Advances in Enzyme-Based Biosensors for Pesticide Detection
Abstract
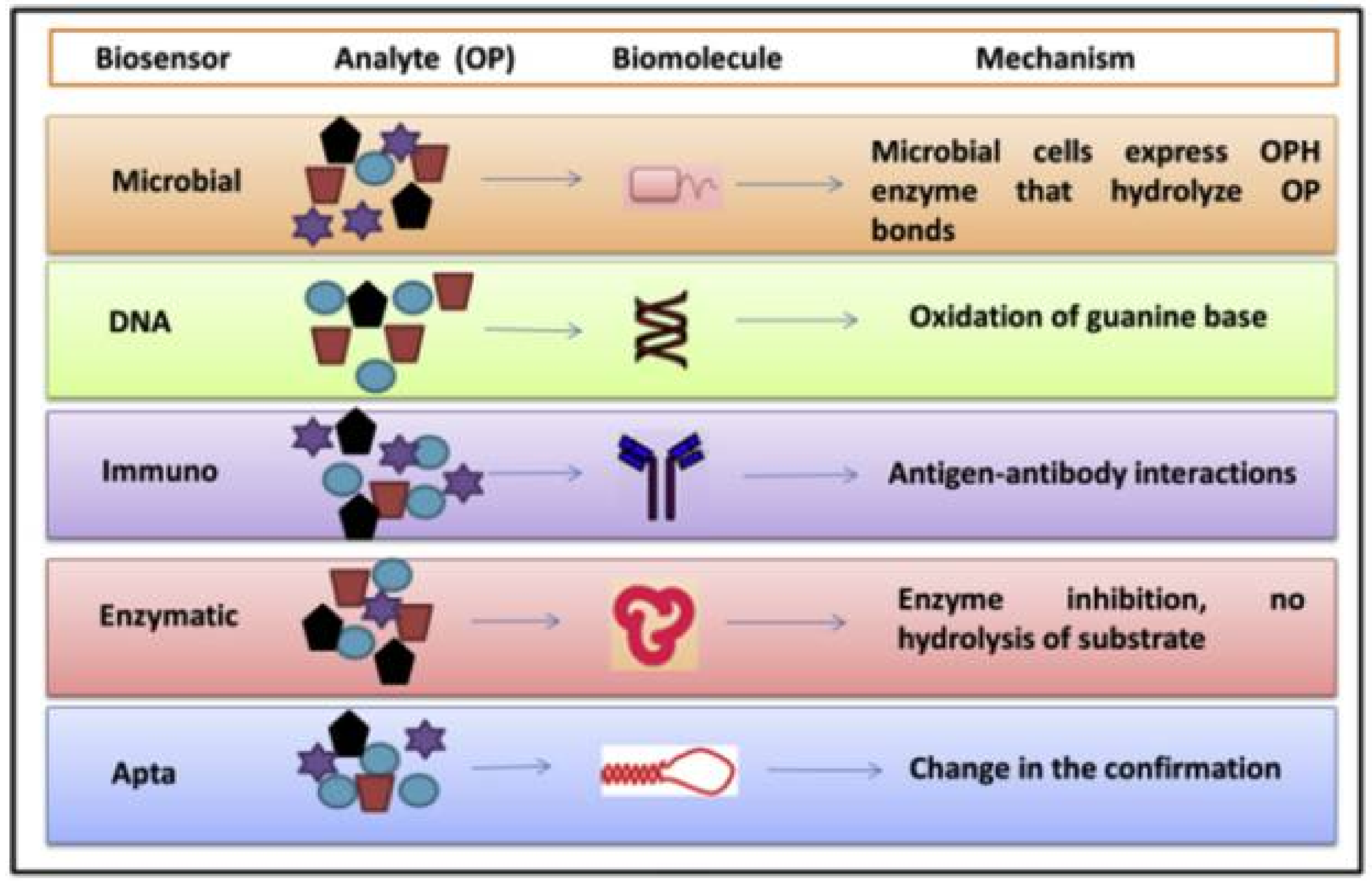
1. Introduction
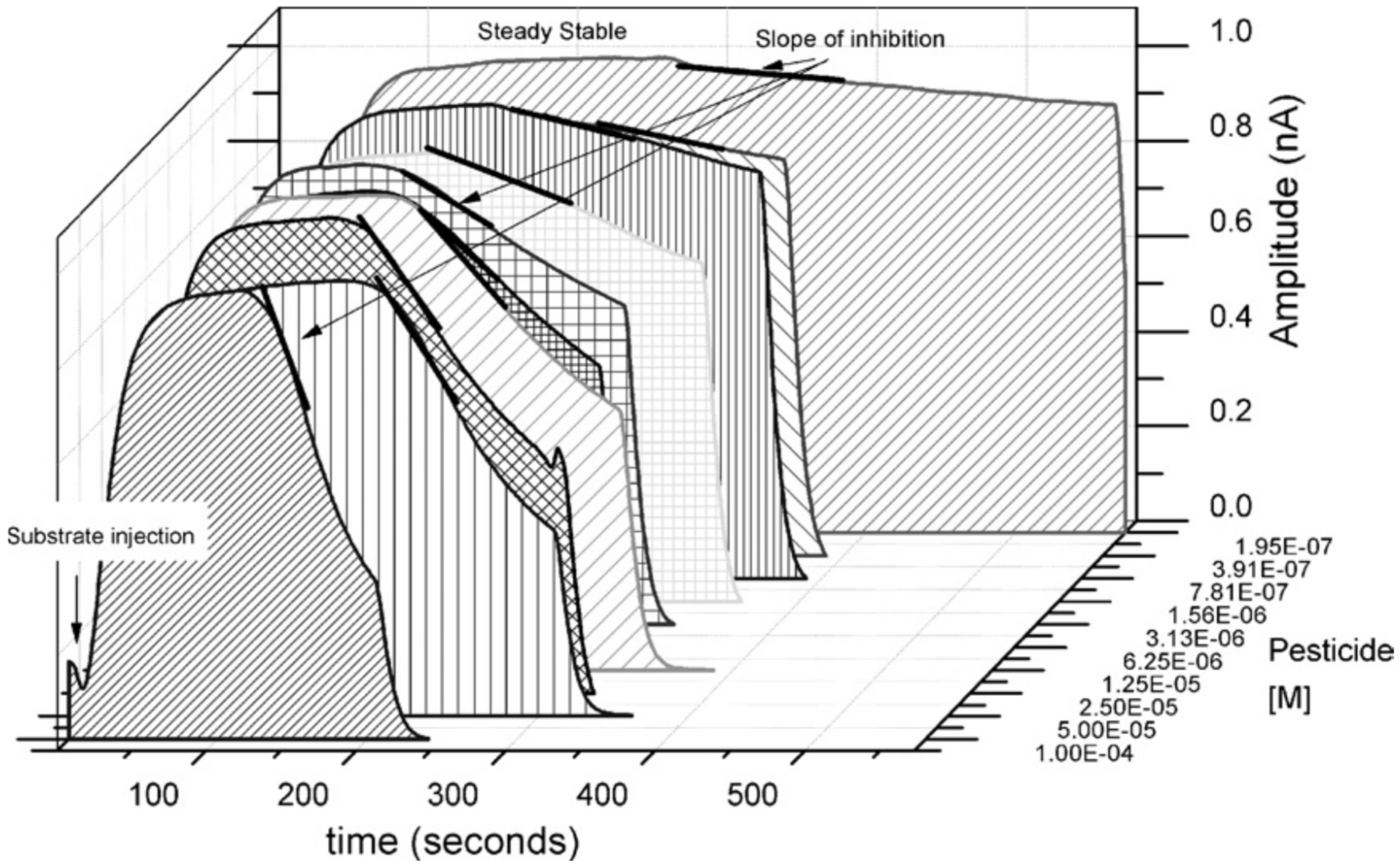
2. Detection of Neurotoxic Insecticides Based on Cholinesterase Inhibition
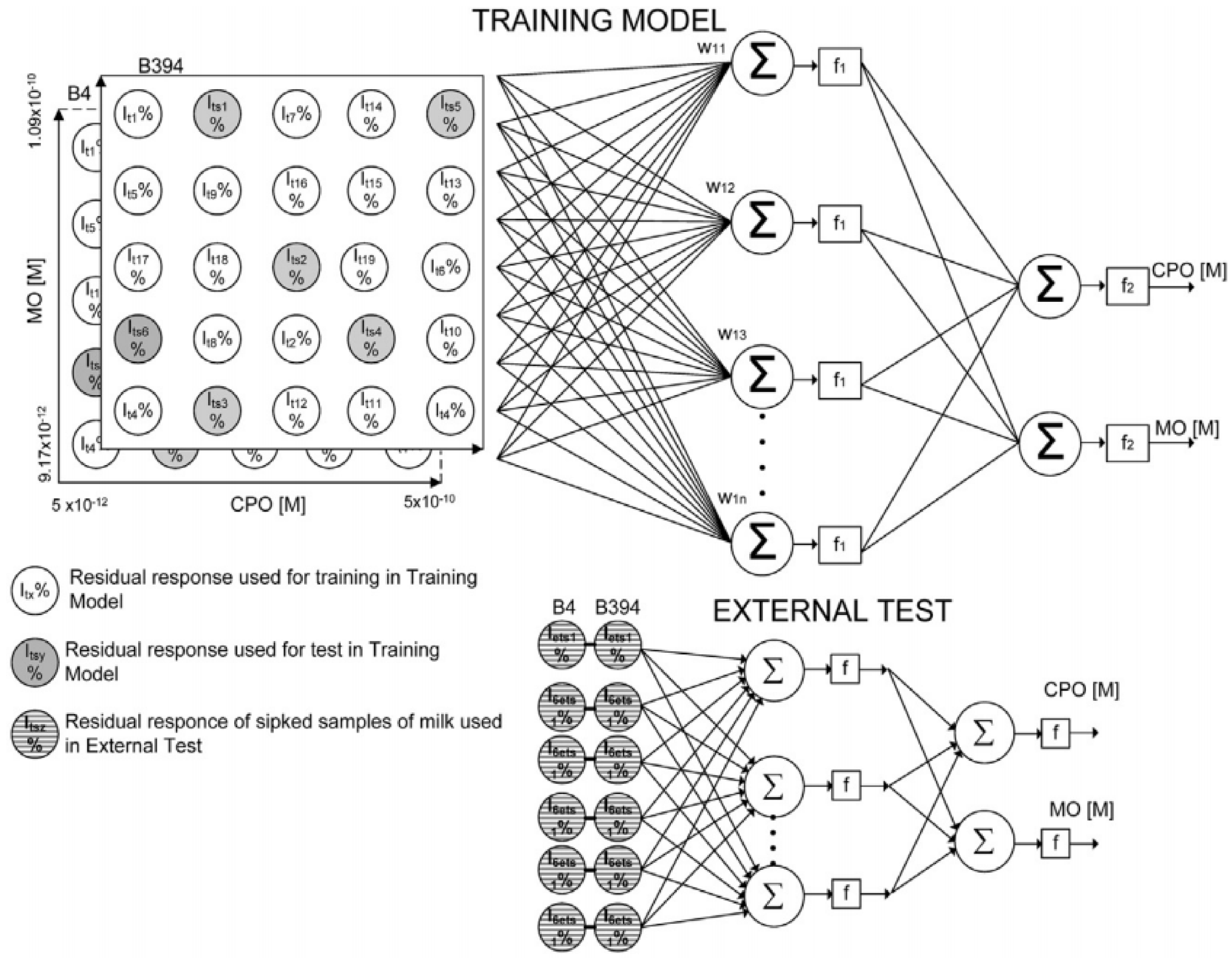
3. Use of Chemometric Methods for Enhancement of AChE Biosensors Performances
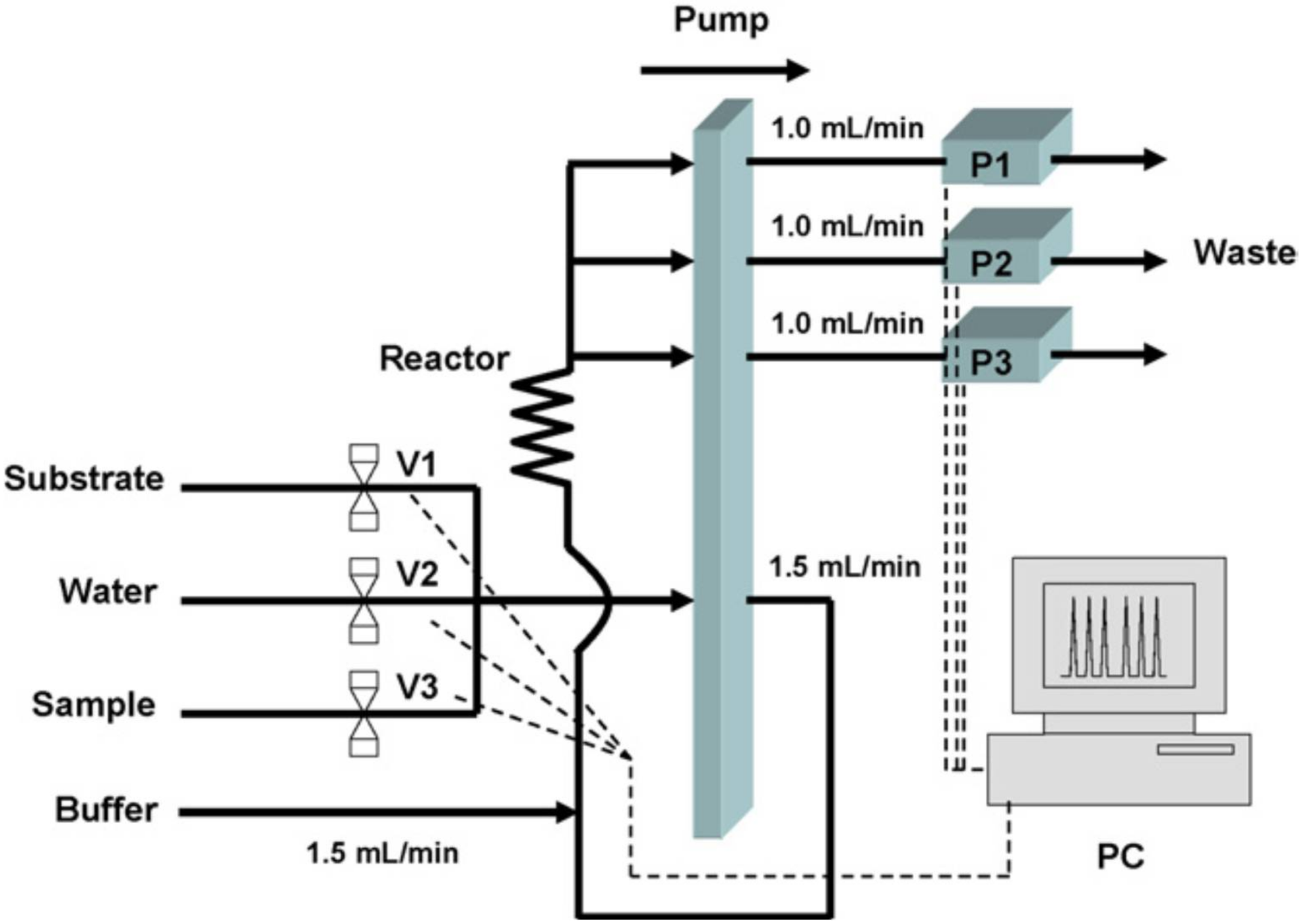
4. (Bio)chemical Sample Treatment in Combination with AChE Biosensors
5. Detection of Photosynthesis-Inhibiting Herbicides
6. Other Enzymes
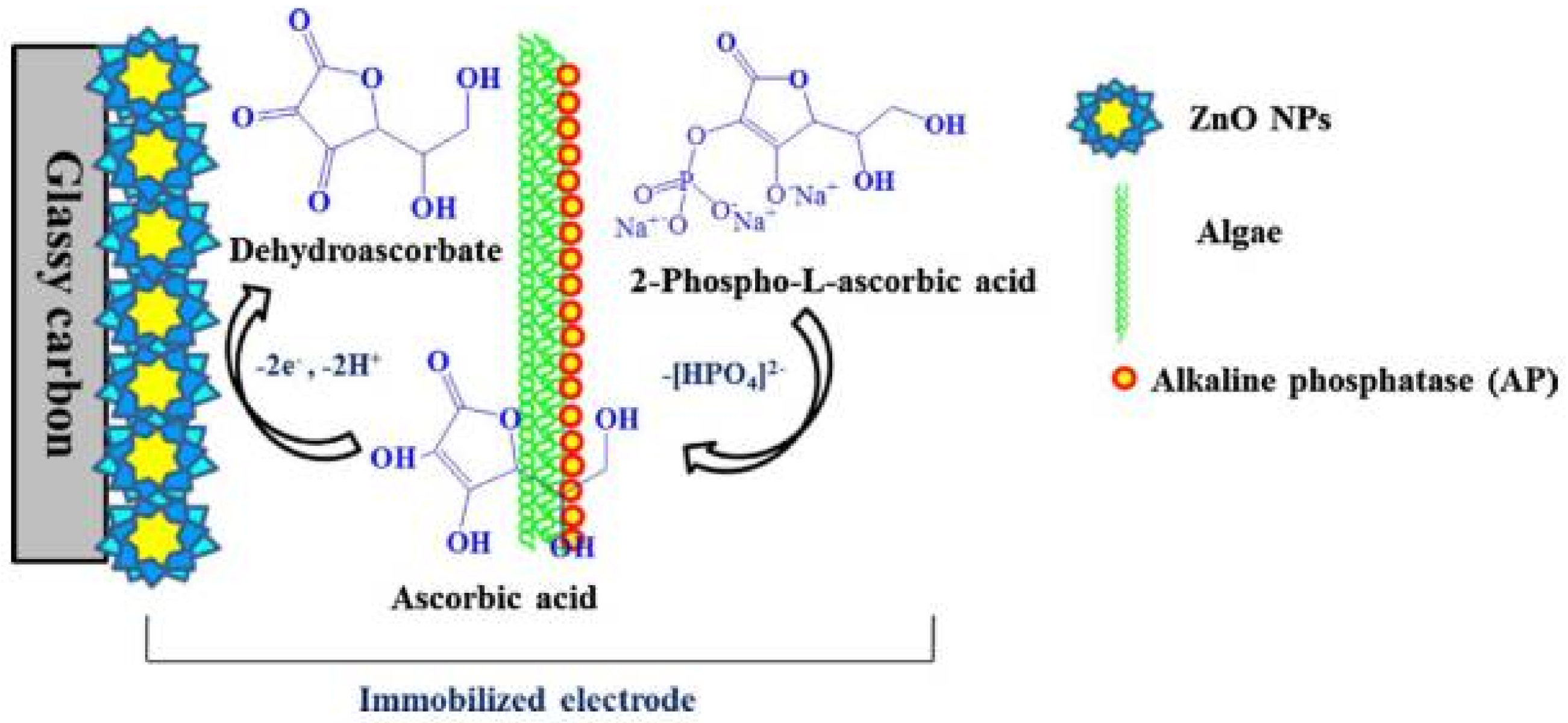
6.1. Alkaline Phosphatase
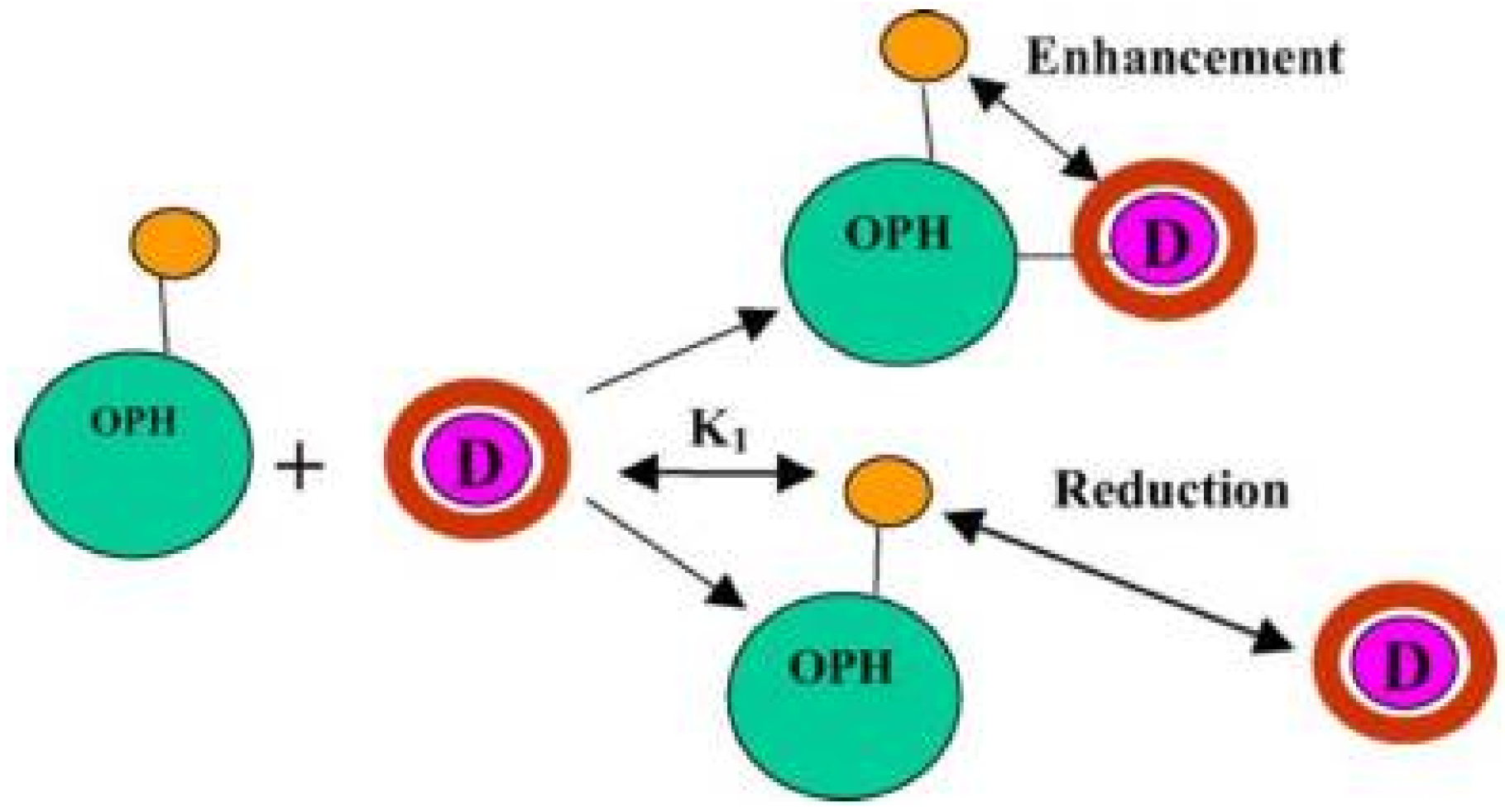
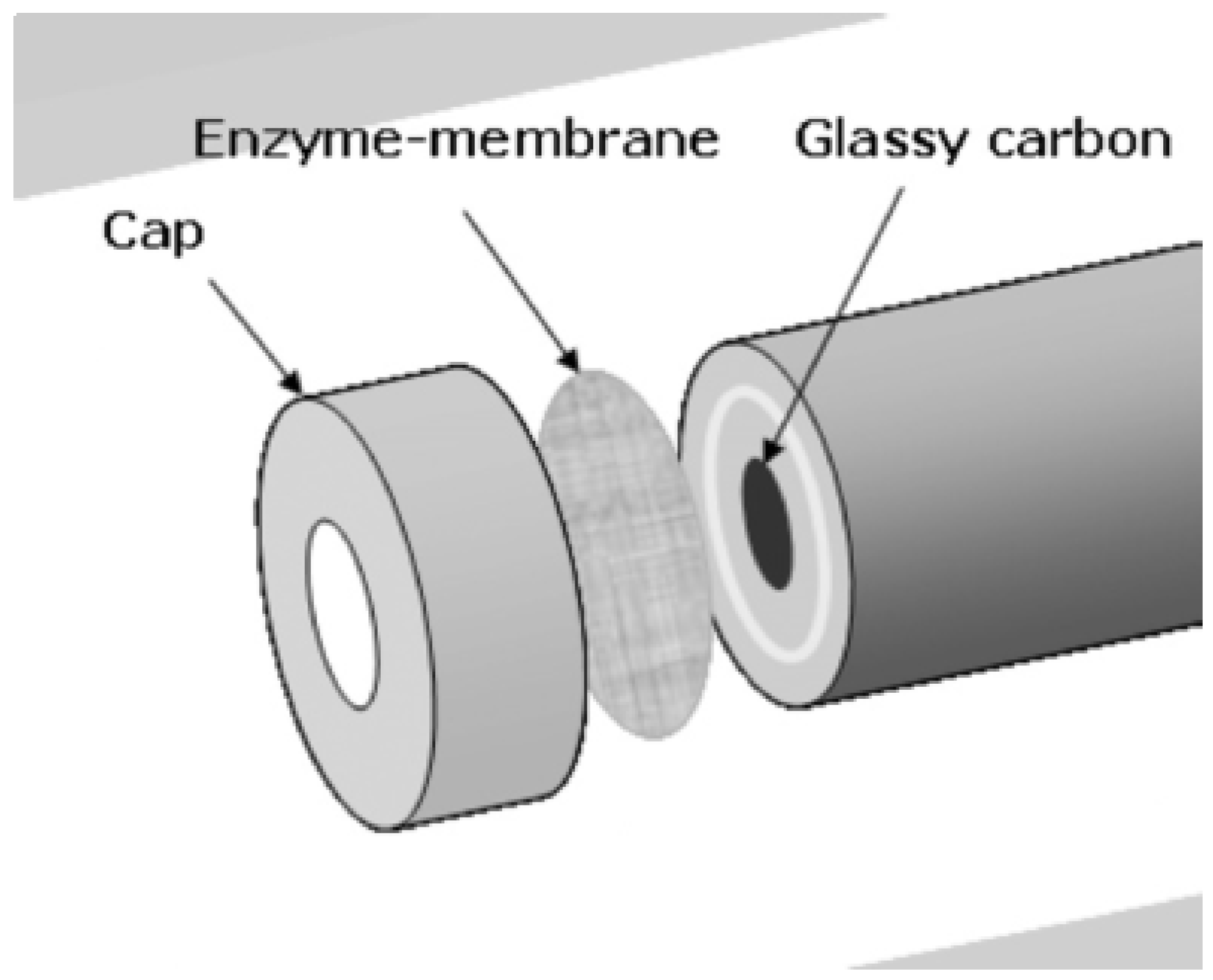
6.2. Organophosphorus Hydrolase
6.3. Tyrosinase
6.4. Laccase
6.5. Heme-Containing Enzymes
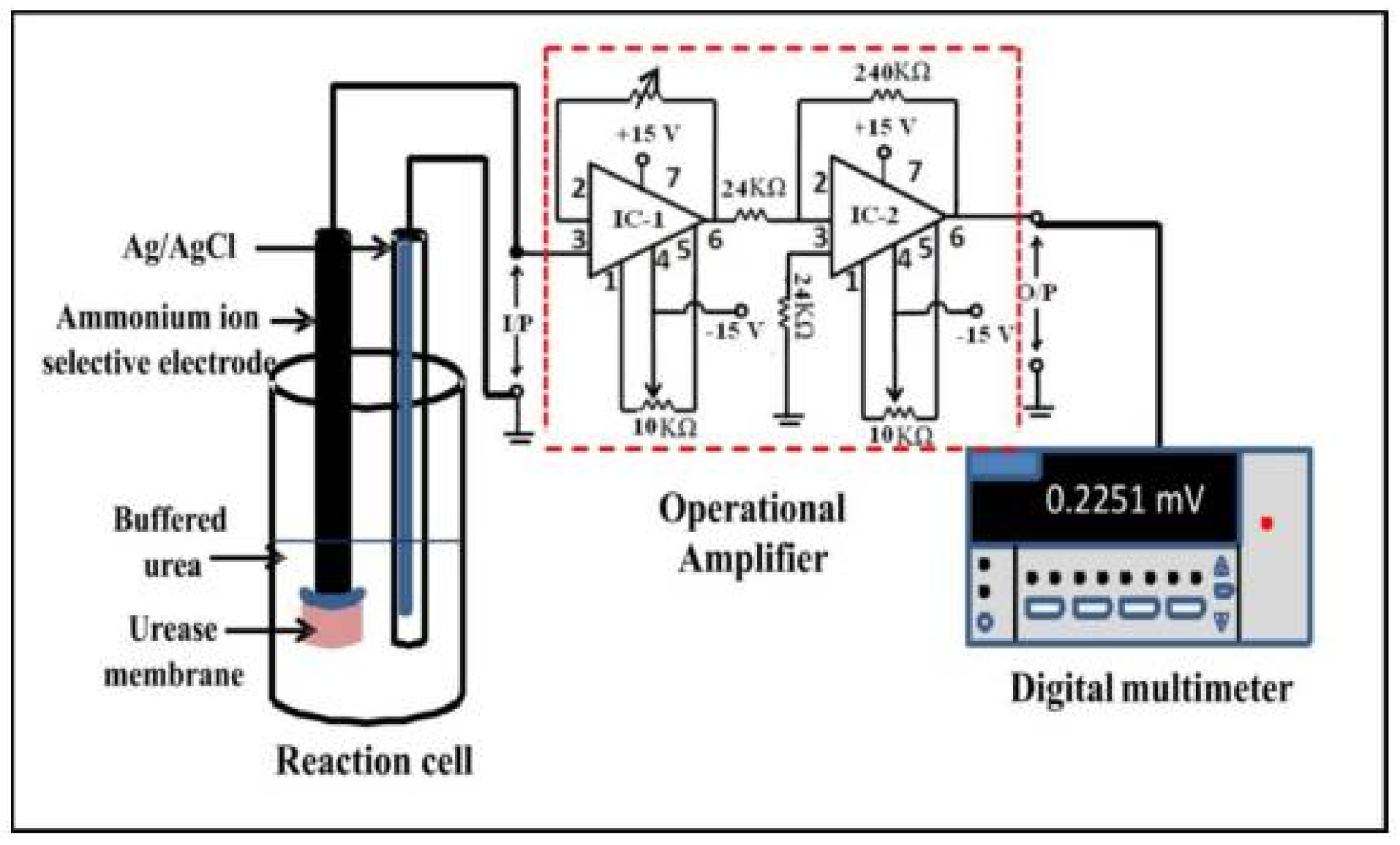
6.6. Urease
6.7. Aldehyde Dehydrogenase
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ongley, E.A. Control of Water Pollution from Agriculture—FAO Irrigation and Drainage Paper 55; Food and Agricultural Organization of the United Nations: Rome, Italy, 1996; ISBN 92-5-103875-9. [Google Scholar]
- Grimalt, S.; Dehouck, P. Review of analytical methods for the determination of pesticide residues in grapes. J. Chromatogr. A 2016, 1433, 1–23. [Google Scholar] [CrossRef] [PubMed]
- LeDoux, M. Analytical methods applied to the determination of pesticide residues in foods of animal origin: A review of the past two decades. J. Chromatogr. A 2011, 1218, 1021–1036. [Google Scholar] [CrossRef] [PubMed]
- Abu-Ali, H.; Nabok, A.; Smith, T.; Al-Shanawa, M. Development of electrochemical inhibition biosensor based on bacteria for detection of environmental pollutants. Sens. Bio-Sens. Res. 2017, 13, 109–114. [Google Scholar] [CrossRef]
- Ayyagari, M.S.; Kamtekar, S.; Pande, R.; Marx, K.A.; Kumar, J.; Tripathy, S.K.; Kaplan, D.L. Biosensors for pesticide detection based on alkaline phosphatase-catalyzed chemiluminescence. Mater. Sci. Eng. C 1995, 2, 191–196. [Google Scholar] [CrossRef]
- Bala, R.; Dhingra, S.; Kumar, M.; Bansal, K.; Mittal, S.; Sharma, R.K.; Wangoo, N. Detection of organophosphorus pesticide—Malathion in environmental samples using peptide and aptamer based nanoprobes. Chem. Eng. J. 2017, 311, 111–116. [Google Scholar] [CrossRef]
- Besombes, J.-L.; Cosnier, S.; Labbé, P.; Reverdy, G. A biosensor as warning device for the detection of cyanide, chlorophenols, atrazine and carbamate pesticides. Anal. Chim. Acta 1995, 311, 255–263. [Google Scholar] [CrossRef]
- Deo, R.P.; Wang, J.; Block, I.; Mulchandani, A.; Joshi, K.A.; Trojanowicz, M.; Scholz, F.; Chen, W.; Lin, Y. Determination of organophosphate pesticides at a carbon nanotube/organophosphorus hydrolase electrochemical biosensor. Anal. Chim. Acta 2005, 530, 185–189. [Google Scholar] [CrossRef]
- Garcı́a Sánchez, F.; Navas Dı́az, A.; Ramos Peinado, M.C.; Belledone, C. Free and sol–gel immobilized alkaline phosphatase-based biosensor for the determination of pesticides and inorganic compounds. Anal. Chim. Acta 2003, 484, 45–51. [Google Scholar] [CrossRef]
- Grawe, G.F.; de Oliveira, T.R.; de Andrade Narciso, E.; Moccelini, S.K.; Terezo, A.J.; Soares, M.A.; Castilho, M. Electrochemical biosensor for carbofuran pesticide based on esterases from eupenicillium shearii FREI-39 endophytic fungus. Biosens. Bioelectron. 2015, 63, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Haddaoui, M.; Raouafi, N. Chlortoluron-induced enzymatic activity inhibition in tyrosinase/ZnO NPs/SPCE biosensor for the detection of ppb levels of herbicide. Sens. Actuators B Chem. 2015, 219, 171–178. [Google Scholar] [CrossRef]
- Kaur, N.; Prabhakar, N. Current scenario in organophosphates detection using electrochemical biosensors. TrAC Trends Anal. Chem. 2017, 92, 62–85. [Google Scholar] [CrossRef]
- Commission, E. Eu Pesticides Database. Available online: http://ec.europa.eu/food/plant/pesticides/eu-pesticides-database/public/?event=activesubstance.selection&language=EN (accessed on 6 February 2018).
- Brancato, A.; Brocca, D.; De Lentdecker, C.; Erdos, Z.; Ferreira, L.; Greco, L.; Jarrah, S.; Kardassi, D.; Leuschner, R.; Lythgo, C.; et al. Review of the existing maximum residue levels for chlorpyrifos-methyl according to article 12 of regulation (EC) no 396/2005. EFSA J. 2017, 15. [Google Scholar] [CrossRef]
- King, A.M.; Aaron, C.K. Organophosphate and carbamate poisoning. Emer. Med. Clin. N. Am. 2015, 33, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Štěpánková, Š.; Vorčáková, K. Cholinesterase-based biosensors. J. Enzyme Inhibition Med. Chem. 2016, 31, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Rajangam, B.; Daniel, D.K.; Krastanov, A.I. Progress in enzyme inhibition based detection of pesticides. Eng. Life Sci. 2018, 18, 4–19. [Google Scholar] [CrossRef]
- Pundir, C.S.; Chauhan, N. Acetylcholinesterase inhibition-based biosensors for pesticide determination: A review. Anal. Biochem. 2012, 429, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Songa, E.A.; Okonkwo, J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: A review. Talanta 2016, 155, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Solé, S.; Merkoçi, A.; Alegret, S. Determination of toxic substances based on enzyme inhibition. Part II. Electrochemical biosensors for the determination of pesticides using flow systems. Crit. Rev. Anal. Chem. 2003, 33, 127–143. [Google Scholar] [CrossRef]
- Prieto-Simón, B.; Campàs, M.; Andreescu, S.; Marty, J.-L. Trends in flow-based biosensing systems for pesticide assessment. Sensors 2006, 6, 1161–1186. [Google Scholar] [CrossRef]
- Marrazza, G. Piezoelectric biosensors for organophosphate and carbamate pesticides: A review. Biosensors 2014, 4, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Arduini, F.; Cinti, S.; Scognamiglio, V.; Moscone, D. Nanomaterials in electrochemical biosensors for pesticide detection: Advances and challenges in food analysis. MicroChim. Acta 2016, 183, 2063–2083. [Google Scholar] [CrossRef]
- Arduini, F.; Cinti, S.; Scognamiglio, V.; Moscone, D.; Palleschi, G. How cutting-edge technologies impact the design of electrochemical (bio)sensors for environmental analysis. A review. Anal. Chim. Acta 2017, 959, 15–42. [Google Scholar] [CrossRef] [PubMed]
- Rotariu, L.; Lagarde, F.; Jaffrezic-Renault, N.; Bala, C. Electrochemical biosensors for fast detection of food contaminants—Trends and perspective. TrAC Trends Anal. Chem. 2016, 79, 80–87. [Google Scholar] [CrossRef]
- Pohanka, M. Electrochemical biosensors based on acetylcholinesterase and butyrylcholinesterase. A review. Int. J. Electrochem. Sci. 2016, 11, 7440–7452. [Google Scholar] [CrossRef]
- Luo, Q.-J.; Li, Y.-X.; Zhang, M.-Q.; Qiu, P.; Deng, Y.-H. A highly sensitive, dual-signal assay based on rhodamine B covered silver nanoparticles for carbamate pesticides. Chin. Chem. Lett. 2017, 28, 345–349. [Google Scholar] [CrossRef]
- Meng, X.; Wei, J.; Ren, X.; Ren, J.; Tang, F. A simple and sensitive fluorescence biosensor for detection of organophosphorus pesticides using H2O2-sensitive quantum dots/bi-enzyme. Biosens. Bioelectron. 2013, 47, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zhou, Y.; Li, X.; Liu, S.; Tang, Z. Highly-sensitive organophosphorous pesticide biosensors based on nanostructured films of acetylcholinesterase and CdTe quantum dots. Biosens. Bioelectron. 2011, 26, 3081–3085. [Google Scholar] [CrossRef] [PubMed]
- Milkani, E.; Lambert, C.R.; McGimpsey, W.G. Direct detection of acetylcholinesterase inhibitor binding with an enzyme-based surface plasmon resonance sensor. Anal. Biochem. 2011, 408, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Kokot, S. Does chemometrics enhance the performance of electroanalysis? Anal. Chim. Acta 2008, 626, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Mwila, K.; Burton, M.H.; Van Dyk, J.S.; Pletschke, B.I. The effect of mixtures of organophosphate and carbamate pesticides on acetylcholinesterase and application of chemometrics to identify pesticides in mixtures. Environ. Monitor. Assess. 2013, 185, 2315–2327. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Cao, D.; Kokot, S. Simultaneous enzymatic kinetic determination of pesticides, carbaryl and phoxim, with the aid of chemometrics. Anal. Chim. Acta 2007, 588, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, T.T.; Schmid, R.D. A disposable multielectrode biosensor for rapid simultaneous detection of the insecticides paraoxon and carbofuran at high resolution. Anal. Chim. Acta 1999, 401, 95–103. [Google Scholar] [CrossRef]
- Bachmann, T.T.; Leca, B.; Vilatte, F.; Marty, J.-L.; Fournier, D.; Schmid, R.D. Improved multianalyte detection of organophosphates and carbamates with disposable multielectrode biosensors using recombinant mutants of drosophila acetylcholinesterase and artificial neural networks. Biosens. Bioelectron. 2000, 15, 193–201. [Google Scholar] [CrossRef]
- Valdés-Ramírez, G.; Gutiérrez, M.; del Valle, M.; Ramírez-Silva, M.T.; Fournier, D.; Marty, J.L. Automated resolution of dichlorvos and methylparaoxon pesticide mixtures employing a flow injection system with an inhibition electronic tongue. Biosens. Bioelectron. 2009, 24, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Alonso, G.A.; Istamboulie, G.; Bhand, S.; Marty, J.-L. Automated flow based biosensor for quantification of binary organophosphates mixture in milk using artificial neural network. Sens. Actuators B Chem. 2015, 208, 228–237. [Google Scholar] [CrossRef]
- Alonso, G.A.; Istamboulie, G.; Noguer, T.; Marty, J.-L.; Muñoz, R. Rapid determination of pesticide mixtures using disposable biosensors based on genetically modified enzymes and artificial neural networks. Sens. Actuators B Chem. 2012, 164, 22–28. [Google Scholar] [CrossRef]
- Bucur, B.; Dondoi, M.; Danet, A.; Marty, J.-L. Insecticide identification using a flow injection analysis system with biosensors based on various cholinesterases. Anal. Chim. Acta 2005, 539, 195–201. [Google Scholar] [CrossRef]
- Li Vigni, M.; Durante, C.; Cocchi, M. Chapter 3—Exploratory data analysis. In Data Handling in Science and Technology; Marini, F., Ed.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 28, pp. 55–126. [Google Scholar]
- Solná, R.; Dock, E.; Christenson, A.; Winther-Nielsen, M.; Carlsson, C.; Emnéus, J.; Ruzgas, T.; Skládal, P. Amperometric screen-printed biosensor arrays with co-immobilised oxidoreductases and cholinesterases. Anal. Chim. Acta 2005, 528, 9–19. [Google Scholar] [CrossRef]
- Dondoi, M.P.; Bucur, B.; Danet, A.F.; Toader, C.N.; Barthelmebs, L.; Marty, J.-L. Organophosphorus insecticides extraction and heterogeneous oxidation on column for analysis with an acetylcholinesterase (AChE) biosensor. Anal. Chim. Acta 2006, 578, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Schulze, H.; Schmid, R.D.; Bachmann, T.T. Rapid detection of neurotoxic insecticides in food using disposable acetyicholinesterase-biosensors and simple solvent extraction. Anal. Bioanal. Chem. 2002, 372, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Arduini, F.; Ricci, F.; Bourais, I.; Amine, A.; Moscone, D.; Palleschi, G. Extraction and detection of pesticides by cholinesterase inhibition in a two-phase system: A strategy to avoid heavy metal interference. Anal. Lett. 2005, 38, 1703–1719. [Google Scholar] [CrossRef]
- Campanella, L.; De Luca, S.; Sammartino, M.P.; Tomassetti, M. A new organic phase enzyme electrode for the analysis of organophosphorus pesticides and carbamates. Anal. Chim. Acta 1999, 385, 59–71. [Google Scholar] [CrossRef]
- Raushel, F.M.; Holden, H.M. Phosphotriesterase: An enzyme in search of its natural substrate. Adv. Enzymol. Relat. Areas Mol. Biol. 2000, 74, 51–93. [Google Scholar] [PubMed]
- Istamboulie, G.; Fournier, D.; Marty, J.-L.; Noguer, T. Phosphotriesterase: A complementary tool for the selective detection of two organophosphate insecticides: Chlorpyrifos and chlorfenvinfos. Talanta 2009, 77, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Rhouati, A.; Istamboulie, G.; Cortina-Puig, M.; Marty, J.-L.; Noguer, T. Selective spectrophotometric detection of insecticides using cholinesterases, phosphotriesterase and chemometric analysis. Enzyme Microb. Technol. 2010, 46, 212–216. [Google Scholar] [CrossRef]
- Iyengar, A.R.S.; Pande, A.H. Organophosphate-hydrolyzing enzymes as first-line of defence against nerve agent-poisoning: Perspectives and the road ahead. Protein J. 2016, 35, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Simonian, A.L.; Flounders, A.W.; Wild, J.R. Fet-based biosensors for the direct detection of organophosphate neurotoxins. Electroanalysis 2004, 16, 1896–1906. [Google Scholar] [CrossRef]
- Simonian, A.L.; Rainina, E.I.; Wild, J.R. A new approach for discriminative detection of organophosphate neurotoxins in the presence of other cholinesterase inhibitors. Anal. Lett. 1997, 30, 2453–2468. [Google Scholar] [CrossRef]
- Giardi, M.T.; Koblı́zek, M.; Masojı́dek, J. Photosystem II-based biosensors for the detection of pollutants. Biosens. Bioelectron. 2001, 16, 1027–1033. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Marco, M.-P.; Lopez de Alda, M.J.; Barceló, D. Biosensors for environmental monitoring of endocrine disruptors: A review article. Anal. Bioanal. Chem. 2004, 378, 588–598. [Google Scholar] [PubMed]
- Rouillon, R.; Piletsky, S.A.; Breton, F.; Piletska, E.V.; Carpentier, R. Photosystem II biosensors for heavy metals monitoring. In Biotechnological Applications of Photosynthetic Proteins: Biochips, Biosensors and Biodevices; Springer: Boston, MA, USA, 2006; pp. 166–174. [Google Scholar]
- Bhalla, V.; Zhao, X.; Zazubovich, V. Detection of explosive compounds using photosystem II-based biosensor. J. Electroanal. Chem. 2011, 657, 84–90. [Google Scholar] [CrossRef]
- Esposito, D.; Margonelli, A.; Pace, E.; Giardi, M.T.; Faraloni, C.; Torzillo, G.; Zanini, A. The effect of ionising radiation on photosynthetic oxygenic microorganisms for survival in space flight revealed by automatic photosystem II-based biosensors. Microgravity Sci. Technol. 2006, 18, 215. [Google Scholar] [CrossRef]
- Campàs, M.; Carpentier, R.; Rouillon, R. Plant tissue-and photosynthesis-based biosensors. Biotechnol. Adv. 2008, 26, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Maly, J.; Masojidek, J.; Masci, A.; Ilie, M.; Cianci, E.; Foglietti, V.; Vastarella, W.; Pilloton, R. Direct mediatorless electron transport between the monolayer of photosystem II and poly(mercapto-p-benzoquinone) modified gold electrode—New design of biosensor for herbicide detection. Biosens. Bioelectron. 2005, 21, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Koblizek, M.; Masojidek, J.; Komenda, J.; Kucera, T.; Pilloton, R.; Mattoo, A.K.; Giardi, M.T. A sensitive photosystem II-based biosensor for detection of a class of herbicides. Biotechnol. Bioeng. 1998, 60, 664–669. [Google Scholar] [CrossRef]
- Koblizek, M.; Maly, J.; Masojidek, J.; Komenda, J.; Kucera, T.; Giardi, M.T.; Mattoo, A.K.; Pilloton, R. A biosensor for the detection of triazine and phenylurea herbicides designed using photosystem ii coupled to a screen-printed electrode. Biotechnol. Bioeng. 2002, 78, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Touloupakis, E.; Giannoudi, L.; Piletsky, S.A.; Guzzella, L.; Pozzoni, F.; Giardi, M.T. A multi-biosensor based on immobilized photosystem II on screen-printed electrodes for the detection of herbicides in river water. Biosens. Bioelectron. 2005, 20, 1984–1992. [Google Scholar] [CrossRef] [PubMed]
- Ventrella, A.; Catucci, L.; Placido, T.; Longobardi, F.; Agostiano, A. Biomaterials based on photosynthetic membranes as potential sensors for herbicides. Biosens. Bioelectron. 2011, 26, 4747–4752. [Google Scholar] [CrossRef] [PubMed]
- Giardi, M.T.; Scognamiglio, V.; Rea, G.; Rodio, G.; Antonacci, A.; Lambreva, M.; Pezzotti, G.; Johanningmeier, U. Optical biosensors for environmental monitoring based on computational and biotechnological tools for engineering the photosynthetic D1 protein of chlamydomonas reinhardtii. Biosens. Bioelectron. 2009, 25, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Husu, I.; Rodio, G.; Touloupakis, E.; Lambreva, M.D.; Buonasera, K.; Litescu, S.C.; Giardi, M.T.; Rea, G. Insights into photo-electrochemical sensing of herbicides driven by chlamydomonas reinhardtii cells. Sens. Actuators B Chem. 2013, 185, 321–330. [Google Scholar] [CrossRef]
- Rasmussen, M.; Minteer, S.D. Self-powered herbicide biosensor utilizing thylakoid membranes. Anal. Methods 2013, 5, 1140–1144. [Google Scholar] [CrossRef]
- Bettazzi, F.; Laschi, S.; Mascini, M. One-shot screen-printed thylakoid membrane-based biosensor for the detection of photosynthetic inhibitors in discrete samples. Anal. Chim. Acta 2007, 589, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, V.; Raffi, D.; Lambreva, M.; Rea, G.; Tibuzzi, A.; Pezzotti, G.; Johanningmeier, U.; Giardi, M.T. Chlamydomonas reinhardtii genetic variants as probes for fluorescence sensing system in detection of pollutants. Anal. Bioanal. Chem. 2009, 394, 1081. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, V.; Zazubovich, V. Self-assembly and sensor response of photosynthetic reaction centers on screen-printed electrodes. Anal. Chim. Acta 2011, 707, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Masojídek, J.; Souček, P.; Máchová, J.; Frolík, J.; Klem, K.; Malý, J. Detection of photosynthetic herbicides: Algal growth inhibition test vs. Electrochemical photosystem II biosensor. Ecotoxicol. Environ. Saf. 2011, 74, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Touloupakis, E.; Boutopoulos, C.; Buonasera, K.; Zergioti, I.; Giardi, M.T. A photosynthetic biosensor with enhanced electron transfer generation realized by laser printing technology. Anal. Bioanal. Chem. 2012, 402, 3237–3244. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, V.; Pezzotti, I.; Pezzotti, G.; Cano, J.; Manfredonia, I.; Buonasera, K.; Rodio, G.; Giardi, M.T. A new embedded biosensor platform based on micro-electrodes array (MEA) technology. Sens. Actuators B Chem. 2013, 176, 275–283. [Google Scholar] [CrossRef]
- Zamaleeva, A.I.; Sharipova, I.R.; Shamagsumova, R.V.; Ivanov, A.N.; Evtugyn, G.A.; Ishmuchametova, D.G.; Fakhrullin, R.F. A whole-cell amperometric herbicide biosensor based on magnetically functionalised microalgae and screen-printed electrodes. Anal. Methods 2011, 3, 509–513. [Google Scholar] [CrossRef]
- Avramescu, A.; Rouillon, R.; Carpentier, R. Potential for use of a cyanobacterium synechocystis sp. Immobilized in poly(vinylalcohol): Application to the detection of pollutants. Biotechnol. Tech. 1999, 13, 559–562. [Google Scholar] [CrossRef]
- Giardi, M.T.; Guzzella, L.; Euzet, P.; Rouillon, R.; Esposito, D. Detection of herbicide subclasses by an optical multibiosensor based on an array of photosystem II mutants. Environ. Sci. Technol. 2005, 39, 5378–5384. [Google Scholar] [CrossRef] [PubMed]
- Moro, L.; Pezzotti, G.; Turemis, M.; Sanchís, J.; Farré, M.; Denaro, R.; Giacobbe, M.G.; Crisafi, F.; Giardi, M.T. Fast pesticide pre-screening in marine environment using a green microalgae-based optical bioassay. Mar. Pollut. Bull. 2018, 129, 212–221. [Google Scholar] [CrossRef]
- Rasmussen, M.; Wingersky, A.; Minteer, S.D. Comparative study of thylakoids from higher plants for solar energy conversion and herbicide detection. ElectroChim. Acta 2014, 140, 304–308. [Google Scholar] [CrossRef]
- Breton, F.; Euzet, P.; Piletsky, S.A.; Giardi, M.T.; Rouillon, R. Integration of photosynthetic biosensor with molecularly imprinted polymer-based solid phase extraction cartridge. Anal. Chim. Acta 2006, 569, 50–57. [Google Scholar] [CrossRef]
- Tibuzzi, A.; Rea, G.; Pezzotti, G.; Esposito, D.; Johanningmeier, U.; Giardi, M.T. A new miniaturized multiarray biosensor system for fluorescence detection. J. Phys. Condens. Matter 2007, 19, 395006. [Google Scholar] [CrossRef]
- European Union. Directive 2000/60/EC of the European parliament and of the council of 23 October 2000 establishing a framework for the community action in the field of water policy (water framework directive). Off. J. Eur. Communities Ser. 2000, 327, 1–7. [Google Scholar]
- Badura, A.; Kothe, T.; Schuhmann, W.; Rogner, M. Wiring photosynthetic enzymes to electrodes. Energy Environ. Sci. 2011, 4, 3263–3274. [Google Scholar] [CrossRef]
- Sawa, M.; Fantuzzi, A.; Bombelli, P.; Howe, C.J.; Hellgardt, K.; Nixon, P.J. Electricity generation from digitally printed cyanobacteria. Nat. Commun. 2017, 8, 1327. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, G.; Liu, M.; Lei, Y.; Li, M. Fabrication of a novel atrazine biosensor and its subpart-per-trillion levels sensitive performance. Environ. Sci. Technol. 2010, 44, 7878–7883. [Google Scholar] [CrossRef] [PubMed]
- Tortolini, C.; Bollella, P.; Antiochia, R.; Favero, G.; Mazzei, F. Inhibition-based biosensor for atrazine detection. Sens. Actuators B Chem. 2016, 224, 552–558. [Google Scholar] [CrossRef]
- Guan, Y.; Liu, L.; Chen, C.; Kang, X.; Xie, Q. Effective immobilization of tyrosinase via enzyme catalytic polymerization of L-DOPA for highly sensitive phenol and atrazine sensing. Talanta 2016, 160, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, F.W.; Barroso, M.F.; Morais, S.; Viswanathan, S.; de Lima-Neto, P.; Correia, A.N.; Oliveira, M.B.; Delerue-Matos, C. Simple laccase-based biosensor for formetanate hydrochloride quantification in fruits. Bioelectrochemistry 2014, 95, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.C.; Moccelini, S.K.; Castilho, M.; Terezo, A.J.; Possavatz, J.; Magalhães, M.R.L.; Dores, E.F.G.C. Biosensor based on atemoya peroxidase immobilised on modified nanoclay for glyphosate biomonitoring. Talanta 2012, 98, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Songa, E.A.; Waryo, T.; Jahed, N.; Baker, P.G.L.; Kgarebe, B.V.; Iwuoha, E.I. Electrochemical nanobiosensor for glyphosate herbicide and its metabolite. Electroanalysis 2009, 21, 671–674. [Google Scholar] [CrossRef]
- Sahin, A.; Dooley, K.; Cropek, D.M.; West, A.C.; Banta, S. A dual enzyme electrochemical assay for the detection of organophosphorus compounds using organophosphorus hydrolase and horseradish peroxidase. Sens. Actuators B Chem. 2011, 158, 353–360. [Google Scholar] [CrossRef]
- Braham, Y.; Barhoumi, H.; Maaref, A. Urease capacitive biosensors using functionalized magnetic nanoparticles for atrazine pesticide detection in environmental samples. Anal. Methods 2013, 5, 4898–4904. [Google Scholar] [CrossRef]
- Mazzei, F.; Botrè, F.; Montilla, S.; Pilloton, R.; Podestà, E.; Botrè, C. Alkaline phosphatase inhibition based electrochemical sensors for the detection of pesticides. J. Electroanal. Chem. 2004, 574, 95–100. [Google Scholar] [CrossRef]
- Pabbi, M.; Kaur, A.; Mittal, S.K.; Jindal, R. A surface expressed alkaline phosphatase biosensor modified with flower shaped ZnO for the detection of chlorpyrifos. Sens. Actuators B Chem. 2018, 258, 215–227. [Google Scholar] [CrossRef]
- Harper, L.L.; McDaniel, C.S.; Miller, C.E.; Wild, J.R. Dissimilar plasmids isolated from pseudomonas diminuta MG and a Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl. Environ. Microbiol. 1988, 54, 2586–2589. [Google Scholar] [PubMed]
- Dumas, D.P.; Durst, H.D.; Landis, W.G.; Raushel, F.M.; Wild, J.R. Inactivation of organophosphorus nerve agents by the phosphotriesterase from pseudomonas diminuta. Arch. Biochem. Biophys. 1990, 277, 155–159. [Google Scholar] [CrossRef]
- Simonian, A.L.; Good, T.A.; Wang, S.S.; Wild, J.R. Nanoparticle-based optical biosensors for the direct detection of organophosphate chemical warfare agents and pesticides. Anal. Chim. Acta 2005, 534, 69–77. [Google Scholar] [CrossRef]
- Kim, G.-Y.; Kang, M.-S.; Shim, J.; Moon, S.-H. Substrate-bound tyrosinase electrode using gold nanoparticles anchored to pyrroloquinoline quinone for a pesticide biosensor. Sens. Actuators B Chem. 2008, 133, 1–4. [Google Scholar] [CrossRef]
- Shim, J.; Woo, J.-J.; Moon, S.-H.; Kim, G.-Y. A preparation of a single-layered enzyme-membrane using asymmetric pBPPO base film for development of pesticide detecting biosensor. J. Membr. Sci. 2009, 330, 341–348. [Google Scholar] [CrossRef]
- Campanella, L.; Lelo, D.; Martini, E.; Tomassetti, M. Organophosphorus and carbamate pesticide analysis using an inhibition tyrosinase organic phase enzyme sensor; comparison by butyrylcholinesterase + choline oxidase OPEE and application to natural waters. Anal. Chim. Acta 2007, 587, 22–32. [Google Scholar] [CrossRef] [PubMed]
- De Albuquerque, Y.D.; Ferreira, L.F. Amperometric biosensing of carbamate and organophosphate pesticides utilizing screen-printed tyrosinase-modified electrodes. Anal. Chim. Acta 2007, 596, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Kuusk, E.; Rinken, T. Transient phase calibration of tyrosinase-based carbaryl biosensor. Enzyme Microb. Technol. 2004, 34, 657–661. [Google Scholar] [CrossRef]
- Rodríguez-Delgado, M.M.; Alemán-Nava, G.S.; Rodríguez-Delgado, J.M.; Dieck-Assad, G.; Martínez-Chapa, S.O.; Barceló, D.; Parra, R. Laccase-based biosensors for detection of phenolic compounds. TrAC Trends Anal. Chem. 2015, 74, 21–45. [Google Scholar] [CrossRef]
- Zapp, E.; Brondani, D.; Vieira, I.C.; Scheeren, C.W.; Dupont, J.; Barbosa, A.M.J.; Ferreira, V.S. Biomonitoring of methomyl pesticide by laccase inhibition on sensor containing platinum nanoparticles in ionic liquid phase supported in montmorillonite. Sens. Actuators B Chem. 2011, 155, 331–339. [Google Scholar] [CrossRef]
- Oliveira, T.M.; Fátima Barroso, M.; Morais, S.; Araújo, M.; Freire, C.; de Lima-Neto, P.; Correia, A.N.; Oliveira, M.B.; Delerue-Matos, C. Laccase–prussian blue film–graphene doped carbon paste modielectrode for carbamate pesticides quantification. Biosens. Bioelectron. 2013, 47, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, T.M.B.F.; Fátima Barroso, M.; Morais, S.; de Lima-Neto, P.; Correia, A.N.; Oliveira, M.B.P.P.; Delerue-Matos, C. Biosensor based on multi-walled carbon nanotubes paste electrode modified with laccase for pirimicarb pesticide quantification. Talanta 2013, 106, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Direct electrochemistry of cytochrome P450 6A1 in mimic bio-membrane and its application for pesticides sensing. Sens. Actuators B Chem. 2011, 156, 773–778. [Google Scholar] [CrossRef]
- Franz, J.E.; Mao, M.K.; Sikorski, J.A. Glyphosate: A Unique Global Herbicide; American Chemical Society: Washington, DC, USA, 1997. [Google Scholar]
- Vaghela, C.; Kulkarni, M.; Haram, S.; Aiyer, R.; Karve, M. A novel inhibition based biosensor using urease nanoconjugate entrapped biocomposite membrane for potentiometric glyphosate detection. Int. J. Biol. Macromol. 2018, 108, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Nomngongo, P.N.; Ngila, J.C.; Nyamori, V.O.; Songa, E.A.; Iwuoha, E.I. Determination of selected heavy metals using amperometric horseradish peroxidase (HRP) inhibition biosensor. J. Anal. Lett. 2011, 44, 2031–2046. [Google Scholar] [CrossRef]
- Sun, H.; Liu, Z.; Wu, C.; Xu, P.; Wang, X. Amperometric inhibitive biosensor based on horseradish peroxidase-nanoporous gold for sulfide determination. Sci. Rep. 2016, 6, 30905. [Google Scholar] [CrossRef] [PubMed]
- Syshchyk, O.; Skryshevsky, V.A.; Soldatkin, O.O.; Soldatkin, A.P. Enzyme biosensor systems based on porous silicon photoluminescence for detection of glucose, urea and heavy metals. Biosens. Bioelectron. 2015, 66, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Noguer, T.; Balasoiu, A.-M.; Avramescu, A.; Marty, J.-L. Development of a disposable biosensor for the detection of metam-sodium and its metabolite mitc. Anal. Lett. 2001, 34, 513–528. [Google Scholar] [CrossRef]
- Noguer, T.; Gradinaru, A.; Ciucu, A.; Marty, J.-L. A new disposable biosensor for the accurate and sensitive detection of ethylenebis(dithiocarbamate) fungicides. Anal. Lett. 1999, 32, 1723–1738. [Google Scholar] [CrossRef]
- Noguer, T.; Leca, B.; Jeanty, G.; Marty, J.-L. Biosensors based on enzyme inhibition: Detection of organophosphorus and carbamate insecticides and dithiocarbamate fungicides. Field Anal. Chem. Technol. 1999, 3, 171–178. [Google Scholar] [CrossRef]
- Noguer, T.; Marty, J.-L. High sensitive bienzymic sensor for the detection of dithiocarbamate fungicides. Anal. Chim. Acta 1997, 347, 63–70. [Google Scholar] [CrossRef]
- FAO. Codex Alimentarius Pesticide Database. 2018. Available online: http://www.fao.org/fao-who-codexalimentarius/codex-texts/dbs/pestres/en/ (accessed on 22 March 2018).
- Fitzmaurice, A.G.; Rhodes, S.L.; Lulla, A.; Murphy, N.P.; Lam, H.A.; O’Donnell, K.C.; Barnhill, L.; Casida, J.E.; Cockburn, M.; Sagasti, A.; et al. Aldehyde dehydrogenase inhibition as a pathogenic mechanism in parkinson disease. Proc. Natl. Acad. Sci. USA 2013, 110, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, Y.; Kazuoka, T.; Yoshida, M.; Yamanaka, K.; Oikawa, T.; Soda, K. Thermostable aldehyde dehydrogenase from psychrophile, cytophaga sp. Kuc-1: Enzymological characteristics and functional properties. Biochem. Biophys. Res. Commun. 2002, 298, 632–637. [Google Scholar] [CrossRef]
- Steffler, F.; Guterl, J.K.; Sieber, V. Improvement of thermostable aldehyde dehydrogenase by directed evolution for application in synthetic cascade biomanufacturing. Enzyme Microb. Technol. 2013, 53, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Rocaboy-Faquet, E.; Barthelmebs, L.; Calas-Blanchard, C.; Noguer, T. A novel amperometric biosensor for ß-triketone herbicides based on hydroxyphenylpyruvate dioxygenase inhibition: A case study for sulcotrione. Talanta 2016, 146, 510–516. [Google Scholar] [CrossRef] [PubMed]
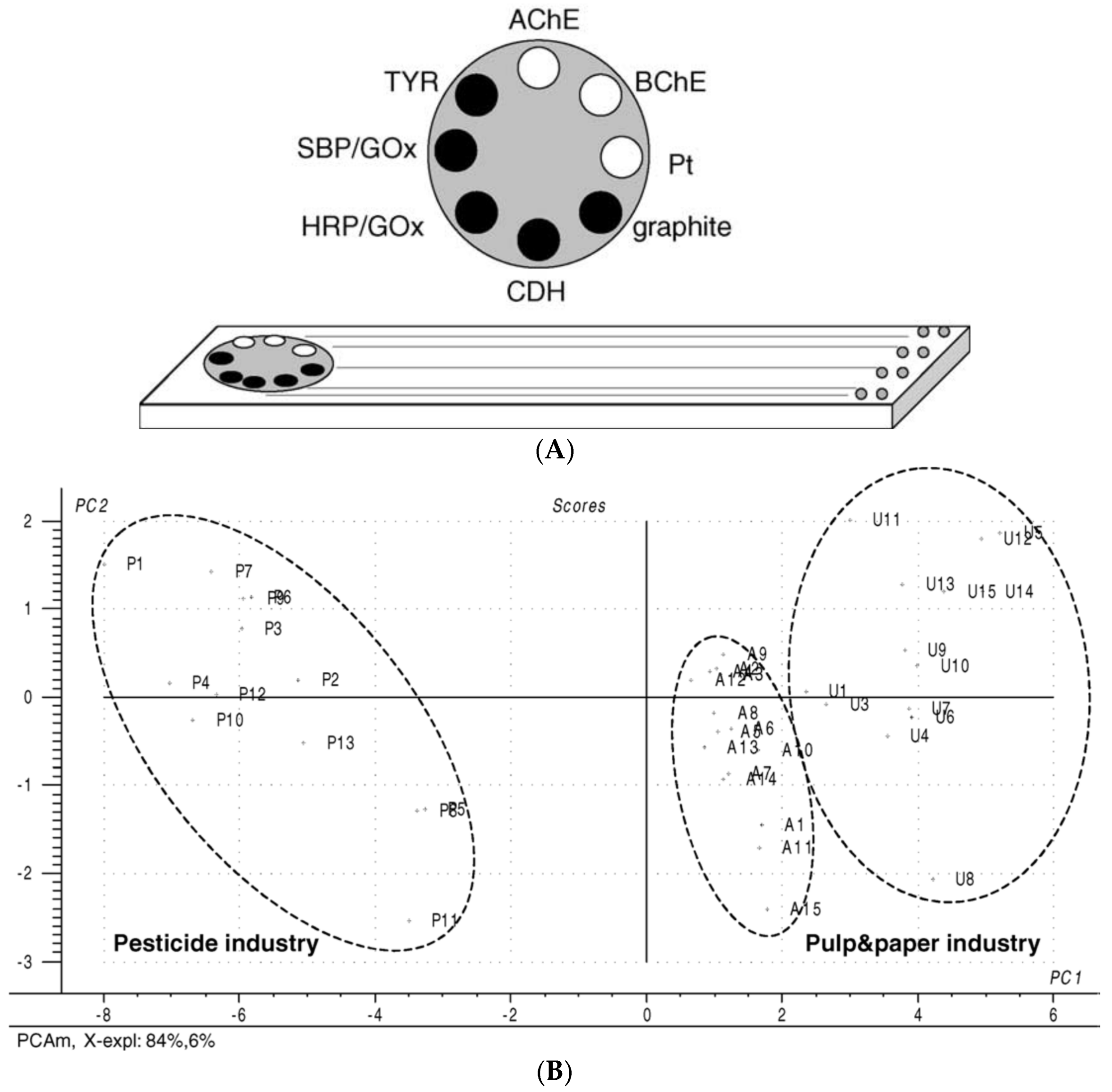
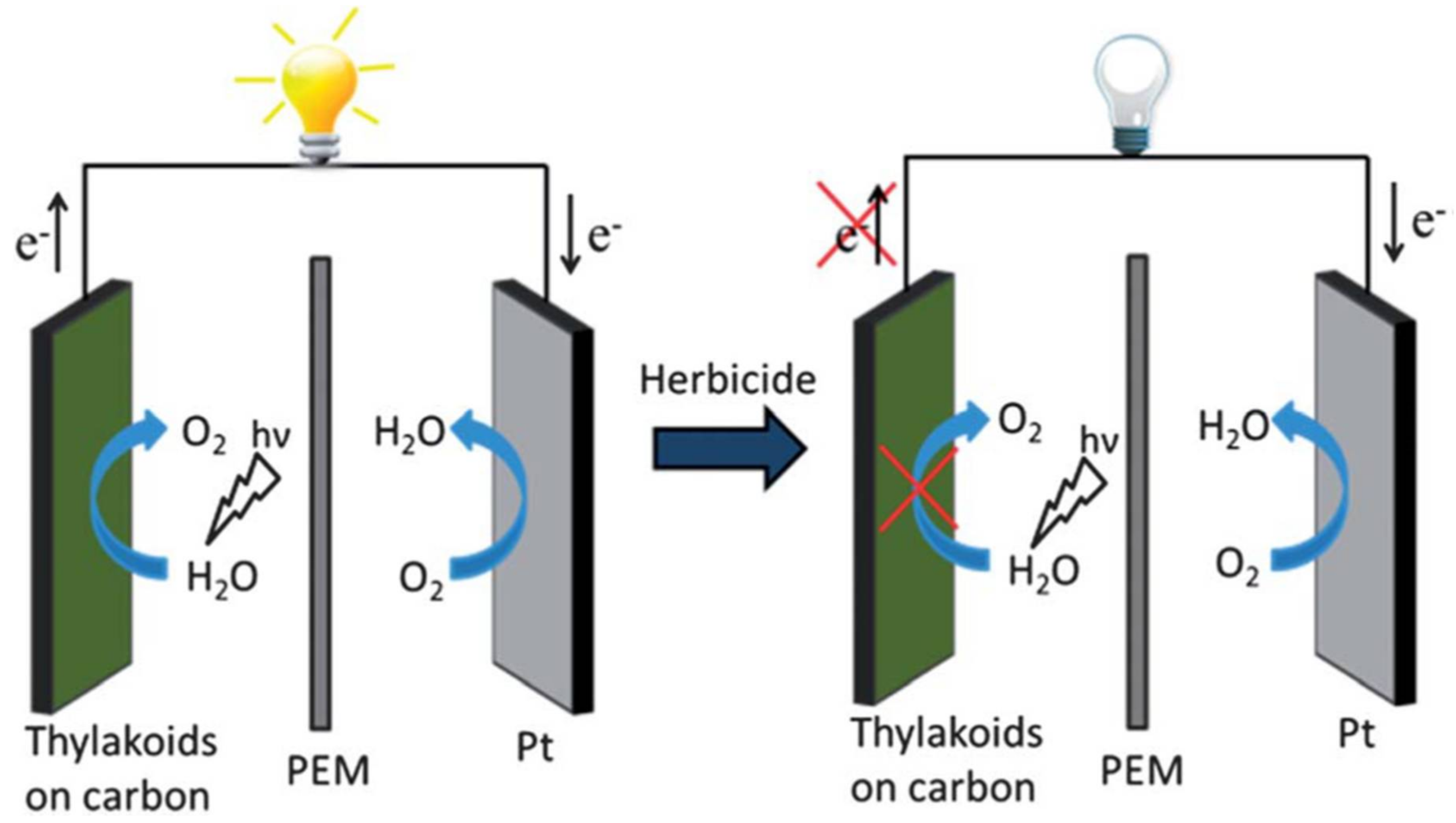
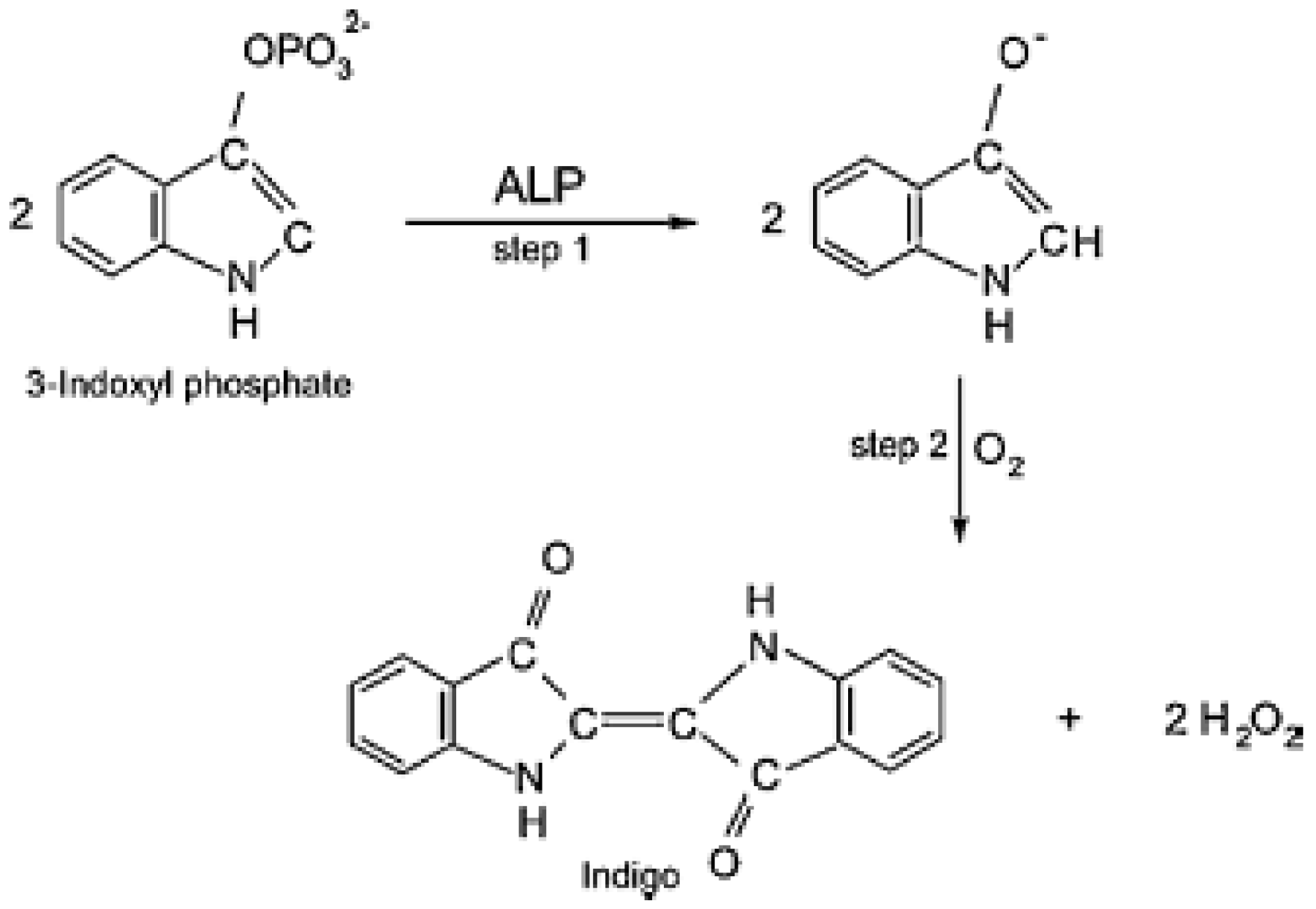
| Pesticide/Real Sample | Photosynthetic Enzyme | Detector | Analytical Performance 1 | Reference |
|---|---|---|---|---|
| Diuron | PSII particles from Synechococcus bigranulatus | Amperometry | I50 = 9 × 10−9 M LOD = 7 × 10−10 M | [58] |
| Diuron Atrazine Simazine Ioxynil Bromoxynil Dinoseb | PSII particles from Synechococcus elongatus | Amperometry | Diuron: LOD: 5 × 10−10 M, I50: 8 × 10−8 M Atrazine: LOD:2 × 10−9 M, I50: 3 × 10−7 M Simazine: LOD: 1 × 10−8 M, I50: 8 × 10−7 M Ioxynil: LOD: 9 × 10−9 M, I50: 4 × 10−7 M Bromoxynil: LOD: 2 × 10−7 M, I50: 8 × 10−6 M Dinoseb: LOD: 6 × 10−8 M, I50: 8 × 10−7 M | [59] |
| Diuron Atrazine Simazine Ioxynil Bromoxynil Dinoseb | PSII isolated from Synechococcus elongatus | Amperometry | Diuron: LOD: 1 × 10−9 M, I50: 7 × 10−8 M Atrazine: LOD: 2 × 10−9 M, I50: 9 × 10−8 M Simazine: LOD: 4 × 10−9 M, I50: 2 × 10−7 M Ioxynil: LOD: 10−7 M Bromoxynil, Dinoseb: LOD: 10−6 M | [60] |
| Diuron Atrazine Simazine Terbuthyl-azine Diethylterbuthylazine | Thylakoid from Spinacia oleracea L., Senecio vulgaris and its mutant resistant to atrazine | Amperometry | LOD: 1.51 × 10−8 M−4.11 × 10−8 M I50: 1 × 10−7 M; for spinach thylakoids; detection in river water | [61] |
| Terbutryin | PSII-enriched thylakoid fractions from spinach | Colorimetry | LOD: 1.58 × 10−7 M | [62] |
| Atrazine Prometryne Terbuthyl-azine Diuron Linuron | Mutant strains of Chlamydomonas reinhardtii with engineered D1 protein | Fluorescence | S268C: LOD: 0.8 × 10−11 M–6.8 × 10−10 M IL: LOD: 1.0 × 10−9 M–3.0 × 10−9 M | [63] |
| Diuron | “BBY”-crude PSII preparation from spinach leaves | Amperometry | LOD: 1.1 × 10−9 M | [55] |
| Linuron Simazine | C. reinhardtii cells | Amperometry | Linuron: LOD: 6 × 10−9 M IC50: 1.2 × 10−7 M Simazine: LOD: 9 × 10−8 M, IC50: 2.3 × 10−6 M | [64] |
| Atrazine Bromacil Diuron | Thylakoids from spinach | Biosolar cell | Atrazine LOD: 0.37 μg L−1 Bromacil LOD: 0.21 μg L−1, Diuron LOD: 0.10 μg L−1; LR: up to ~15 μg L−1 | [65] |
| Diuron Atrazine Ioxynil | Thylakoids from spinach | Amperometry | Diuron: LOD 1.3 ± 0.5 µg L−1, IC50: 2.1 µg L−1 Atrazine: LOD: 2.8 ± 0.3 µg L−1; IC50: 5.6 µg L−1 Ioxynil: LOD: 2.1 ± 0.3 µg L−1 IC50: 3.4 µg L−1; analysis of spiked water | [66] |
| Atrazine Prometryn Diuron | C. reinhardtii mutants | Fluorescence | LOD: 10−10 M for all mutants, Exception: F255N mutant is resistant to urea herbicides | [67] |
| Atrazine | Pure PS II cores and BBY particles from spinach | Amperometry | LOD: 1.15 × 10−9 M | [68] |
| Atrazine Isoproturon Diuron | PSII complex from Synechococcus elongatus f. thermalis | Amperometry | Atrazine: LOD: 6.4 × 10−10 M; IC50: 8.9 × 10−7 M Isoproturon: LOD : 5.5 × 10−9 M; IC50: 7.2 × 10−7 M Diuron: LOD: 4.6 10−10 M; IC50: 2.3 × 10−7 M | [69] |
| Diuron Linuron | Thylakoids from spinach | Amperometry | Diuron: LOD: 8.0 × 10−9 M; I50: 1.87 × 10−7 M; Linuron: LOD: 4.0 × 10−9 M; I50: 5.65 × 10−8 M | [70] |
| Atrazine Prometryn Diuron | Whole cells of Chlamydomonas reinhardtii | Fluorescence | Atrazine LOD: 5 × 10−10 M, Prometryn: 3.1 × 10−10 M Diuron: 4.81 × 10−10 M | [71] |
| Atrazine Propazine | Chlorella pyrenoidosa microalgae | Amperometry | Atrazine: LOD: 7 × 10−7 M; LR: 9 × 10−7 M–7.4 × 10−5 M Propazine: LOD: 4 × 10−7 M; LR: 6 × 10−7 M–1.2 × 10−4 M | [72] |
| Diuron | Synechocystis sp. PCC6803 cyanobacteria | Amperometry, | Diuron LOD: 5 × 10−8 M (cells in solution) and 5 × 10−7 M (immobilized cells) | [73] |
| Urea, diamine, triazine, phenols | Thylakoids from mutant spinach plants | Fluorescence | LOD: 3 × 10−9 M (in river water) | [74] |
| Diuron, Simazine Irgarol | Chlorella mirabilis algae | Fluorescence | Diuron: LOD: 0.067 µg L−1; LR: 0.50–25.0 µg L−1 Simazine: LOD: 0.705 µg L−1; LR: 1.00–50.0 µg L−1 Irgarol; LOD: 0.135 µg L−1 LR: 1.00–50.0 µg L−1; detection in seawater | [75] |
| Diuron | Thylakoids from spinach | Biosolar cell | I50: 67 ± 2 ng L−1 | [76] |
| Pesticide | Detection Method | Limit of Detection | Linear Range | Reference |
|---|---|---|---|---|
| Alkaline phosphatase | ||||
| Metham-sodium | Fluorimetry | 36.5 µM | 75–480 µM | [7] |
| Tetradifon | 4.1 µM | 5–35 µM | ||
| Fenitrothion | 45.5 µM | 135–270 µM | ||
| 2,4-dichlorophenoxyacetic acid | Amperometry | 0.5 ppb | 1.5–60.0 µg L−1 | [70] |
| Malathion | 0.1 ppb | 0.2–45.0 µg L−1 | ||
| Paraoxon | Chemilumines-cence | 50 ppb | n.d. * | [3] |
| Chlorpyrifos | Voltammetry | 10−9 M | 0.05–0.55 mM | [71] |
| Organophosphate hydrolase | ||||
| Paraoxon | Fluorescence | 20 µM | up to 240 µM | [72] |
| Tyrosinase | ||||
| 2,4-dichlorophenoxyacetic acid | Amperometry | 0.6 ppt | 0–10 ppt | [73] |
| Parathion | Amperometry | 0.005 ppb | 0.01–1 ppb | [74] |
| Carbaryl | 0.008 ppb | 0.01–10 ppb | ||
| Atrazine | Amperometry | 0.1 ppt | 0.2 ppt–2 ppb | [82] |
| Atrazine | Amperometry | 0.3 ppm | 0.5–20 ppm | [83] |
| Atrazine | Amperometry | 10 ppb | 50 ppb–30 ppm | [84] |
| Laccase | ||||
| Methomyl | Square wave voltammetry | 2.35 × 10−7 M | 9.8 × 10−7–9.0 × 10−6 M | [75] |
| Carbofuran | Square wave voltammetry | 0.022 mg/kg | 4.98 × 10−7–5.88 × 10−6 M | [76] |
| Carbaryl | 0.02 mg/kg | 7.44 × 10−8–8.47 × 10−7 M | ||
| Formetanate | 0.21 mg/kg | 2.49 × 10−7–7.46 × 10−6 M | ||
| Pirimicarb | 0.23 mg/kg | 2.99 × 10−7–5.66 × 10−6 M | ||
| Ziram | 0.02 mg/kg | 2.49 × 10−7–5.66 × 10−6 M | ||
| Pirimicarb | Square wave voltammetry | 1.8 × 10−7 M | 9.95 × 10−7–1.15 × 10−5 M | [77] |
| Formetanate | Square wave voltammetry | 95 nM | n.d. * | [85] |
| Heme-containing enzymes | ||||
| Aldrin | Amperometry | 8 × 10−6 M | 9.08 × 10−6–4.54 × 10−5 M | [78] |
| Heptachlor | n.d. * | 8.91 × 10−6–4.46 × 10−5 M | ||
| Glyphosate | SWV | 30 µg L−1 | 0.1–4.5 mg L−1 | [86] |
| Aminomethylphosphonic acid | Amperometry | 1.µg L−1 | 1.5–7.5 mg L−1 | [87] |
| Glyphosate | 0.16 μg L−1 | 2.0–14.0 µg L−1 | ||
| Dichlofenthion | Amperometry | 24 µM | 5–100 µM | [88] |
| Urease | ||||
| Atrazine | Enzyme Field Effect Capacitive System | 0.12 µM | 0.1 µM–10 mM | [89] |
| Glyphosate | Potentiometric | 0.5 ppm | 0.5 ppm–50 ppm | [79] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bucur, B.; Munteanu, F.-D.; Marty, J.-L.; Vasilescu, A. Advances in Enzyme-Based Biosensors for Pesticide Detection. Biosensors 2018, 8, 27. https://doi.org/10.3390/bios8020027
Bucur B, Munteanu F-D, Marty J-L, Vasilescu A. Advances in Enzyme-Based Biosensors for Pesticide Detection. Biosensors. 2018; 8(2):27. https://doi.org/10.3390/bios8020027
Chicago/Turabian StyleBucur, Bogdan, Florentina-Daniela Munteanu, Jean-Louis Marty, and Alina Vasilescu. 2018. "Advances in Enzyme-Based Biosensors for Pesticide Detection" Biosensors 8, no. 2: 27. https://doi.org/10.3390/bios8020027
APA StyleBucur, B., Munteanu, F.-D., Marty, J.-L., & Vasilescu, A. (2018). Advances in Enzyme-Based Biosensors for Pesticide Detection. Biosensors, 8(2), 27. https://doi.org/10.3390/bios8020027







