensors for biomedical applications: Strategies for safeguarding analytical performances in biological fluids. Sensors (Switzerland) 2016, 16, 780. [Google Scholar] [CrossRef] [PubMed]
- Songa, E.A.; Okonkwo, J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: A review. Talanta 2016, 155, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, Z.F.; Su, Y.; Kitto, R.Z.; Hammond, M.C. Engineering and In Vivo Applications of Riboswitches. Annu. Rev. Biochem. 2017, 86, 515–539. [Google Scholar] [CrossRef] [PubMed]
- Bazin, I.; Tria, S.A.; Hayat, A.; Marty, J.L. New biorecognition molecules in biosensors for the detection of toxins. Biosens. Bioelectron. 2017, 87, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Kintzios, S.; Prabhakarpandian, B. Biotoxin detection using cell-based sensors. Toxins (Basel) 2013, 5, 2366–2383. [Google Scholar] [CrossRef] [PubMed]
- Löfblom, J.; Feldwisch, J.; Tolmachev, V.; Carlsson, J.; Ståhl, S.; Frejd, F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010, 584, 2670–2680. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Tamboli, V.; Harniman, R.L.; Estrela, P.; Allender, C.J.; Bowen, J.L. Aptamer-MIP hybrid receptor for highly sensitive electrochemical detection of prostate specific antigen. Biosens. Bioelectron. 2016, 75, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ren, J.; Su, L.; Gao, X.; Tang, Y.; Ma, T.; Zhu, L.; Li, J. Novel hybrid probe based on double recognition of aptamer-molecularly imprinted polymer grafted on upconversion nanoparticles for enrofloxacin sensing. Biosens. Bioelectron. 2017, 87, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, J. Molecularly Imprinted Polymers with DNA Aptamer Fragments as Macromonomers. ACS Appl. Mater. Interfaces 2016, 8, 6371–6378. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Guerreiro, A.; Whitcombe, M.J.; Piletska, E.V.; Turner, A.P.F.; Piletsky, S.A. Solid-Phase Synthesis of Molecularly Imprinted Polymer Nanoparticles with a Reusable Template—“Plastic Antibodies”. Adv. Funct. Mater. 2013, 23, 2821–2827. [Google Scholar] [CrossRef] [PubMed]
- Smolinska-Kempisty, K.; Guerreiro, A.; Canfarotta, F.; Cáceres, C.; Whitcombe, M.J.; Piletsky, S. A comparison of the performance of molecularly imprinted polymer nanoparticles for small molecule targets and antibodies in the ELISA format. Sci. Rep. 2016, 6, 37638. [Google Scholar] [CrossRef] [PubMed]
- Moczko, E.; Poma, A.; Guerreiro, A.; Perez, I.; Sansalvador, D.V.; Caygill, S.; Canfarotta, F.; Whitcombe, M.J.; Piletsky, S. Surface-modified multifunctional MIP nanoparticles. Nanoscale 2016, 5, 3733–3741. [Google Scholar] [CrossRef] [PubMed]
- Basozabal, I.; Guerreiro, A.; Gomez-Caballero, A.; Aranzazu Goicolea, M.; Barrio, R.J. Direct potentiometric quantification of histamine using solid-phase imprinted nanoparticles as recognition elements. Biosens. Bioelectron. 2014, 58, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Korposh, S.; Chianella, I.; Guerreiro, A.; Caygill, S.; Piletsky, S.; James, S.W.; Tatam, R.P. Selective vancomycin detection using optical fibre long period gratings functionalised with molecularly imprinted polymer nanoparticles. Analyst 2014, 139, 2229–2236. [Google Scholar] [CrossRef] [PubMed]
- Chianella, I.; Guerreiro, A.; Moczko, E.; Caygill, J.S.; Piletska, E.V.; De Vargas Sansalvador, I.M.P.; Whitcombe, M.J.; Piletsky, S.A. Direct replacement of antibodies with molecularly imprinted polymer nanoparticles in ELISA—Development of a novel assay for vancomycin. Anal. Chem. 2013, 85, 8462–8468. [Google Scholar] [CrossRef] [PubMed]
- Altintas, Z.; Gittens, M.; Guerreiro, A.; Thompson, K.-A.; Walker, J.; Piletsky, S.; Tothill, I.E. Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers. Anal. Chem. 2015, 87, 6801–6807. [Google Scholar] [CrossRef] [PubMed]
- Poma, A.; Guerreiro, A.; Caygill, S.; Moczko, E.; Piletsky, S. Automatic reactor for solid-phase synthesis of molecularly imprinted polymeric nanoparticles (MIP NPs) in water. RSC Adv. 2014, 4, 4203–4206. [Google Scholar] [CrossRef] [PubMed]
- Canfarotta, F.; Poma, A.; Guerreiro, A.; Piletsky, S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 2016, 11, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Karim, K.; Cowen, T.; Guerreiro, A.; Piletska, E.; Whitcombe, M.J.; Piletsky, S.A. A Protocol for the Computational Design of High Affi nity Molecularly Imprinted Polymer Synthetic Receptors. Glob. J. Biotechnol. Biomater. Sci. 2017, 1, 001–007. [Google Scholar] [CrossRef]
- Rohloff, J.C.; Gelinas, A.D.; Jarvis, T.C.; Ochsner, U.A.; Schneider, D.J.; Gold, L.; Janjic, N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol. Ther. Nucleic Acids 2014, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Egholm, M.; Buchardt, O.; Christensen, L.; Behrens, C.; Freier, S.M.; Driver, D.A.; Berg, R.H.; Kim, S.K.; Norden, B.; Nielsen, P.E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 1993, 365, 566–568. [Google Scholar] [CrossRef] [PubMed]
- Brudno, Y.; Birnbaum, M.E.; Kleiner, R.E.; Liu, D.R. An in vitro translation, selection and amplification system for peptide nucleic acids. Nat. Chem. Biol. 2010, 6, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhang, S.; Chaput, J.C. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012, 4, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Hili, R.; Liu, D.R. Enzyme-free translation of DNA into sequence-defined synthetic polymers structurally unrelated to nucleic acids. Nat. Chem. 2013, 5, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Menezes, A.A.; Cumbers, J.; Hogan, J.A.; Arkin, A.P. Towards synthetic biological approaches to resource utilization on space missions. J. R. Soc. Interface 2015, 12, 20140715. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, L.J. Synthetic biology meets bioprinting: Enabling technologies for humans on Mars (and Earth). Biochem. Soc. Trans. 2016, 44, 1158–1164. [Google Scholar] [CrossRef] [PubMed]
- Lasseur, C.; Brunet, J.; Weever, H.D.; Dixon, M.; Dussap, G.; Godia, F.; Leys, N.; Mergeay, M.; Straeten, D. Van Der Melissa: The European project of closed life support system. Gravitational Sp. Biol. 2010, 23, 3–12. [Google Scholar]
- Verseux, C.N.; Paulino-Lima, I.G.; Baqué, M.; Billi, D.; Rothschild, L.J. Synthetic Biology for Space Exploration: Promises and Societal Implications. Ambivalences Creat. Life Soc. Philos. Dimens. Synth. Biol. 2016, 73–100. [Google Scholar] [CrossRef]
- Lu, Y. Cell-free synthetic biology: Engineering in an open world. Synth. Syst. Biotechnol. 2017, 2, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Bundy, B.C.; Swartz, J.R. Efficient disulfide bond formation in virus-like particles. J. Biotechnol. 2011, 154, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Slomovic, S.; Nguyen, P.Q.; Lee, J.W.; Donghia, N.; Burrill, D.; Ferrante, T.; McSorley, F.R.; Furuta, Y.; Vernet, A.; et al. Portable, On-Demand Biomolecular Manufacturing. Cell 2016, 167, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Groff, D.; Armstrong, S.; Rivers, P.J.; Zhang, J.; Yang, J.; Green, E.; Rozzelle, J.; Liang, S.; Kittle, J.D.; Steiner, A.R.; et al. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. MAbs 2014, 6, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Z.; Hayes, C.A.; Shin, J.; Caschera, F.; Murray, R.M.; Noireaux, V. Protocols for Implementing an Escherichia coli Based TX-TL Cell-Free Expression System for Synthetic Biology. J. Vis. Exp. 2013, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Resource Paper-Based Synthetic Gene Networks. Cell 2014, 159, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.T.; Berkheimer, S.D.; Werner, C.J.; Bundy, B.C. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques 2014, 56, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Roda, A.; Mirasoli, M.; Guardigli, M.; Zangheri, M.; Caliceti, C.; Calabria, D.; Simoni, P. Advanced biosensors for monitoring astronauts’ health during long-duration space missions. Biosens. Bioelectron. 2018, 111, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Goud, K.Y.; Hayat, A.; Catanante, G.; Satyanarayana, S.M.; Gobi, K.V.; Marty, J.L. An electrochemical aptasensor based on functionalized graphene oxide assisted electrocatalytic signal amplification of methylene blue for aflatoxin B1 detection. Electrochim. Acta 2017, 244, 96–103. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).